New Alkaloids from the Mediterranean Sponge Hamigera hamigera
Abstract
:Introduction
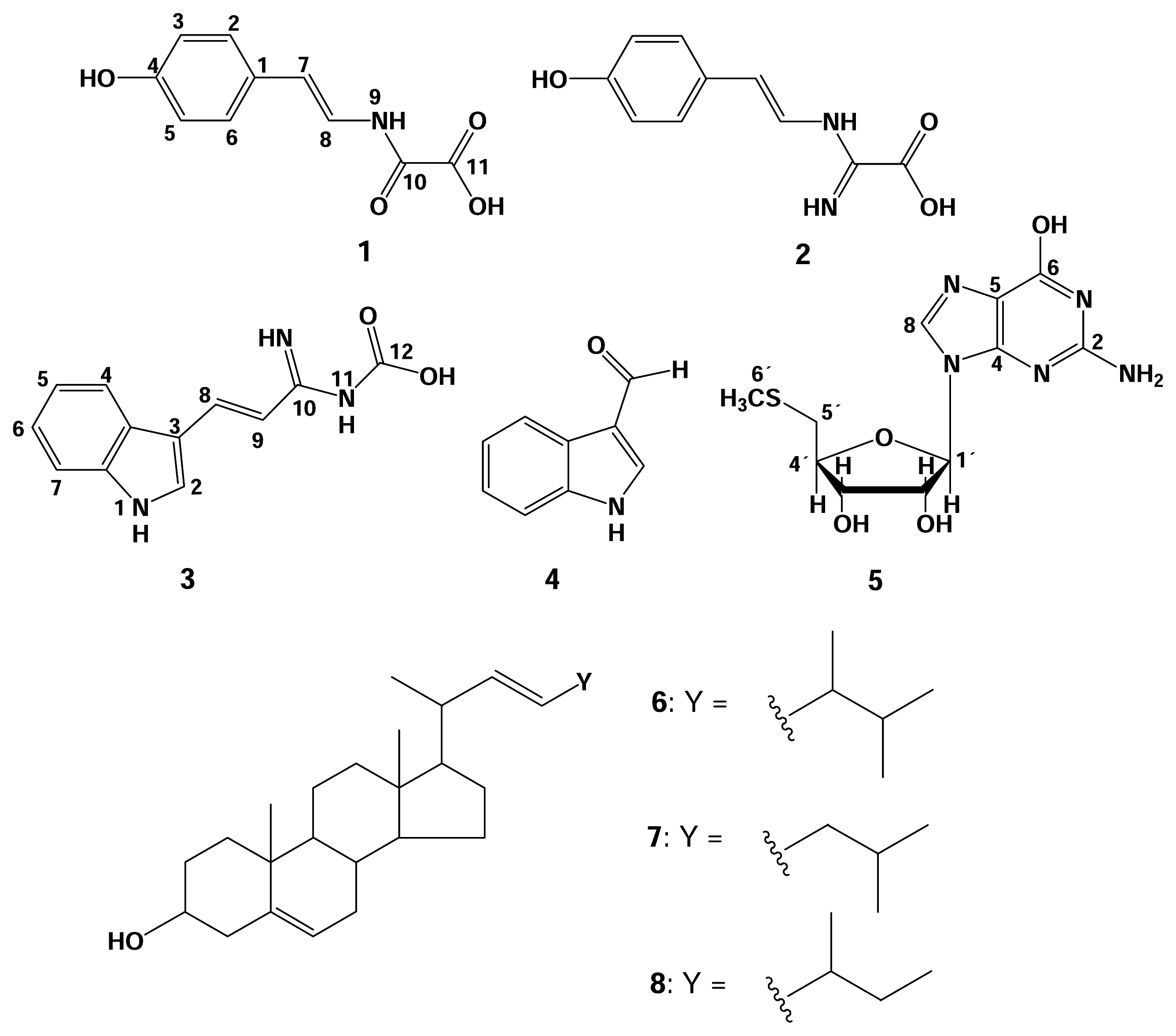
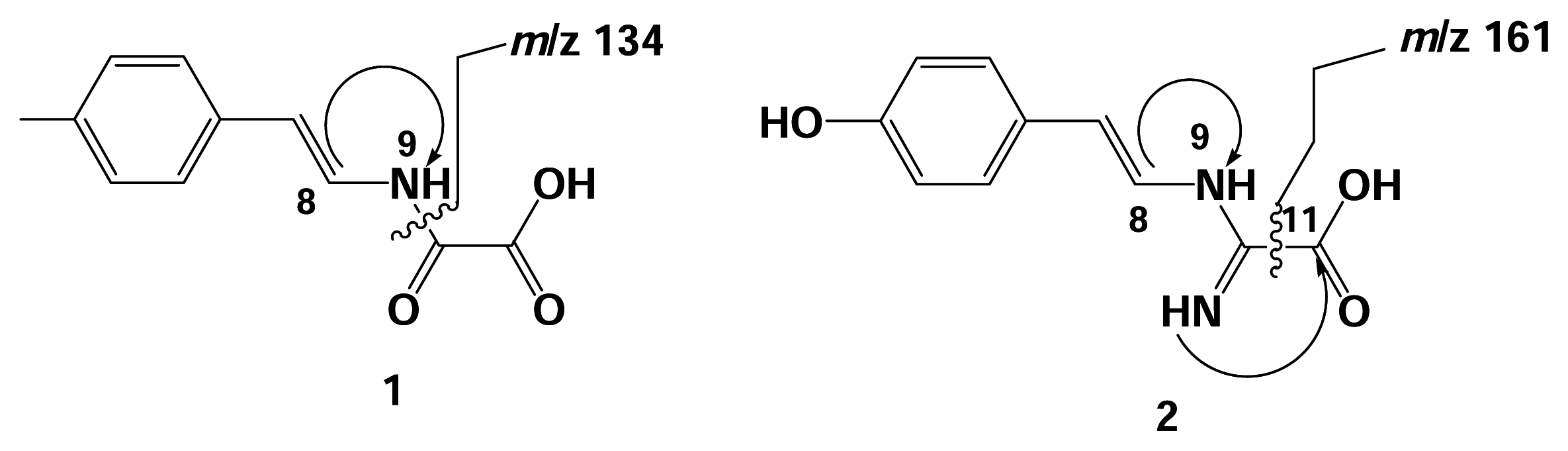
Results and Discussion

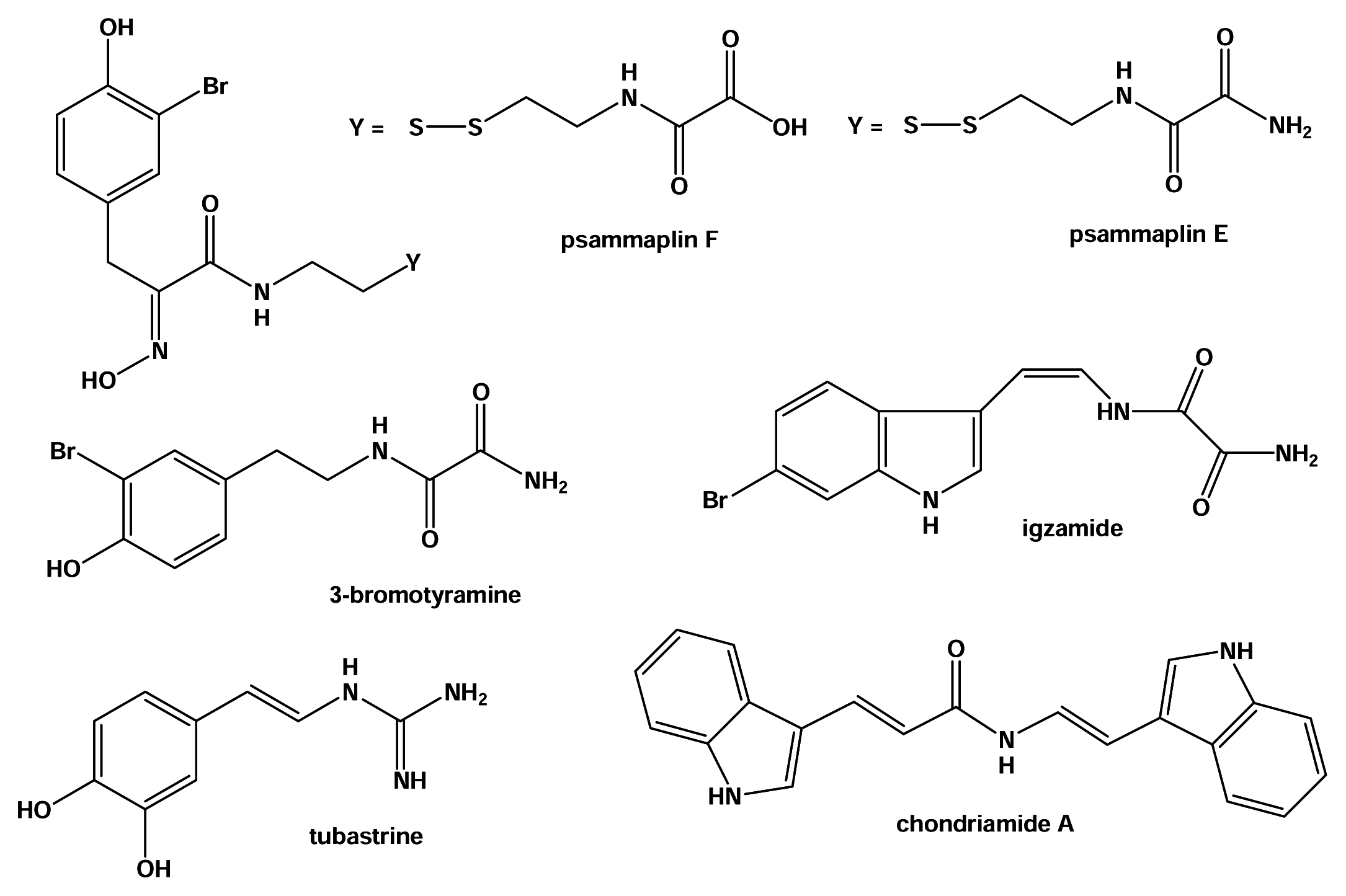
Material and Methods
General experimental procedures
Animal Material
Extraction and isolation
Spectroscopic Data
Feeding experiments with Blennius sphinx

| 1 | 2 | |||||
|---|---|---|---|---|---|---|
| No | δH (m, J in Hz) | δC | HMBC (δH to δC) | δH (m, J in Hz) | δC | HMBC (δH to δC) |
| 1 | 128.2 s | 125.9 s | ||||
| 2 | 7.09 (d, 8.5) | 127.9 d | C-1, C-4, C-7 | 7.25 (d, 8.4) | 127.5 d | C-1, C-6, C-7 |
| 3 | 6.67 (d, 8.5) | 116.5 d | C-1, C-4, C-5 | 6.75 (d, 8.4) | 115.6 d | C-1, C-4, C-5 |
| 4 | 9.31 (OH, br s) | 156.4 s | C-3, C-4, C-5 | 9.62 (OH, br s) | 157.3 s | C-3, C-4, C-5 |
| 5 | 6.67 (d, 8.5) | 116.5 d | C-1, C-3, C-4 | 6.75 (d, 8.4) | 115.6 d | C-1, C-3, C-4 |
