Thirtieth Anniversary of the Discovery of Laxaphycins. Intriguing Peptides Keeping a Part of Their Mystery
Abstract
1. Introduction
2. Discovery and Characterization of Laxaphycins and Their Derivatives
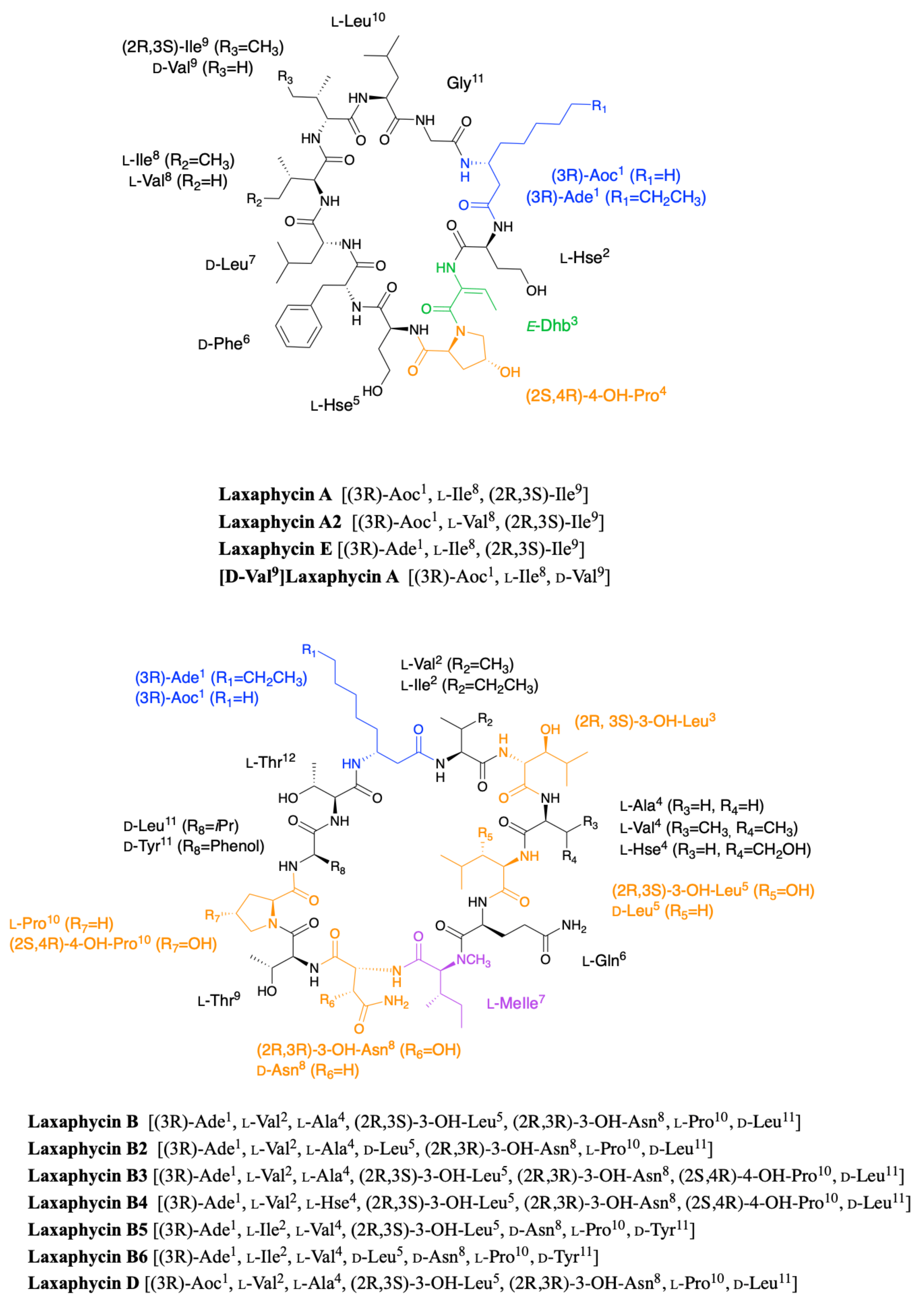
2.1. Laxaphycins
