Antimicrobial Biosynthetic Potential and Phylogenetic Analysis of Culturable Bacteria Associated with the Sponge Ophlitaspongia sp. from the Yellow Sea, China
Abstract
1. Introduction
2. Results
2.1. Antimicrobial Activity of Sponge-Associated Bacteria
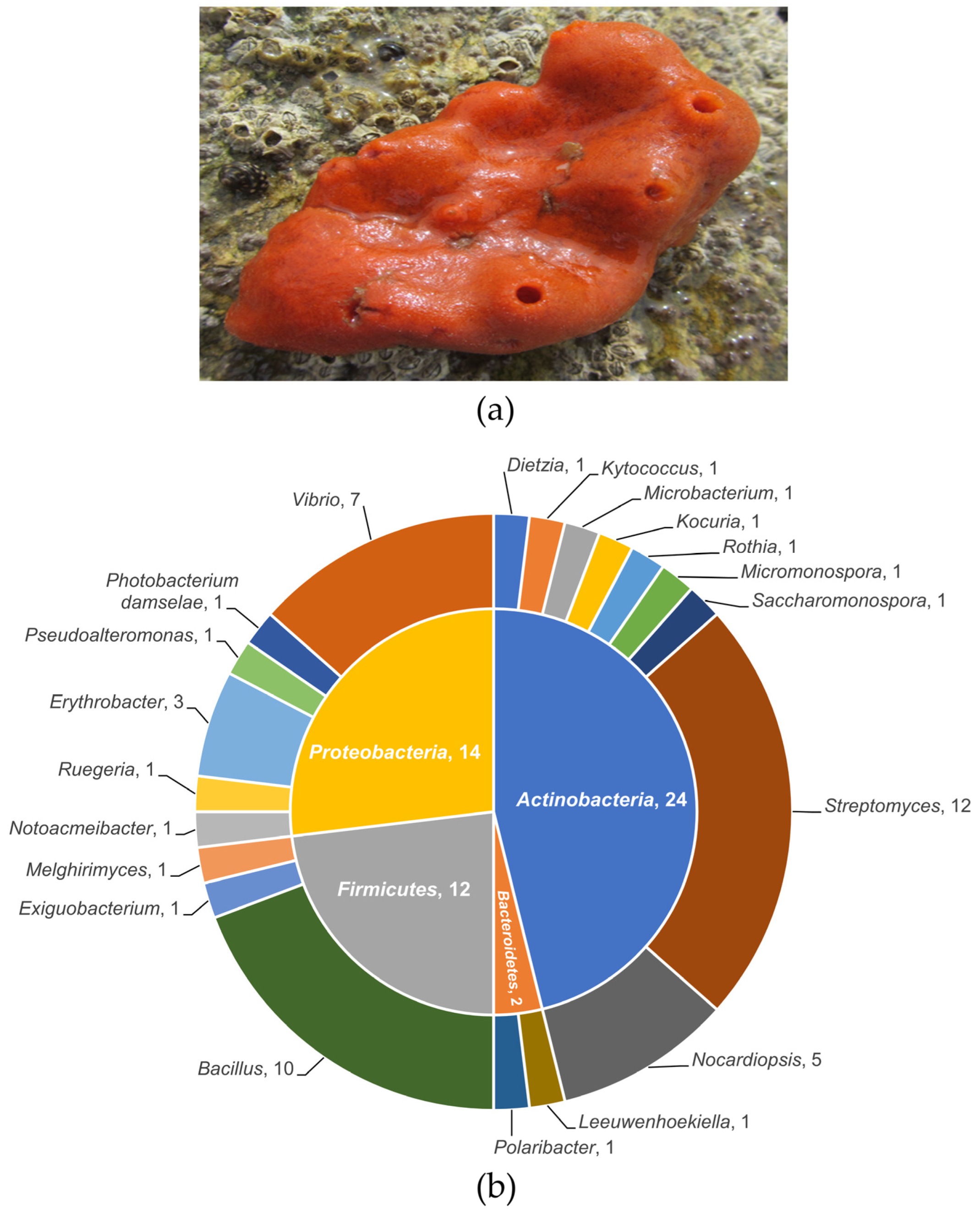
2.2. Phylogenetics of Sponge-Associated Bacteria
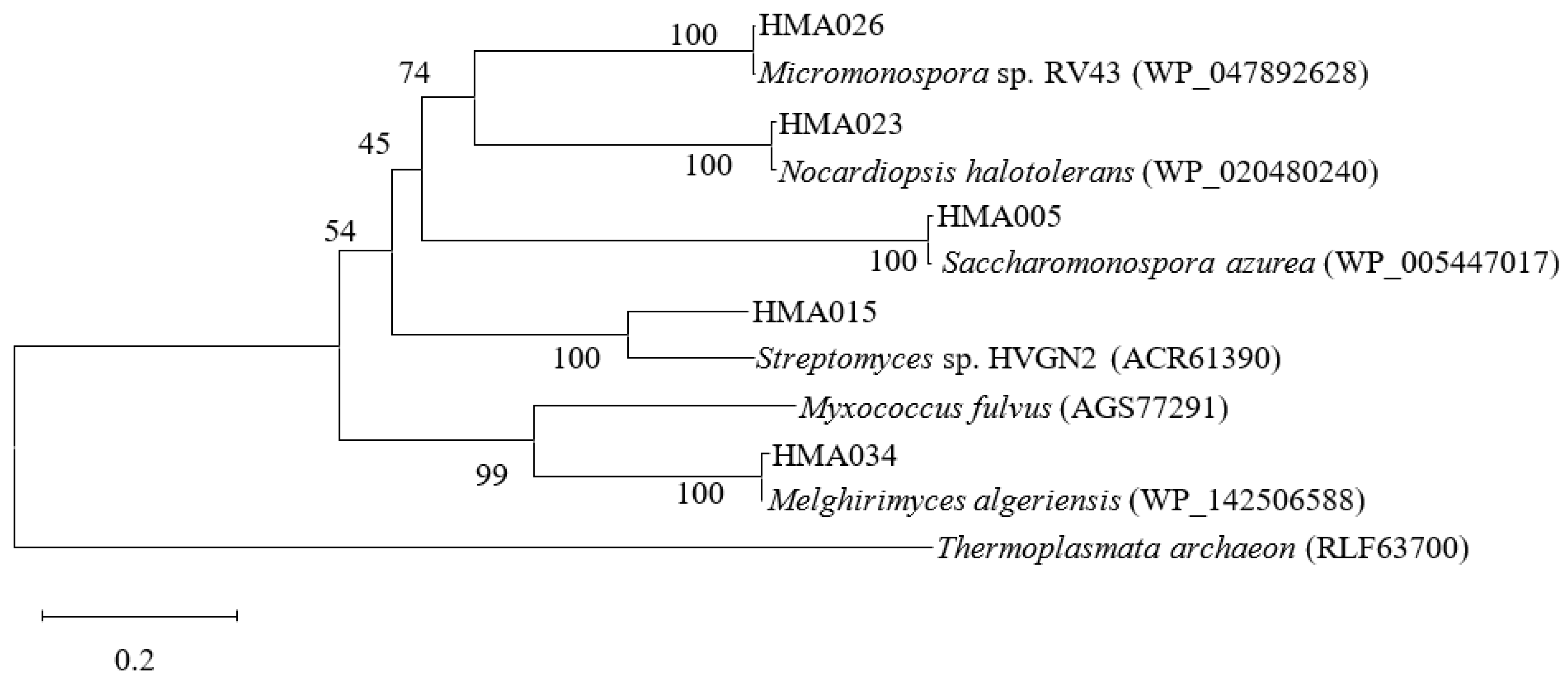
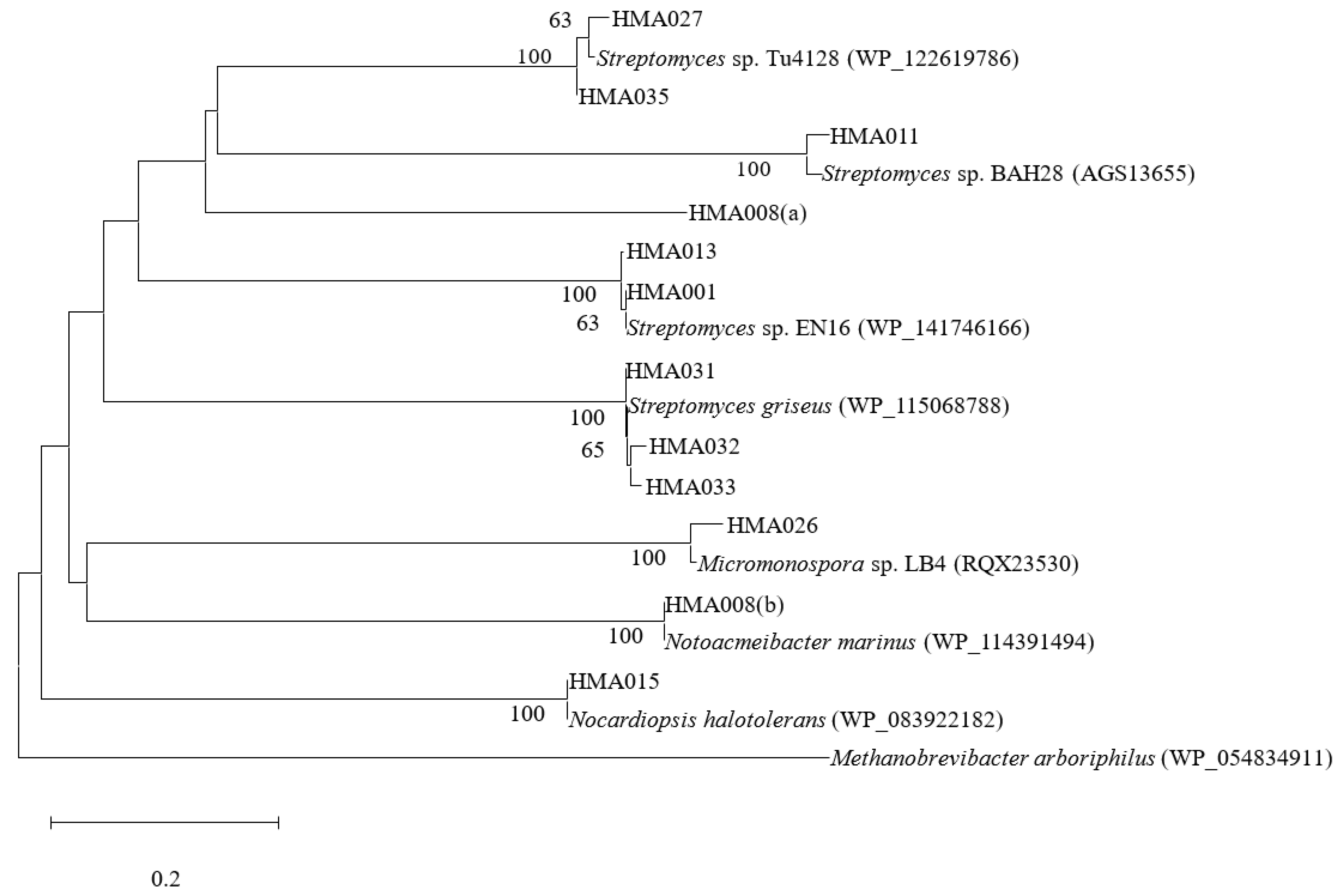
2.3. Phylogenetic Analyses of PKS and NRPS Encoding Genes
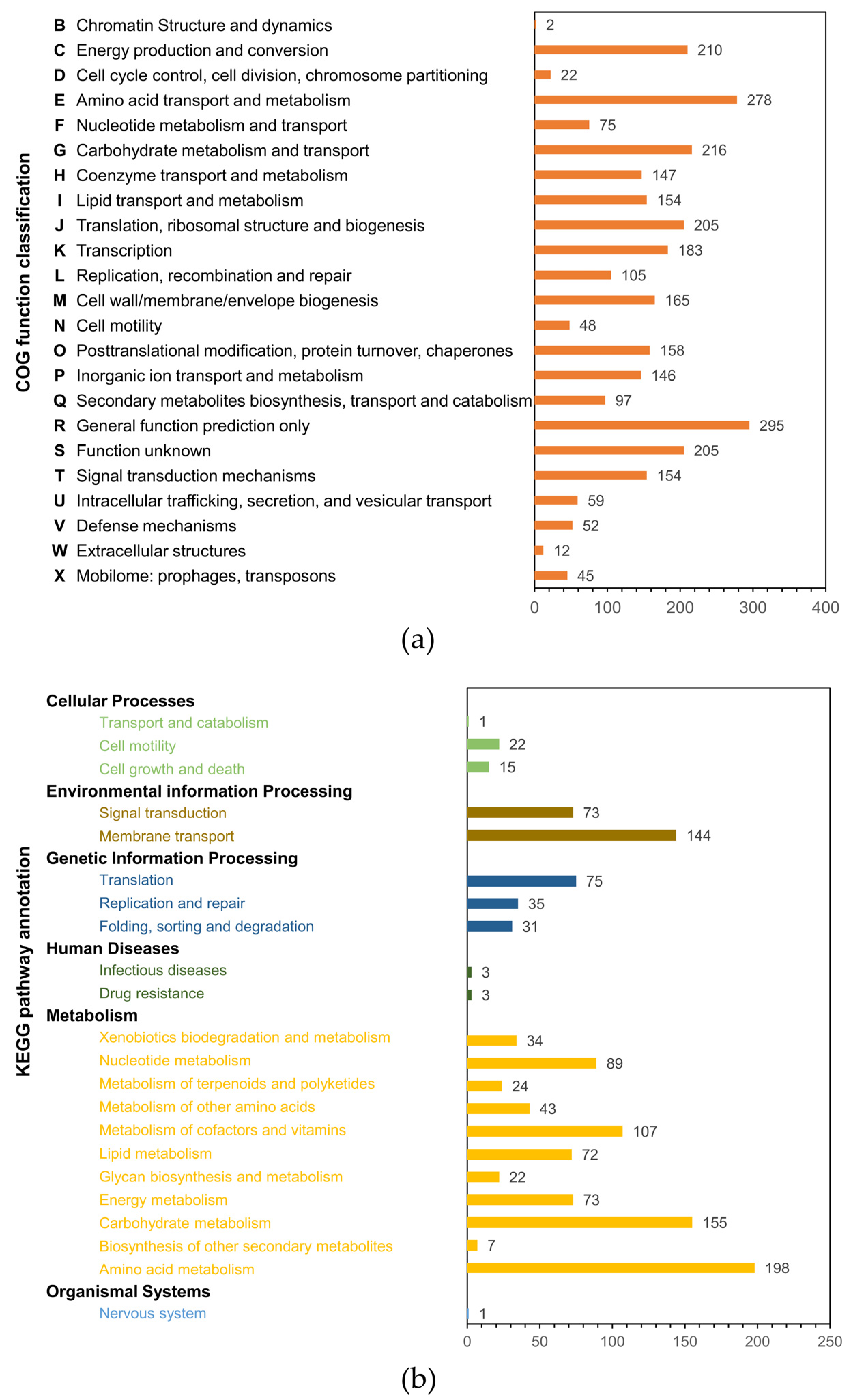
2.4. HMA008 Genome Sequence Annotation and Mining for Secondary Metabolite Biosynthetic Gene Clusters
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Preparation
4.2. Isolation of Culturable Bacteria
4.3. Selection and Preservation of Collected Strains
4.4. Preparation of Crude Extracts and Antibacterial Activity Screening
4.5. 16S rRNA Gene Sequencing and Phylogenetic Analyses
4.6. Genome Sequencing and Secondary Metabolite Cluster Analysis
4.7. Screening for PKS and NRPS Genes
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zuppa, A.; Costantini, S.; Costantini, M. Comparative sequence analysis of bacterial symbionts from the marine sponges Geodia cydonium and Ircinia muscarum. Bioinformation 2014, 10, 196–200. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thomas, T.; Moitinho-Silva, L.; Lurgi, M.; Björk, J.; Easson, C.; Garcia, M.D.C.A.; Olson, J.B.; Erwin, P.M.; Lopez-Legentil, S.; Luter, H.; et al. Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 2016, 7, 11870. [Google Scholar] [CrossRef] [PubMed]
- Lackner, G.; Peters, E.E.; Helfrich, E.J.; Piel, J. Insights into the lifestyle of uncultured bacterial natural product factories associated with marine sponges. Proc. Natl. Acad. Sci. USA 2017, 114, E347–E356. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wu, S.; Zhang, R.; Wang, D.; Chen, J.; Zhao, J. Diversity and antimicrobial potential of Actinobacteria isolated from diverse marine sponges along the Beibu Gulf of the South China Sea. FEMS Microbiol. Ecol. 2019, 95, fiz089. [Google Scholar] [CrossRef] [PubMed]
- Dat, T.T.H.; Cuc, N.T.K.; Cuong, P.V.; Smidt, H.; Sipkema, D. Diversity and antimicrobial activity of Vietnamese sponge-associated bacteria. Mar. Drugs 2021, 19, 353. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Zhou, F.; Al-Kareef, A.M.; Wang, H. Anticancer agents from marine sponges. J. Asian Nat. Prod. Res. 2015, 17, 64–88. [Google Scholar] [CrossRef]
- Jimenez, P.C.; Wilke, D.V.; Branco, P.C.; Bauermeister, A.; Rezende-Teixeira, P.; Gaudêncio, S.P.; Costa-Lotufo, L.V. Enriching cancer pharmacology with drugs of marine origin. Br. J. Pharmacol. 2020, 177, 3–27. [Google Scholar] [CrossRef]
- Swami, U.; Shah, U.; Goel, S. Eribulin in cancer treatment. Mar. Drugs 2015, 13, 5016–5058. [Google Scholar] [CrossRef]
- Bringmann, G.; Lang, G.; Mühlbacher, J.; Schaumann, K.; Steffens, S.; Rytik, P.G.; Hentschel, U.; Morschhäuser, J.; Müller, W.E. Sorbicillactone A: A structurally unprecedented bioactive novel-type alkaloid from a sponge-derived fungus. Prog. Mol. Subcell. Biol. 2003, 37, 231–253. [Google Scholar]
- Indraningrat, A.A.; Smidt, H.; Sipkema, D. Bioprospecting sponge-associated microbes for antimicrobial compounds. Mar. Drugs 2016, 14, 87. [Google Scholar] [CrossRef]