| 6 | 7.09 (d, 8.5) | 127.9 d | C-1, C-4, C-7 | 7.25 (d, 8.4) | 127.5 d | C-1, C-2, C-7 |
| 7 | 6.35 (d,14.5) | 114.0 d | C-2, C-6, C-8 | 6.81 (d, 13.9) | 121.3 d | C-2, C-6, C-8 |
| 8 | 7.10 (dd, 14.5, 10.7) | 122.1 d | C-1, C-7 | 7.29 (br d, 13.9) | 119.5 d | C-7, C-9, C-10 |
| 9 | 10.07 (NH, d, 10.7) | 10.97 (NH, br s) | ||||
| 10 | 163.0 s | 9.29 (NH, br s)a | 155.1 s | C-11 | ||
| 11 | 158.3 s | 9.10 (OH, br s)a | 155.7 s | |||
| No. | δH (m, J in Hz) | δCa | HMBC (δH to δC) |
|---|---|---|---|
| 1 | 11.20 (NH, br s) | ||
| 2 | 7.50 (s) | 127.3 d | C-3, C-3a, C-7a, C-8 |
| 3 | 111.2 s | ||
| 3a | 124.5 s | ||
| 4 | 7.90 (d, 7.8) | 120.1 d | C-6, C-3a, C-7a |
| 5 | 7.08 (t, 7.8) | 119.0 d | C-7, C-3a |
| 6 | 7.15 (t, 7.8) | 121.5 d | C-7a |
| 7 | 7.40 (d, 7.8) | 112.3 d | C-5, C-3a |
| 7a | 138.4 s | ||
| 8 | 7.10 (d,14.0) | 117.3 d | C-2, C-3a |