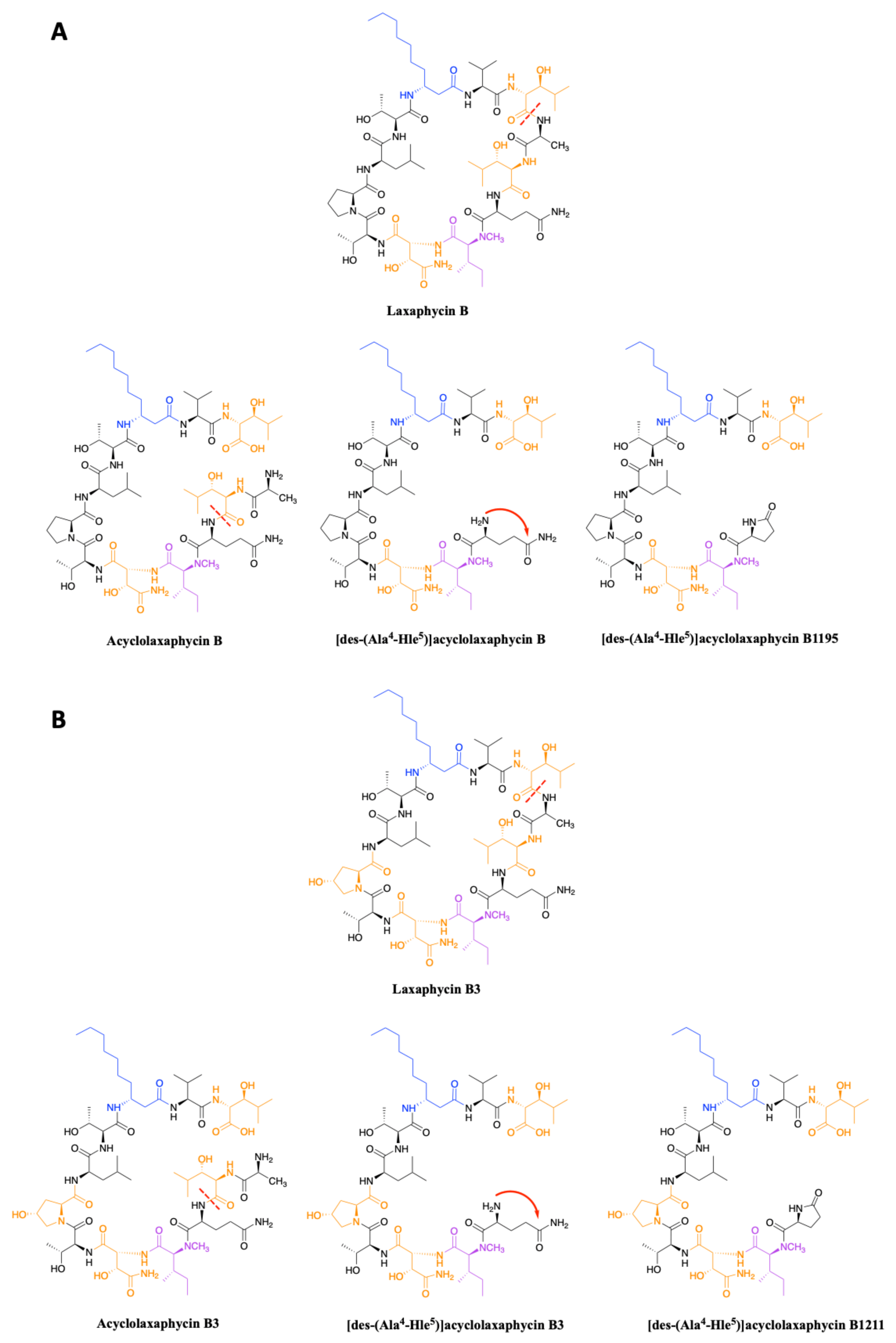
2.1.1. Cyclic Laxaphycins
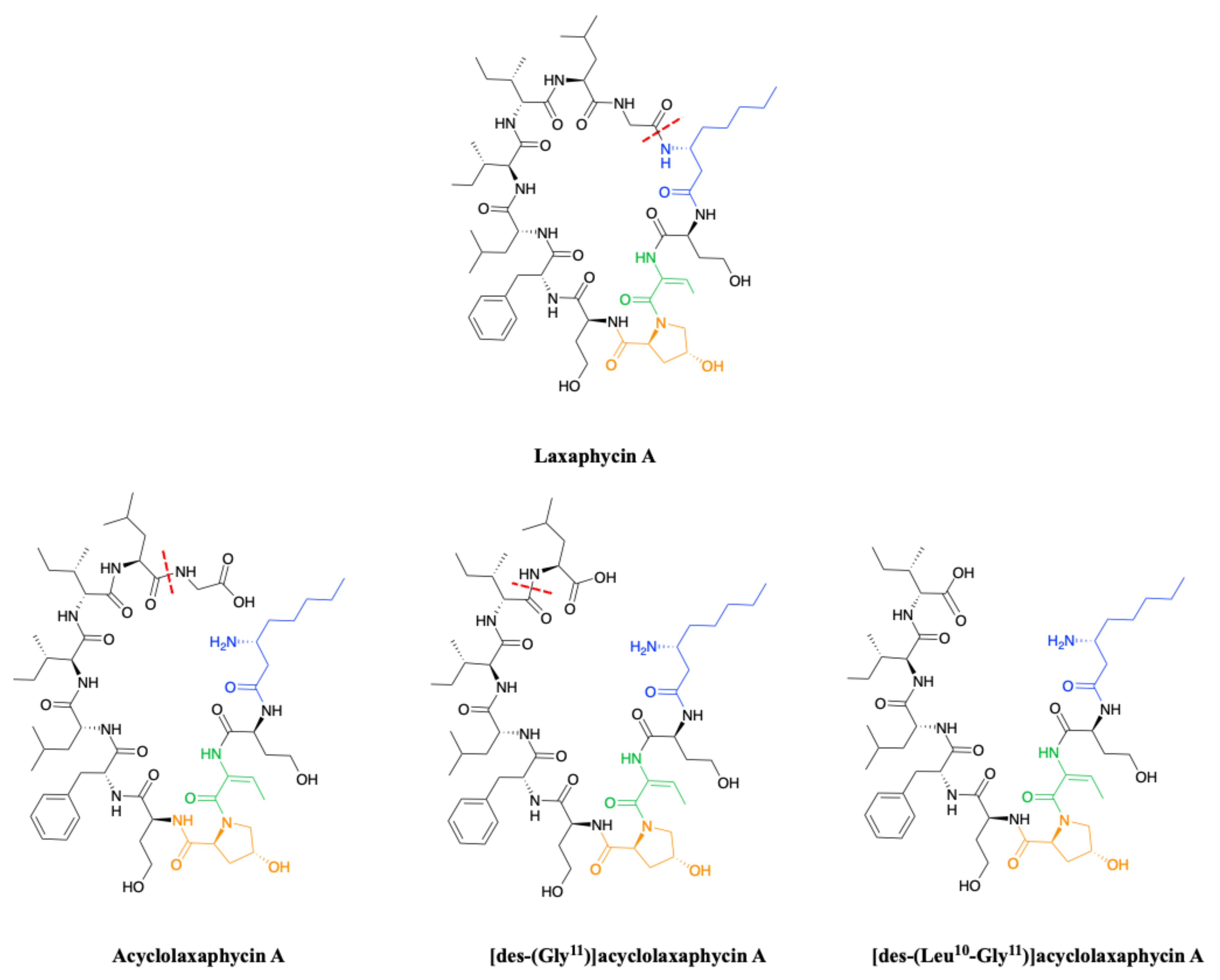