- Palomo, S.; González, I.; de la Cruz, M.; Martín, J.; Tormo, J.R.; Anderson, M.; Hill, R.T.; Vicente, F.; Reyes, F.; Genilloud, O. Sponge-derived Kocuria and Micrococcus spp. as sources of the new thiazolyl peptide antibiotic kocurin. Mar. Drugs 2013, 11, 1071–1086. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Xu, Y.; Shao, C.-L.; Yang, R.-Y.; Zheng, C.-J.; Chen, Y.-Y.; Fu, X.-M.; Qian, P.-Y.; She, Z.-G.; De Voogd, N.J.; et al. Antibacterial bisabolane-type sesquiterpenoids from the sponge-derived fungus Aspergillus sp. Mar. Drugs 2012, 10, 234–241. [Google Scholar] [CrossRef]
- Pruksakorn, P.; Arai, M.; Kotoku, N.; Vilchèze, C.; Baughn, A.D.; Moodley, P.; Jacobs, W.R., Jr.; Kobayashi, M. Trichoderins, novel aminolipopeptides from a marine sponge-derived Trichoderma sp., are active against dormant mycobacteria. Bioorganic Med. Chem. Lett. 2010, 20, 3658–3663. [Google Scholar] [CrossRef] [PubMed]
- El-Gendy, M.M.; El-Bondkly, A.M. Production and genetic improvement of a novel antimycotic agent, saadamycin, against dermatophytes and other clinical fungi from endophytic Streptomyces sp. Hedaya48. J. Ind. Microbiol. Biotechnol. 2010, 37, 831–841. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.J.; Steiniger, C.; Cairns, T.C.; Wisecaver, J.H.; Lind, A.L.; Pohl, C.; Regner, C.; Rokas, A.; Meyer, V. Beyond the biosynthetic gene cluster paradigm: Genome-wide coexpression networks connect clustered and unclustered transcription factors to secondary metabolic pathways. Microbiol. Spectr. 2021, 9, e0089821. [Google Scholar] [CrossRef] [PubMed]
- Katz, L.; Baltz, R.H. Natural product discovery: Past, present, and future. J. Ind. Microbiol. Biotechnol. 2016, 43, 155–176. [Google Scholar] [CrossRef]
- Pimentel-Elardo, S.M.; Grozdanov, L.; Proksch, S.; Hentschel, U. Diversity of nonribosomal peptide synthetase genes in the microbial metagenomes of marine sponges. Mar. Drugs 2012, 10, 1192–1202. [Google Scholar] [CrossRef]
- Liu, H.; Zhu, G.; Zhao, S.; Fu, P.; Zhu, W. Bioactive natural products from the marine sponge-derived Nocardiopsis dassonvillei OUCMDZ-4534. Chin. J. Org. Chem. 2019, 39, 507–514. [Google Scholar] [CrossRef]
- Ameen, F.; AlNadhari, S.; Al-Homaidan, A.A. Marine microorganisms as an untapped source of bioactive compounds. Saudi J. Biol. Sci. 2021, 28, 224–231. [Google Scholar] [CrossRef]
- Riyanti; Balansa, W.; Liu, Y.; Sharma, A.; Mihajlovic, S.; Hartwig, C.; Leis, B.; Rieuwpassa, F.J.; Ijong, F.G.; Wägele, H.; et al. Selection of sponge-associated bacteria with high potential for the production of antibacterial compounds. Sci. Rep. 2020, 10, 19614. [Google Scholar] [CrossRef]
- Dat, T.T.H.; Steinert, G.; Cuc, N.T.K.; Smidt, H.; Sipkema, D. Bacteria cultivated from sponges and bacteria not yet cultivated from sponges—A review. Front. Microbiol. 2021, 12, 737925. [Google Scholar] [CrossRef] [PubMed]
- Laport, M.S. Isolating bacteria from sponges: Why and how? Curr. Pharm. Biotechnol. 2017, 18, 1224–1236. [Google Scholar] [CrossRef] [PubMed]
- Arulprakasam, K.R.; Dharumadurai, D. Genome mining of biosynthetic gene clusters intended for secondary metabolites conservation in actinobacteria. Microb. Pathog. 2021, 161 Pt A, 105252. [Google Scholar] [CrossRef]
- Nicault, M.; Tidjani, A.R.; Gauthier, A.; Dumarcay, S.; Gelhaye, E.; Bontemps, C.; Leblond, P. Mining the biosynthetic potential for specialized metabolism of a Streptomyces soil community. Antibiotics 2020, 9, 271. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.; Bai, X.; Chen, J.; Zhang, H.; Wang, H. Exploring structural diversity of microbe secondary metabolites using OSMAC strategy: A literature review. Front. Microbiol. 2019, 10, 294. [Google Scholar] [CrossRef]