| 9 | 7.18 (d, 14.0) | 118.2 d | C-3 |
| 10 | b | ||
| 11 | 8.91 (NH, br s) | ||
| 12 | b | b |
| No. | δH (m, J in Hz) | δC | 1H-1H COSY | HMBC (δH to δC) |
|---|---|---|---|---|
| 2 | (NH2) 6.40 (br s) | 153.5 s | ||
| 4 | 153.0 s | |||
| 5 | 118.0 s | |||
| 6 | (OH) 10.57 (br s) | 157.2 s | ||
| 8 | 7.90 s | 135.0 d | C-5, C-4, C-6 | |
| 1′ | 5.60 (d, 5.7) | 87.0 d | H-2′ | C-8, C-4, C-3′ |
| 2′ | 4.50 (q, 5.7) | 74.0 d | H-1′, H-3′ | C-1′, C-4′ |
| 2′-OH | 5.40 (d, 6.3) | C-1′, C-3′ | ||
| 3′ | 3.95 (m) | 71.5 d | H-2′, H-4′ | C-1′ |
| 3′-OH | 5.20 (d, 5.1) | C-2′, C-4′ | ||
| 4′ | 4.05 (t, 6.9) | 84.0 d | H-3′, H-5′A, H-5′B | C-3′ |
| 5′A | 2.80 (dd, 13.9, 6.9) | 35.0 t | H-5′ B, H-4′ | C-6′, C-3′, C-4′ |
| 5′B | 2.70 (dd, 13.9, 6.9) | |||
| 6′ | 2.05 (s) | 16.0 q | H-5′A, H-4′ | C-5′ |
Acknowledgments
- Sample Availability: Samples are available from the authors.