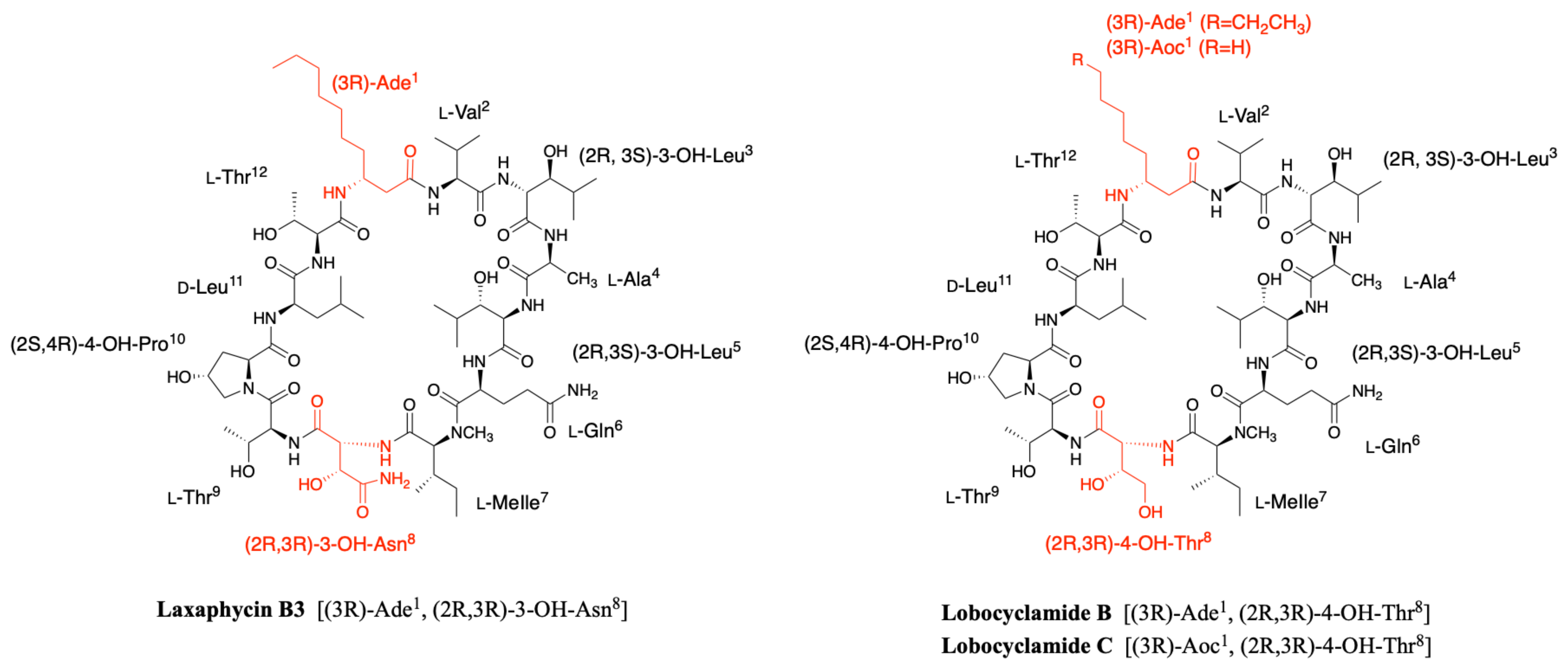
2.1.2. Acyclic Laxaphycins