- Wang, W.; Drott, M.; Greco, C.; Luciano-Rosario, D.; Wang, P.; Keller, N.P. Transcription factor repurposing offers insights into evolution of biosynthetic gene cluster regulation. mBio 2021, 12, e0139921. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Hwang, S.; Lee, Y.; Cho, S.; Palsson, B.; Cho, B.K. Synthetic biology tools for novel secondary metabolite discovery in Streptomyces. J. Microbiol. Biotechnol. 2019, 29, 667–686. [Google Scholar] [CrossRef]
- Alex, A.; Antunes, A. Whole-genome comparisons among the genus Shewanella reveal the enrichment of genes encoding ankyrin-repeats containing proteins in sponge-associated bacteria. Front. Microbiol. 2019, 10, 5. [Google Scholar] [CrossRef]
- Howson, C.M.; Chambers, S.J. Ophlitaspongia and Ophlitaspongia papilla reinstated, and a new species of Ophlitaspongia described (Porifera: Demospongiae: Microcionidae). J. Mar. Biol. Assoc. 1999, 79, 609–620. [Google Scholar] [CrossRef]
- Ferreira, M.; Cabado, A.G.; Chapela, M.J.; Fajardo, P.; Atanassova, M.; Garrido, A.; Vieites, J.M.; Lago, J. Cytotoxic activity of extracts of marine sponges from NW Spain on a neuroblastoma cell line. Environ. Toxicol. Pharmacol. 2011, 32, 430–437. [Google Scholar] [CrossRef]
- Alex, A.; Antunes, A. Comparative genomics reveals metabolic specificity of endozoicomonas isolated from a marine sponge and the genomic repertoire for host-bacteria symbioses. Microorganisms 2019, 7, 635. [Google Scholar] [CrossRef]
- Moitinho-Silva, L.; Nielsen, S.; Amir, A.; Gonzalez, A.; Ackermann, G.L.; Cerrano, C.; Astudillo-Garcia, C.; Easson, C.; Sipkema, D.; Liu, F.; et al. The sponge microbiome project. Gigascience 2017, 6, gix077. [Google Scholar] [CrossRef]
- Su, J.; Zhang, F.; Sun, W.; Karuppiah, V.; Zhang, G.; Li, Z.; Jiang, Q. A new alkaline lipase obtained from the metagenome of marine sponge Ircinia sp. World J. Microbiol. Biotechnol. 2015, 31, 1093–1102. [Google Scholar] [CrossRef]
- Gaikwad, S.; Shouche, Y.S.; Gade, W.N. Microbial community structure of two freshwater sponges using Illumina MiSeq sequencing revealed high microbial diversity. AMB Express 2016, 6, 40. [Google Scholar] [CrossRef]
- Yang, S.; Sun, W.; Tang, C.; Jin, L.; Zhang, F.; Li, Z. Phylogenetic diversity of actinobacteria associated with soft coral Alcyonium gracllimum and stony coral Tubastraea coccinea in the East China Sea. Microb. Ecol. 2013, 66, 189–199. [Google Scholar] [CrossRef]
- Mitova, M.; Popov, S.; De Rosa, S. Cyclic peptides from a Ruegeria strain of bacteria associated with the sponge Suberites domuncula. J. Nat. Prod. 2004, 67, 1178–1181. [Google Scholar] [CrossRef]
- Mitra, S.; Matsuo, Y.; Haga, T.; Yasumoto-Hirose, M.; Yoon, J.; Kasai, H.; Yokota, A. Leptobacterium flavescens gen. nov., sp. nov., a marine member of the family Flavobacteriaceae, isolated from marine sponge and seawater. Int. J. Syst. Evol. Microbiol. 2009, 59 Pt 2, 207–212. [Google Scholar] [CrossRef]
- Selvin, J.; Shanmugha Priya, S.; Seghal Kiran, G.; Thangavelu, T.; Sapna Bai, N. Sponge-associated marine bacteria as indicators of heavy metal pollution. Microbiol. Res. 2009, 164, 352–363. [Google Scholar] [CrossRef]