References and Notes
- Casapullo, A.; Finamore, E.; Minale, L.; Zollo, F. A dimeric peptide alkaloid of a completely new type, Anchinopeptolide A, from the marine sponge Anchinoe tenacior. Tetrahedron Lett 1993, 34, 6297–6300. [Google Scholar]
- Casapullo, A.; Minale, L.; Zollo, F. The unique 6-(p-hydroxyphenyl)-2-H-3,4-dihydro-1,1-dioxo-1,4-thiazine and the new tripeptide L-glu-gly-4-hydroxystirylamine from the marine sponge Anchinoe tenacior. Tetrahedron Lett 1994, 35, 2421–2422. [Google Scholar]
- Casapullo, A.; Minale, L.; Zollo, F. Four new dimeric peptide alkaloids, anchinopeptolides B-D, and cycloanchinopeptolide C, congeners of anchinopeptolide A, from the Mediterrenean marine sponge Anchinoe tenacior. J. Nat. Prod 1994, 57, 1227–1233. [Google Scholar]
- Rudi, A.; Stein, Z.; Green, S.; Goldberg, I.; Kashman, Y. Phorbazoles A-D, novel chlorinated phenylpyrrolyloxazoles from the marine sponge Phorbas aff. clathrata. Tetrahedron Lett 1994, 35, 2589–2492. [Google Scholar]
- Searle, A. S.; Molinski, T. F. Absolute configuration of Phorboxazole A and B from the marine sponge Phorbas sp. 1. Macrolide and hemiketal rings. J. Am. Chem. Soc 1996, 118, 9422–9423. [Google Scholar]
- Molinski, T. F. Absolute configuration of Phorboxazole A and B from the marine sponge Phorbas sp. 2. C-43 and complete stereochemistry. Tetrahedron Lett 1996, 37, 7879–7880. [Google Scholar]
- Wellington, K. D.; Cambie, R. C.; Rutledge, P. S.; Bergquist, R. Chemistry of sponges. 19. Novel bioactive metabolites from Hamigera tarangensis. J. Nat. Prod 2000, 63, 79–85. [Google Scholar]
- Cambie, R. C.; Lai, A. R.; Kernan, M. R.; Bergquist, P. R. Chemistry of sponges. A novel brominated benzocyclooctane derivative from Hamigera hamigera. J. Nat. Prod 1995, 58, 940–942. [Google Scholar]
- Guella, G.; Mancini, I.; Zibrowius, H.; Pietra, F. 160. Aplysinopsin-type alkaloids from Dendrophyllia sp., a Scleractinian coral of the family Dendrophylliidae of the Philippines. Facile photochemical (Z/E). Helv. Chim. Acta 1989, 72, 1444–1450. [Google Scholar]
- Anjaneyulu, V.; Rao, K. N. R.; Kobayashi, M. (24S)-24-Methylcholesta-4-ene-3β,6β-diol from a gorgonian (Rumphella aggregata) of the Andaman and Nicobar Islands. Ind. J. Chem 1995, 34B, 78–80. [Google Scholar]
- Wright, J.; McInnes, A.; Shmizu, S.; Smith, D.; Walter, J. Identification of C-24 alkylepimers of marine sterols by 13C nuclear magnetic resonance spectroscopy. Can. J. Chem 1978, 56, 1898–1903. [Google Scholar]
- Itoh, T.; Sica, D.; Djerassi, C. Minor and trace sterols in marine invertebrate. Part 1. 35. Isolation and structure elucidation of seventy four sterols from the sponge Axinella cannabina. J. Chem. Soc. Perkin Trans 1983, 1, 147–153. [Google Scholar]
- Palermo, J. A.; Brasco, M. F. R.; Seldes, A. M. Stroniamides A-D: Alkaloids from a Patagonian sponge Cliona sp. Tetrahedron 1996, 52, 2727–2734. [Google Scholar]
- Bokesch, H. R.; Pannell, L. K.; McKee, T. C.; Boyd, M. R. Coscinamides A, B, and C, three new indole alkaloids from the marine sponge Coscinoderma sp. Tetrahedron Lett 2000, 41, 6305–6308. [Google Scholar]