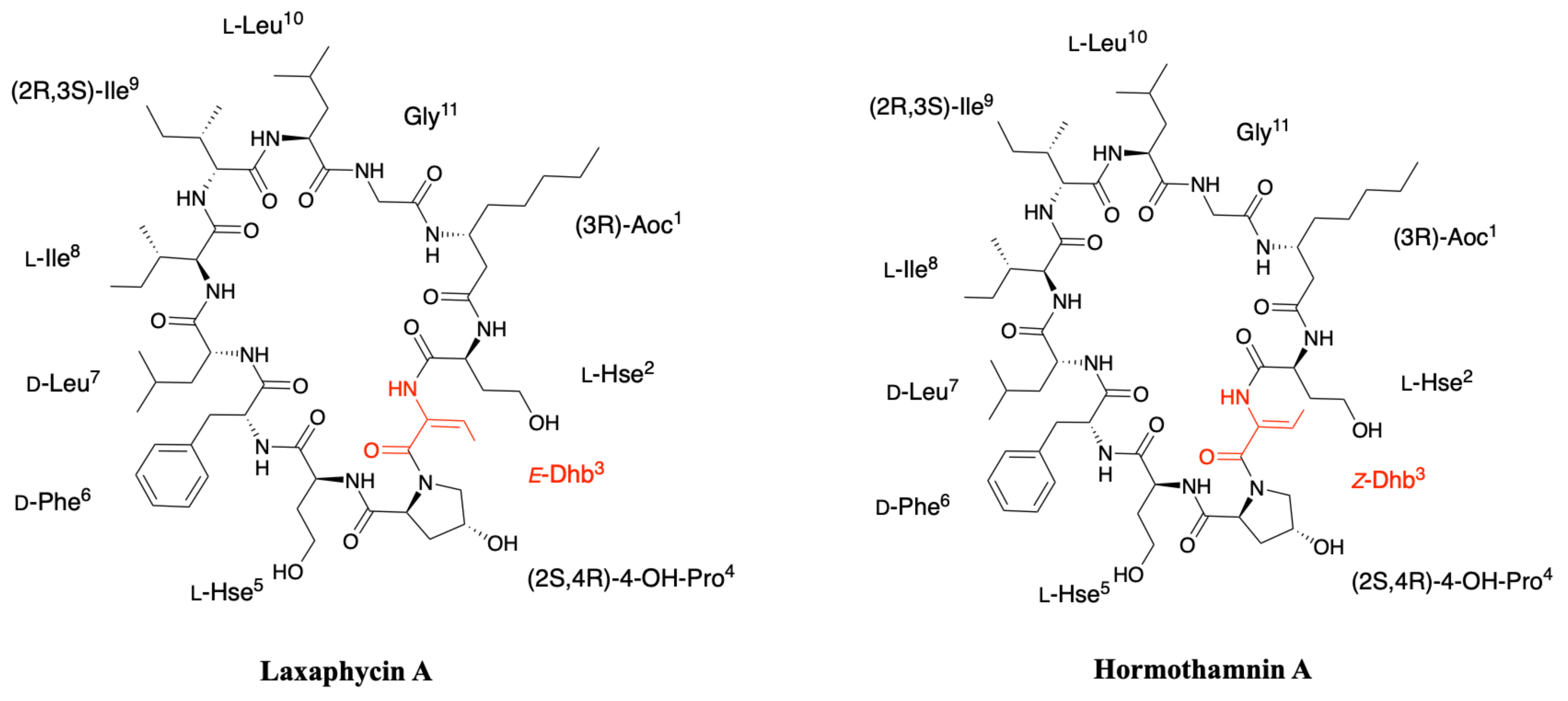
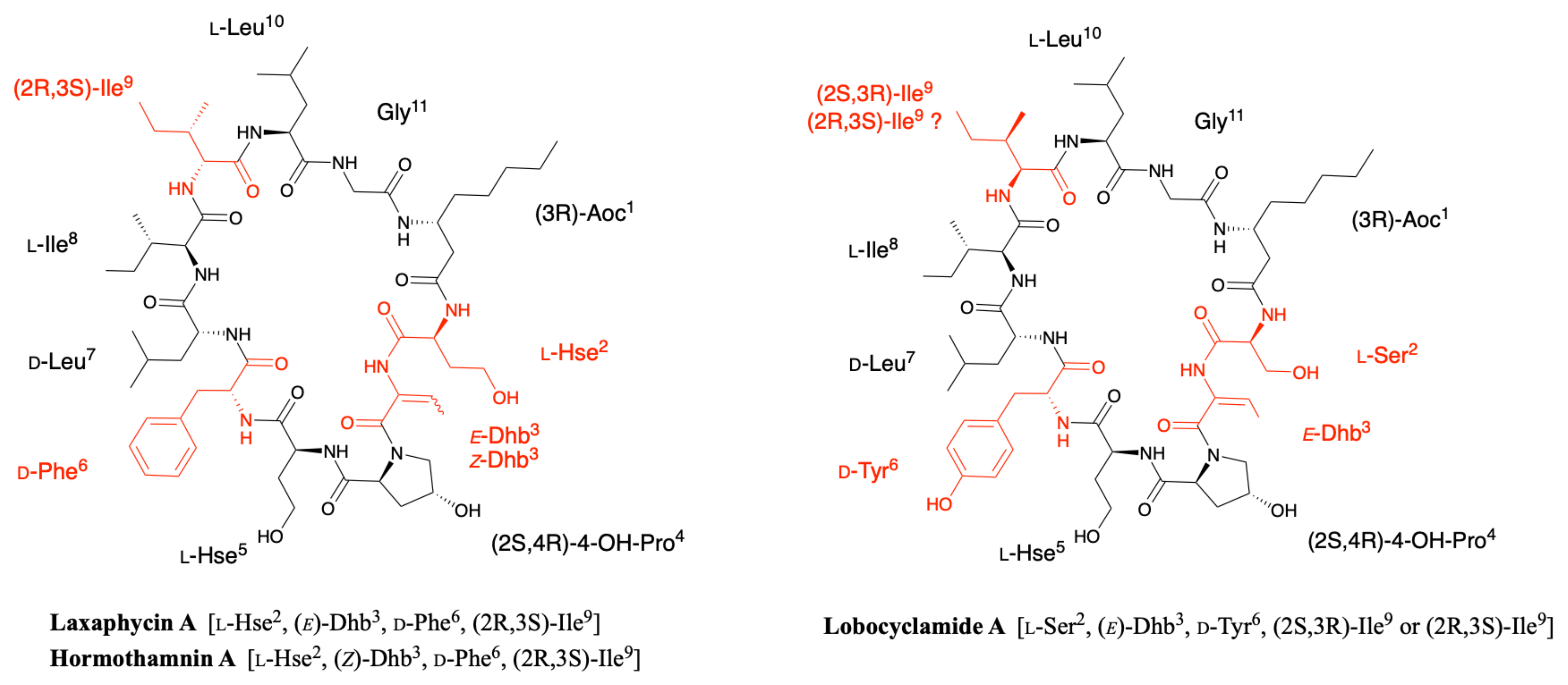
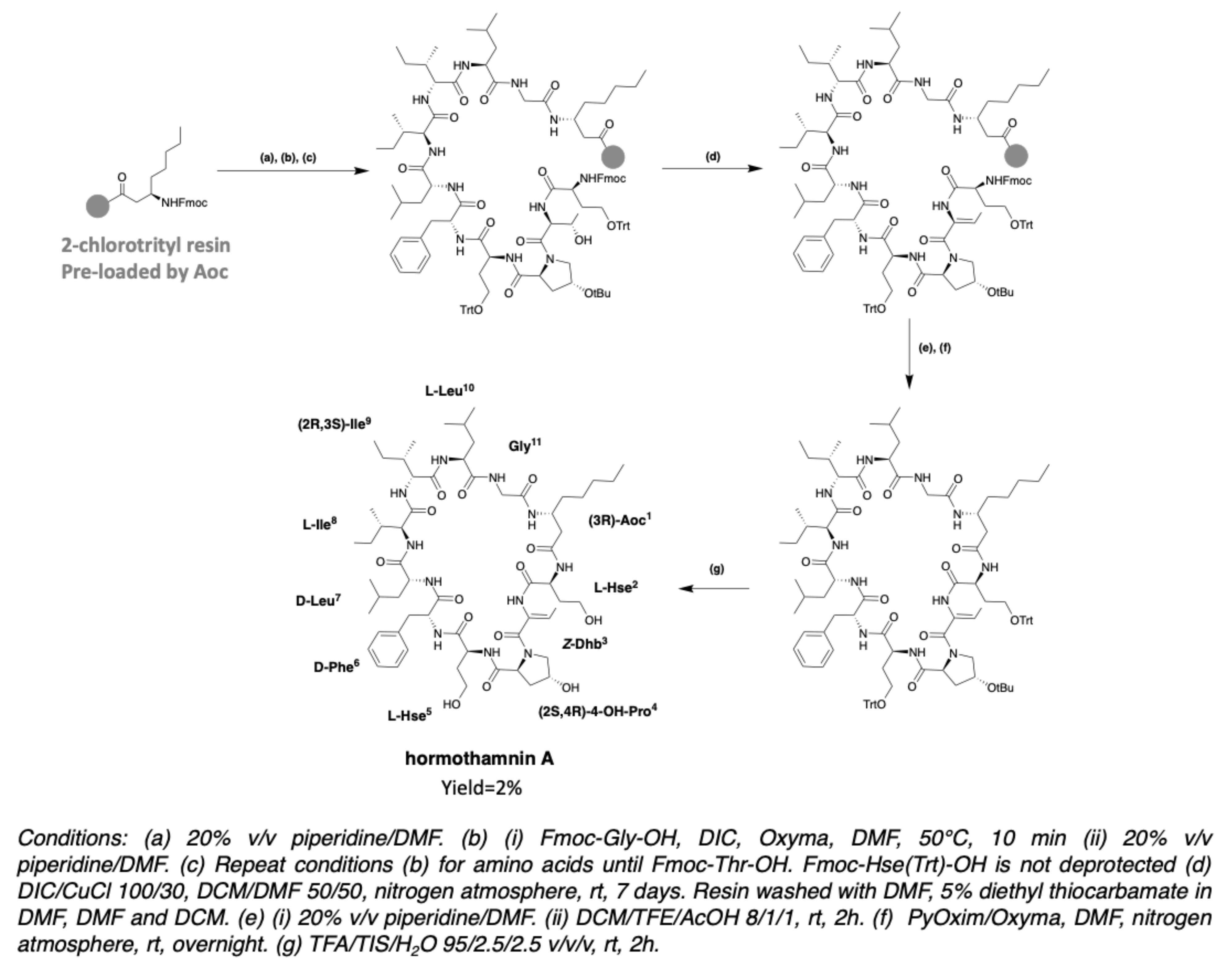