- Abbamondi, G.R.; De Rosa, S.; Iodice, C.; Tommonaro, G. Cyclic dipeptides produced by marine sponge-associated bacteria as quorum sensing signals. Nat. Prod. Commun. 2014, 9, 229–232. [Google Scholar] [CrossRef]
- Sun, W.; Zhang, F.; He, L.; Karthik, L.; Li, Z. Actinomycetes from the South China Sea sponges: Isolation, diversity, and potential for aromatic polyketides discovery. Front. Microbiol. 2015, 6, 1048. [Google Scholar] [CrossRef]
- Amelia, T.S.M.; Amirul, A.A.; Saidin, J.; Bhubalan, K. Identification of cultivable bacteria from tropical marine sponges and their biotechnological potentials. Trop. Life Sci. Res. 2018, 29, 187–199. [Google Scholar] [CrossRef]
- Batista, D.; Costa, R.; Carvalho, A.P.; Batista, W.R.; Rua, C.; de Oliveira, L.; Leomil, L.; Fróes, A.M.; Thompson, F.L.; Coutinho, R.; et al. Environmental conditions affect activity and associated microorganisms of marine sponges. Mar. Environ. Res. 2018, 142, 59–68. [Google Scholar] [CrossRef]
- Zhuang, L.; Lin, B.; Xu, L.; Li, G.; Wu, C.J.; Luo, L. Erythrobacter spongiae sp. nov., isolated from marine sponge. Int. J. Syst. Evol. Microbiol. 2019, 69, 1111–1116. [Google Scholar] [CrossRef]
- Mahmoud, H.M.; Kalendar, A.A. Coral-associated actinobacteria: Diversity, abundance, and biotechnological potentials. Front. Microbiol. 2016, 7, 204. [Google Scholar] [CrossRef]
- Chen, J.; Xu, L.; Zhou, Y.; Han, B. Natural products from actinomycetes associated with marine organisms. Mar. Drugs 2021, 19, 629. [Google Scholar] [CrossRef]
- Chen, L.; Wang, X.N.; Fu, C.M.; Wang, G.Y. Phylogenetic analysis and screening of antimicrobial and antiproliferative activities of culturable bacteria associated with the ascidian Styela clava from the Yellow Sea, China. BioMed Res. Int. 2019, 2019, 7851251. [Google Scholar] [CrossRef]
- Francis, A.; Chakraborty, K. Marine macroalga-associated heterotroph Bacillus velezensis as prospective therapeutic agent. Arch. Microbiol. 2021, 203, 1671–1682. [Google Scholar] [CrossRef]
- Savi, D.C.; Shaaban, K.A.; Gos, F.M.W.; Thorson, J.S.; Glienke, C.; Rohr, J. Secondary metabolites produced by Microbacterium sp. LGMB471 with antifungal activity against the phytopathogen Phyllosticta citricarpa. Folia Microbiol. 2019, 64, 453–460. [Google Scholar] [CrossRef]
- Bennur, T.; Kumar, A.R.; Zinjarde, S.; Javdekar, V. Nocardiopsis species: Incidence, ecological roles and adaptations. Microbiol. Res. 2015, 174, 33–47. [Google Scholar] [CrossRef]
- Paulsen, S.S.; Strube, M.L.; Bech, P.K.; Gram, L.; Sonnenschein, E.C. Marine chitinolytic Pseudoalteromonas represents an untapped reservoir of bioactive potential. mSystems 2019, 4, e00060-19. [Google Scholar] [CrossRef]
- Le, T.C.; Lee, E.J.; Lee, J.; Hong, A.; Yim, C.-Y.; Yang, I.; Choi, H.; Chin, J.; Cho, S.J.; Ko, J.; et al. Saccharoquinoline, a cytotoxic alkaloidal meroterpenoid from marine-derived bacterium Saccharomonospora sp. Mar. Drugs 2019, 17, 98. [Google Scholar]
- Čihák, M.; Kameník, Z.; Šmídová, K.; Bergman, N.; Benada, O.; Kofroňová, O.; Petříčková, K.; Bobek, J. Secondary metabolites produced during the germination of Streptomyces coelicolor. Front. Microbiol. 2017, 8, 2495. [Google Scholar] [CrossRef]
- Bérdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef]
- Maloney, K.N.; Macmillan, J.B.; Kauffman, C.A.; Jensen, P.R.; Dipasquale, A.G.; Rheingold, A.L.; Fenical, W. Lodopyridone, a structurally unprecedented alkaloid from a marine actinomycete. Org. Lett. 2009, 11, 5422–5424. [Google Scholar]