- Piña, I. C.; Gautschi, J. T.; Wang, G.-Y.-S.; Sanders, M. L.; Schmitz, F. J.; France, D.; Cornell-Kennon, S.; Sambucetti, L. C.; Remiszewski, S. W.; Perez, L. B.; Bair, K. W.; Crews, P. Psammaplins from the sponge Pseudoceratina purpurea: Inhibition of both histone deacetylase and DNA methyltransferase. J. Org. Chem 2003, 68, 3866–3873. [Google Scholar]
- Pettit, G. R.; Butler, M. S.; Williams, M. D.; Filiatrault, M. J.; Pettit, R. K. Isolation and structure of hemibastadinols 1–3 from the Papua New Guinea marine sponge Ianthella basta. J. Nat. Prod 1996, 59, 327–934. [Google Scholar]
- Dumdei, E.; Andersen, R. J. Igzamide, a metabolite of the marine sponge Plocamissma igzo. J. Nat. Prod 1993, 56, 792–794. [Google Scholar]
- Sakai, R.; Higa, T. Tubastrine, a new guanidostyrene from the coral Tubastrea aurea. Chem. Lett 1987, 122–128. [Google Scholar]
- Sperry, S.; Crews, P. Dihydrotubastrines: Phenethylguanidine analogues from the Indo-Pacific marine sponge Petrosia cf. contignata. J. Nat. Prod 1998, 61, 859–861. [Google Scholar]
- Davyt, D.; Entz, W.; Fernandez, R.; Mariezcurrena, R.; Mombrú, A. W.; Saldaña, J.; Domínguez, L.; Coll, J.; Manta, E. A new indole derivative from the red alga Chondria atropurpurea. Isolation, structure determination, and anthelmintic activity. J. Nat. Prod 1998, 61, 1560–1563. [Google Scholar]
- Pretsch, E.; Seibl, J.; Simon, W.; Clerc, T. Tabellen zur Strukturaufklärung organischer Verbindungen mit spektroskopischen Methoden; 1990; Heidelberg; Springer Verlag; p. C212. [Google Scholar]
- Balzarini, J.; Haller-Meier, F.; De Clercq, E.; Meier, C. Antiviral activity of cyclosaligenyl prodrugs of acyclovir, carbovir and abacavir. Antivir. Chem. Chemother 2001, 12, 301–306. [Google Scholar]
- Hay, M. E.; Stachowicz, J. J.; Cruz-Rivera, E.; Bullard, S.; Deal, M. S.; Linquist, N. Bioassays with marine and freshwater macroorganisms. Haynes, KF, Millar, J. G., Eds.; In Methods in Chemical Ecology; 1998; Volume 2, Bioassay methods; Chapman and Hall: New York; pp. 39–141. [Google Scholar]
© 2004 by MDPI Reproduction is permitted for noncommercial purposes.
Share and Cite
Hassan, W.; Edrada, R.; Ebel, R.; Wray, V.; Proksch, P. New Alkaloids from the Mediterranean Sponge Hamigera hamigera. Mar. Drugs 2004, 2, 88-100. https://doi.org/10.3390/md203088
Hassan W, Edrada R, Ebel R, Wray V, Proksch P. New Alkaloids from the Mediterranean Sponge Hamigera hamigera. Marine Drugs. 2004; 2(3):88-100. https://doi.org/10.3390/md203088
Chicago/Turabian StyleHassan, Wafaa, RuAngelie Edrada, Rainer Ebel, Victor Wray, and Peter Proksch. 2004. "New Alkaloids from the Mediterranean Sponge Hamigera hamigera" Marine Drugs 2, no. 3: 88-100. https://doi.org/10.3390/md203088
APA StyleHassan, W., Edrada, R., Ebel, R., Wray, V., & Proksch, P. (2004). New Alkaloids from the Mediterranean Sponge Hamigera hamigera. Marine Drugs, 2(3), 88-100. https://doi.org/10.3390/md203088