2.2. Hormothamnin
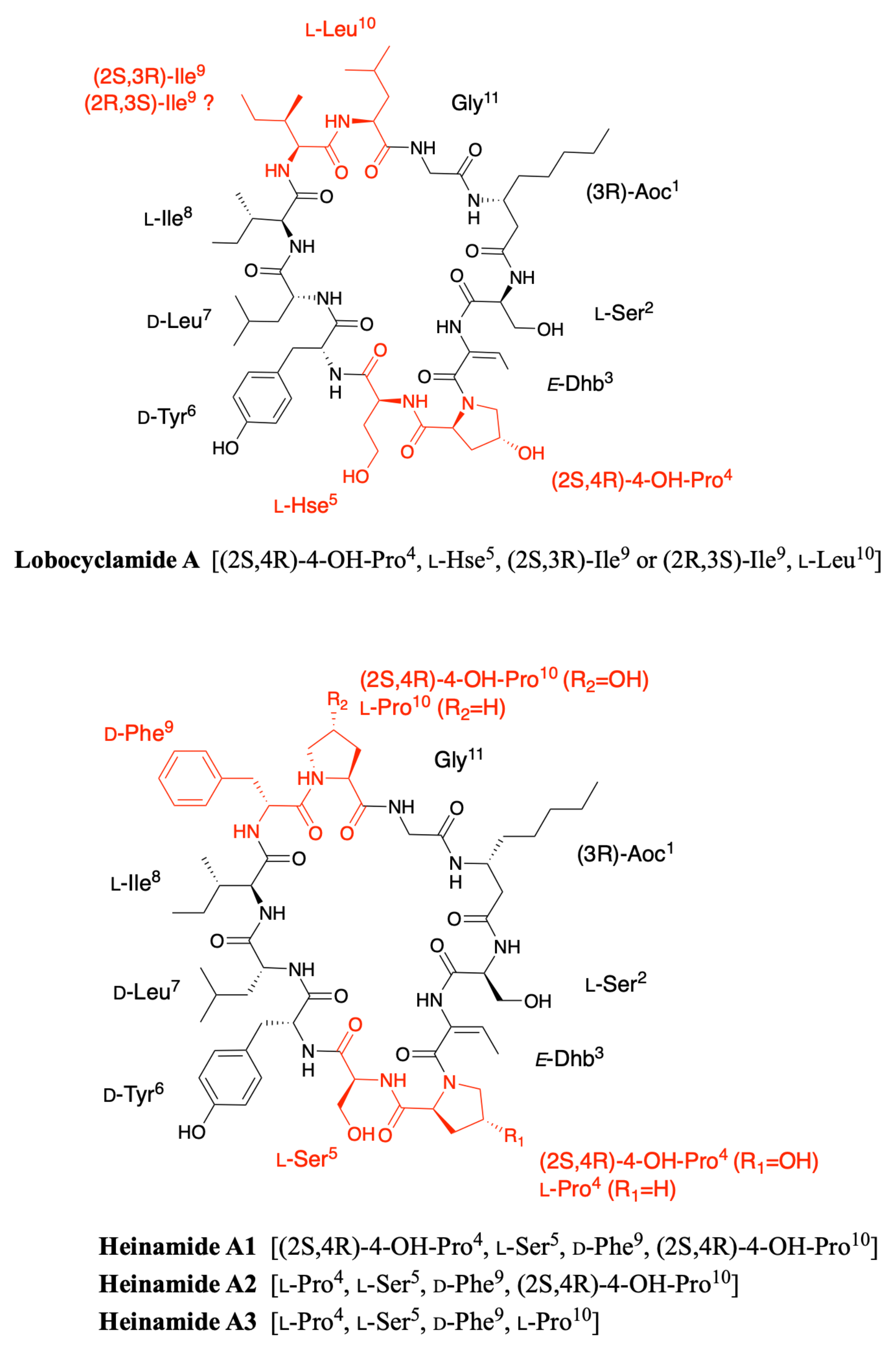