- Yim, C.Y.; Le, T.C.; Lee, T.G.; Yang, I.; Choi, H.; Lee, J.; Kang, K.Y.; Lee, J.S.; Lim, K.M.; Yee, S.T.; et al. Saccharomonopyrones A–C, new α-pyrones from a marine sediment-derived bacterium Saccharomonospora sp. CNQ-490. Mar. Drugs 2017, 15, 239. [Google Scholar] [CrossRef]
- Ausuri, J.; Vitale, G.A.; Coppola, D.; Palma Esposito, F.; Buonocore, C.; de Pascale, D. Assessment of the degradation potential and genomic insights towards phenanthrene by Dietzia psychralcaliphila JI1D. Microorganisms 2021, 9, 1327. [Google Scholar]
- Uranga, C.C.; Arroyo, P., Jr.; Duggan, B.M.; Gerwick, W.H.; Edlund, A. Commensal oral Rothia mucilaginosa produces enterobactin, a metal-chelating siderophore. Msystems 2020, 5, e00161-20. [Google Scholar] [CrossRef]
- Tahon, G.; Lebbe, L.; De Troch, M.; Sabbe, K.; Willems, A. Leeuwenhoekiella aestuarii sp. nov., isolated from salt-water sediment and first insights in the genomes of Leeuwenhoekiella species. Int. J. Syst. Evol. Microbiol. 2020, 70, 1706–1719. [Google Scholar] [CrossRef]
- Xing, P.; Hahnke, R.L.; Unfried, F.; Markert, S.; Huang, S.; Barbeyron, T.; Harder, J.; Becher, D.; Schweder, T.; Glöckner, F.O.; et al. Niches of two polysaccharide-degrading Polaribacter isolates from the North Sea during a spring diatom bloom. ISME J. 2015, 9, 1410–1422. [Google Scholar] [CrossRef]
- Gao, B.; Gallagher, T.; Zhang, Y.; Elbadawi-Sidhu, M.; Lai, Z.; Fiehn, O.; Whiteson, K.L. Tracking polymicrobial metabolism in cystic fibrosis airways: Pseudomonas aeruginosa metabolism and physiology are influenced by Rothia mucilaginosa-derived metabolites. mSphere 2018, 3, e00151-18. [Google Scholar] [CrossRef]
- Morris, J.J.; Kirkegaard, R.; Szul, M.J.; Johnson, Z.I.; Zinser, E.R. Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by “helper” heterotrophic bacteria. Appl. Environ. Microbiol. 2008, 74, 4530–4534. [Google Scholar] [CrossRef]
- Wang, X.; Isbrandt, T.; Strube, M.L.; Paulsen, S.S.; Nielsen, M.W.; Buijs, Y.; Schoof, E.M.; Larsen, T.O.; Gram, L.; Zhang, S.D. Chitin Degradation machinery and secondary metabolite profiles in the marine bacterium Pseudoalteromonas rubra S4059. Mar. Drugs 2021, 19, 108. [Google Scholar] [CrossRef]
- Risdian, C.; Landwehr, W.; Rohde, M.; Schumann, P.; Hahnke, R.L.; Spröer, C.; Bunk, B.; Kämpfer, P.; Schupp, P.J.; Wink, J. Streptomyces bathyalis sp. nov., an actinobacterium isolated from the sponge in a deep sea. Antonie Leeuwenhoek 2021, 114, 425–435. [Google Scholar] [CrossRef]
- Li, L.; Zhu, H.R.; Xu, Q.H.; Lin, H.W.; Lu, Y.H. Micromonospora craniellae sp. nov., isolated from a marine sponge, and reclassification of Jishengella endophytica as Micromonospora endophytica comb. nov. Int. J. Syst. Evol. Microbiol. 2019, 69, 715–720. [Google Scholar] [CrossRef]
- Lee, G.E.; Im, W.T.; Park, J.S. Bacillus spongiae sp. nov. isolated from sponge of Jeju Island. J. Microbiol. 2018, 56, 217–222. [Google Scholar] [CrossRef]
- Alex, A.; Silva, V.; Vasconcelos, V.; Antunes, A. Evidence of unique and generalist microbes in distantly related sympatric intertidal marine sponges (Porifera: Demospongiae). PLoS ONE 2013, 8, e80653. [Google Scholar] [CrossRef]
- Thompson, F.L.; Iida, T.; Swings, J. Biodiversity of Vibrios. Microbiol. Mol. Biol. Rev. 2004, 68, 403–431. [Google Scholar] [CrossRef]
- Fiore, C.L.; Jarett, J.K.; Olson, N.D.; Lesser, M.P. Nitrogen fixation and nitrogen transformations in marine symbioses. Trends Microbiol. 2010, 18, 455–463. [Google Scholar] [CrossRef]
- Hameş-Kocabaş, E.E.; Uzel, A. Isolation strategies of marine-derived actinomycetes from sponge and sediment samples. J. Microbiol. Methods 2012, 88, 342–347. [Google Scholar] [CrossRef]
- Froelich, B.A.; Daines, D.A. In hot water: Effects of climate change on Vibrio-human interactions. Environ. Microbiol. 2020, 22, 4101–4111. [Google Scholar] [CrossRef]
- Bibi, F.; Faheem, M.; Azhar, E.I.; Yasir, M.; Alvi, S.A.; Kamal, M.A.; Ullah, I.; Naseer, M.I. Bacteria from marine sponges: A source of new drugs. Curr. Drug Metab. 2017, 18, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, C.; Liu, X.; Wang, G.Y. Phylogenetic analysis and screening of antimicrobial and cytotoxic activities of culturable bacteria associated with the ascidian Botryllus schlosseri. J. Appl. Microbiol. 2020, 129, 892–905. [Google Scholar] [CrossRef] [PubMed]
- Alibert, S.; N’Gompaza Diarra, J.; Hernandez, J.; Stutzmann, A.; Fouad, M.; Boyer, G.; Pagès, J.M. Multidrug efflux pumps and their role in antibiotic and antiseptic resistance: A pharmacodynamic perspective. Expert Opin. Drug Metab. Toxicol. 2017, 13, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Cita, Y.P.; Suhermanto, A.; Radjasa, O.K.; Sudharmono, P. Antibacterial activity of marine bacteria isolated from sponge Xestospongia testudinaria from Sorong, Papua. Asian Pac. J. Trop. Biomed. 2017, 7, 450–454. [Google Scholar] [CrossRef]
- Jackson, S.A.; Crossman, L.; Almeida, E.L.; Margassery, L.M.; Kennedy, J.; Dobson, A.D.W. Diverse and abundant secondary metabolism biosynthetic gene clusters in the genomes of marine sponge derived Streptomyces spp. isolates. Mar. Drugs 2018, 16, 67. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Garzón, J.F.; Zehl, M.; Schneider, O.; Rückert, C.; Busche, T.; Kalinowski, J.; Bredholt, H.; Zotchev, S.B. Streptomyces spp. from the marine sponge Antho dichotoma: Analyses of secondary metabolite biosynthesis gene clusters and some of their products. Front. Microbiol. 2020, 11, 437. [Google Scholar] [CrossRef]
- Valasek, A.; Kiss, Í.; Fodor, I.; Kovács, M.; Urbán, P.; Jámbor, É.; Fekete, C.; Kerepesi, I. Proteomic insight into the primycin fermentation process of Saccharomonospora azurea. Acta Biol. Hung. 2016, 67, 424–430. [Google Scholar] [CrossRef]
- Zhang, Z.Q.; Zhang, J.L.; Song, R.T.; Guo, Z.X.; Wang, H.X.; Zhu, J.; Lu, C.H.; Shen, Y. Ansavaricins A–E: Five new streptovaricin derivatives from Streptomyces sp. S012. Rsc. Adv. 2017, 7, 5684–5693. [Google Scholar] [CrossRef]
- Luo, Y.; Huang, H.; Liang, J.; Wang, M.; Lu, L.; Shao, Z.; Cobb, R.E.; Zhao, H. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nat. Commun. 2013, 4, 2894. [Google Scholar] [CrossRef]
- Liu, L.; Salam, N.; Jiao, J.Y.; Jiang, H.C.; Zhou, E.M.; Yin, Y.R.; Ming, H.; Li, W.J. Diversity of culturable thermophilic actinobacteria in hot springs in Tengchong, China and studies of their biosynthetic gene profiles. Microb. Ecol. 2016, 72, 150–162. [Google Scholar] [CrossRef]
- Bull, A.T.; Stach, J.E. Marine actinobacteria: New opportunities for natural product search and discovery. Trends Microbiol. 2007, 15, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Guo, F.; Lai, Q.; Shao, Z. Notoacmeibacter marinus gen. nov., sp. nov., isolated from the gut of a limpet and proposal of Notoacmeibacteraceae fam. nov. in the order Rhizobiales of the class Alphaproteobacteria. Int. J. Syst. Evol. Microbiol. 2017, 67, 2527–2531. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.R.; Tuo, L. Notoacmeibacter ruber sp. nov., a novel endophytic bacterium isolated from leaf of Rhizophora stylosa. Antonie Leeuwenhoek 2019, 112, 919–925. [Google Scholar] [CrossRef] [PubMed]
- Gokarn, K.; Pal, R.B. Activity of siderophores against drug-resistant Gram-positive and Gram-negative bacteria. Infect. Drug Resist. 2018, 11, 61–75. [Google Scholar] [CrossRef]
- Robinson, S.L.; Christenson, J.K.; Wackett, L.P. Biosynthesis and chemical diversity of β-lactone natural products. Nat. Prod. Rep. 2019, 36, 458–475. [Google Scholar] [CrossRef]
- Danevčič, T.; Borić Vezjak, M.; Zorec, M.; Stopar, D. Prodigiosin—A multifaceted Escherichia coli antimicrobial agent. PLoS ONE 2016, 11, e0162412. [Google Scholar] [CrossRef]
- Suryawanshi, R.K.; Patil, C.D.; Koli, S.H.; Hallsworth, J.E.; Patil, S.V. Antimicrobial activity of prodigiosin is attributable to plasma-membrane damage. Nat. Prod. Res. 2017, 31, 572–577. [Google Scholar] [CrossRef]
- Dubourg, G.; Lagier, J.C.; Robert, C.; Armstrong, N.; Couderc, C.; Fournier, P.E.; Raoult, D. Risungbinella massiliensis sp. nov., a new member of Thermoactinomycetaceae isolated from human gut. Antonie Leeuwenhoek 2016, 109, 773–784. [Google Scholar] [CrossRef]
- Frikha Dammak, D.; Zarai, Z.; Najah, S.; Abdennabi, R.; Belbahri, L.; Rateb, M.E.; Mejdoub, H.; Maalej, S. Antagonistic properties of some halophilic Thermoactinomycetes isolated from superficial sediment of a solar saltern and production of cyclic antimicrobial peptides by the novel isolate Paludifilum halophilum. Bio. Med. Res. Int. 2017, 2017, 1205258. [Google Scholar] [CrossRef]
- Jiang, Z.; Xiao, M.; Yang, L.L.; Zhi, X.Y.; Li, W.J. Genome-based taxonomic classification within the family Thermoactinomycetaceae. Int. J. Syst. Evol. Microbiol. 2019, 69, 2028–2036. [Google Scholar] [CrossRef]
- Hassane, C.S.; Fouillaud, M.; Le Goff, G.; Sklirou, A.D.; Boyer, J.B.; Trougakos, I.P.; Jerabek, M.; Bignon, J.; De Voogd, N.J.; Ouazzani, J.; et al. Microorganisms associated with the marine sponge Scopalina hapalia: A reservoir of bioactive molecules to slow down the aging process. Microorganisms 2020, 8, 1262. [Google Scholar] [CrossRef] [PubMed]
- Karuppiah, V.; Li, Y.; Sun, W.; Feng, G.; Li, Z. Functional gene-based discovery of phenazines from the actinobacteria associated with marine sponges in the South China Sea. Appl Microbiol Biotechnol. 2015, 99, 5939–5950. [Google Scholar] [CrossRef] [PubMed]
- Dhiman, A.; Nanda, A.; Ahmad, S.; Narasimhan, B. In vitro antimicrobial activity of methanolic leaf extract of Psidium guajava L. J. Pharm. Bioallied Sci. 2011, 3, 226–229. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Kristensen, D.M.; Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Microbial genome analysis: The COG approach. Brief. Bioinform. 2017, 20, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef]
- Barrios-Llerena, M.E.; Burja, A.M.; Wright, P.C. Genetic analysis of polyketide synthase and peptide synthetase genes in cyanobacteria as a mining tool for secondary metabolites. J. Ind. Microbiol. Biotechnol. 2007, 34, 443–456. [Google Scholar] [CrossRef]
- Li, J.; Dong, J.D.; Yang, J.; Luo, X.M.; Zhang, S. Detection of polyketide synthase and nonribosomal peptide synthetase biosynthetic genes from antimicrobial coral-associated actinomycetes. Antonie Leeuwenhoek 2014, 106, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Du, S.; Qu, W.Y.; Guo, F.R.; Wang, G.Y. Biosynthetic potential of culturable bacteria associated with Apostichopus japonicus. J. Appl. Microbiol. 2019, 127, 1686–1697. [Google Scholar] [CrossRef] [PubMed]
| Test Strain | 16S rRNA Gene Homology (%) with Type Strains | PKS Amino Acid Sequence Homology (%) | NRPS Amino Acid Sequence Homology (%) |
|---|---|---|---|
| HMA001 | Streptomyces pratensis NRRLB 24916T/JQ806215/99.88 | - | Streptomyces sp. EN16/WP_141746166/100 |
| HMA005 | Saccharomonospora azurea DSM 44631T/Z38017/99.71 | Saccharomonospora azurea/WP_005447017/99.11 | - |
| HMA008 | Notoacmeibacter marinus MCCC 1A01882T/NR157752/99.93 | - | NRPS a: Notoacmeibacter marinus/WP_114391483/100 |
| NRPS b: Notoacmeibacter marinus/WP_114391494/100 | |||
| HMA011 | Streptomyces iakyrus DSM 40482T/NR041231/100 | - | Streptomyces sp. BAH28/AGS13655/95.51 |
| HMA013 | Streptomyces griseolus NBRC 3415T/NR041207/99.87 | - | Streptomyces sp. EN16 WP_141746166/98.72 |
| HMA015 | Nocardiopsis halotolerans DSM 44410T/AJ290448/100 | Streptomyces sp. HVGN2/ACR61390/76.92 | Nocardiopsis halotolerans/WP_083922182/100 |
| HMA023 | Nocardiopsis halotolerans DSM 44410T/AJ290448/99.71 | Nocardiopsis halotolerans DSM4410/WP_020480240/99 | - |
| HMA026 | Micromonospora aurantiaca ATCC 27029T/NR074415/100 | Micromonospora sp. RV43/WP_047892628/99.56 | Micromonospora sp. LB4/RQX23530/96.52 |
| HMA027 | Streptomyces malachitospinus NBRC 101004T/AB249954/99.86 | - | Streptomyces sp. Tu4128/WP_122619786/97.5 |
| HMA031 | Streptomyces gougerotii NBRC 3198T/NR041201/100 | - | Streptomyces griseus/WP_115068788/98.25 |
| HMA032 | Streptomyces gougerotii NBRC 3198T/NR041201/100 | - | Streptomyces griseus/WP_115068788/98.69 |
| HMA033 | Streptomyces gougerotii NBRC 3198T/NR041201/100 | - | Streptomyces griseus/SUP57408/98.69 |
| HMA034 | Melghirimyces algeriensis DSM 45474T/HQ383683/99.65 | Melghirimyces algeriensis/WP_142506588/99.55 | - |
| HMA035 | Streptomyces malachitospinus NBRC 101004T/AB249954/99.87 | - | Streptomyces sp. Tu4128/WP_122619786/96.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Wang, X.-N.; Bi, H.-Y.; Wang, G.-Y. Antimicrobial Biosynthetic Potential and Phylogenetic Analysis of Culturable Bacteria Associated with the Sponge Ophlitaspongia sp. from the Yellow Sea, China. Mar. Drugs 2022, 20, 588. https://doi.org/10.3390/md20100588
Chen L, Wang X-N, Bi H-Y, Wang G-Y. Antimicrobial Biosynthetic Potential and Phylogenetic Analysis of Culturable Bacteria Associated with the Sponge Ophlitaspongia sp. from the Yellow Sea, China. Marine Drugs. 2022; 20(10):588. https://doi.org/10.3390/md20100588
Chicago/Turabian StyleChen, Lei, Xue-Ning Wang, Hong-Yu Bi, and Guang-Yu Wang. 2022. "Antimicrobial Biosynthetic Potential and Phylogenetic Analysis of Culturable Bacteria Associated with the Sponge Ophlitaspongia sp. from the Yellow Sea, China" Marine Drugs 20, no. 10: 588. https://doi.org/10.3390/md20100588
APA StyleChen, L., Wang, X.-N., Bi, H.-Y., & Wang, G.-Y. (2022). Antimicrobial Biosynthetic Potential and Phylogenetic Analysis of Culturable Bacteria Associated with the Sponge Ophlitaspongia sp. from the Yellow Sea, China. Marine Drugs, 20(10), 588. https://doi.org/10.3390/md20100588