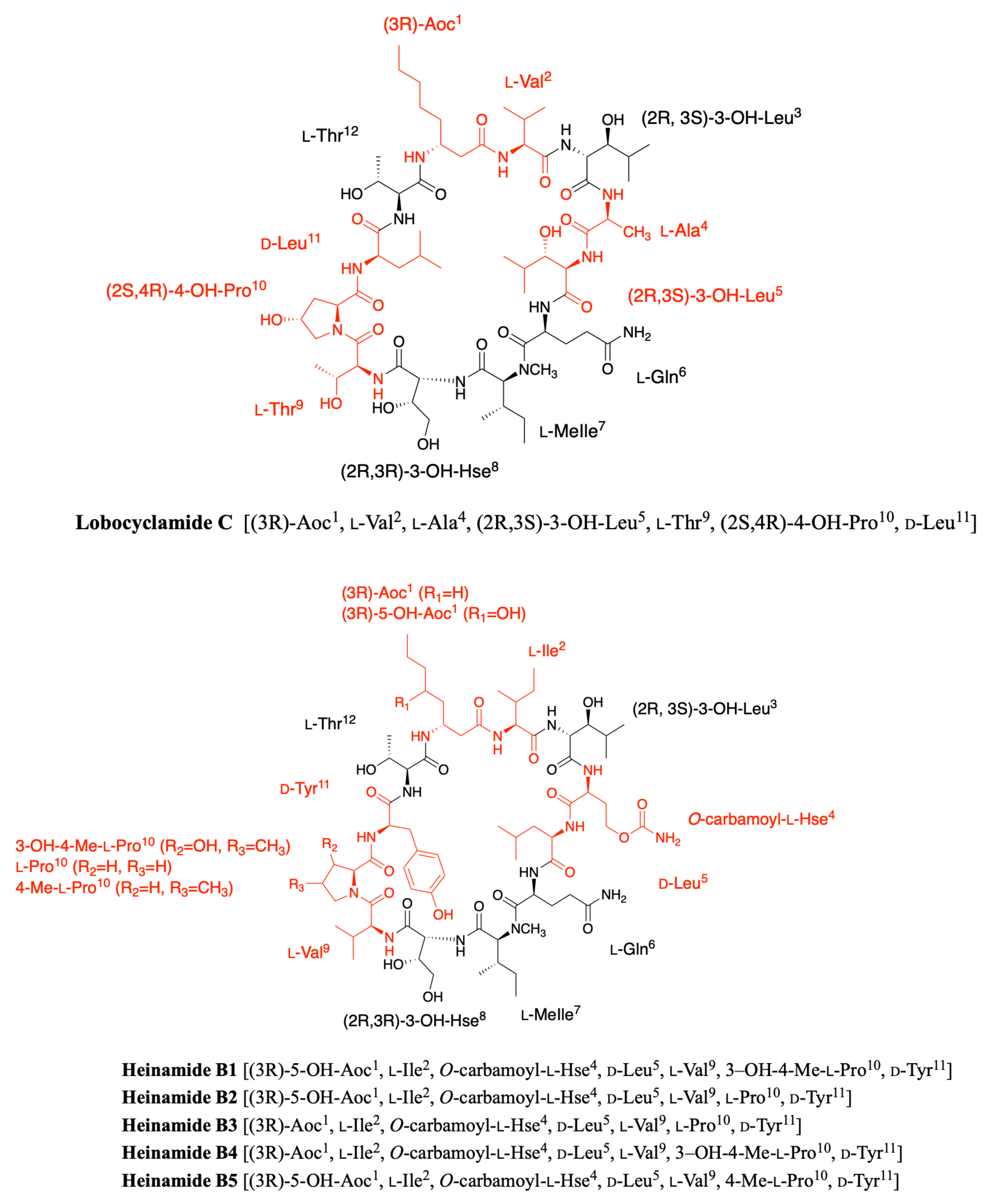
2.3. Lobocyclamides
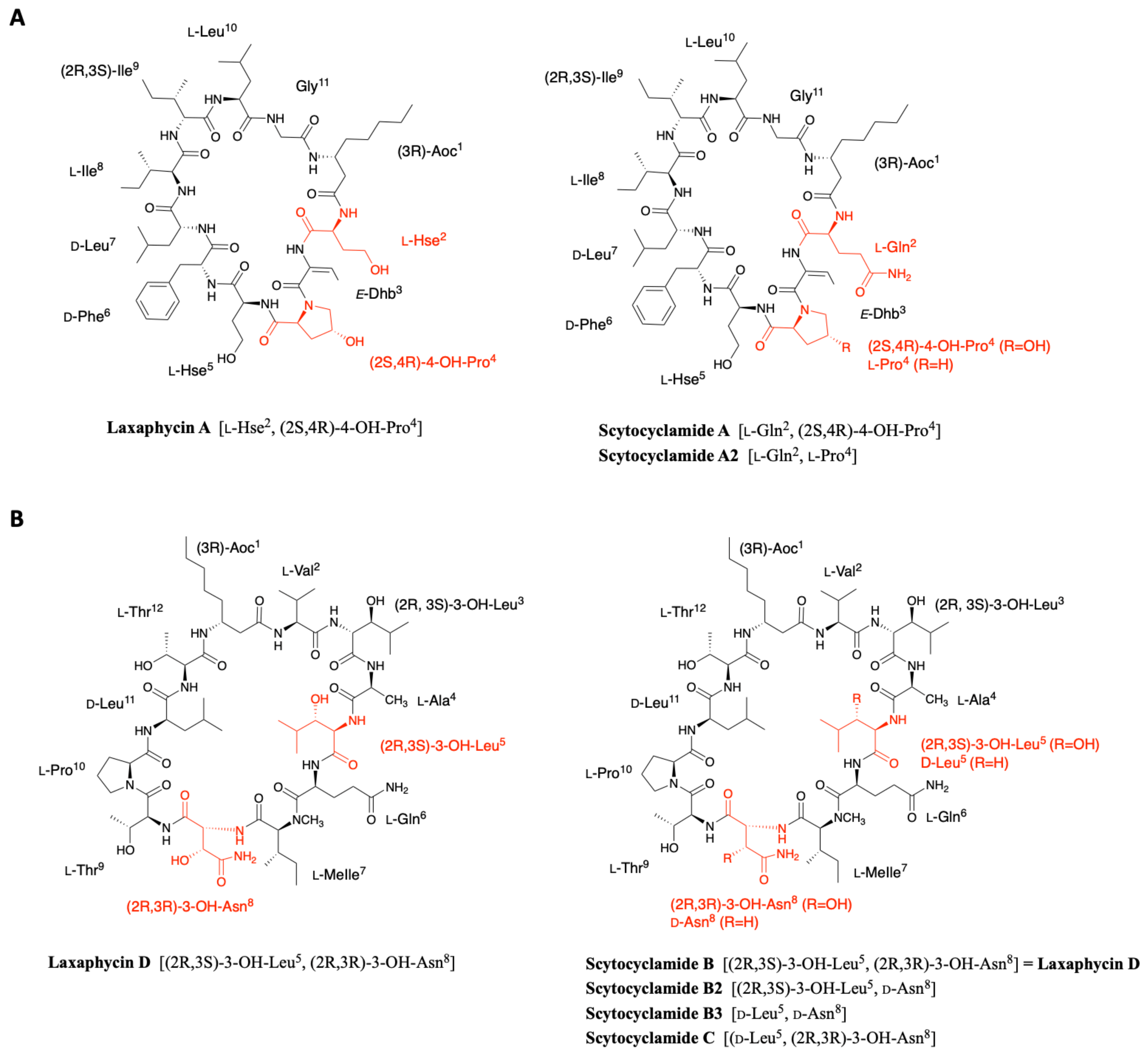
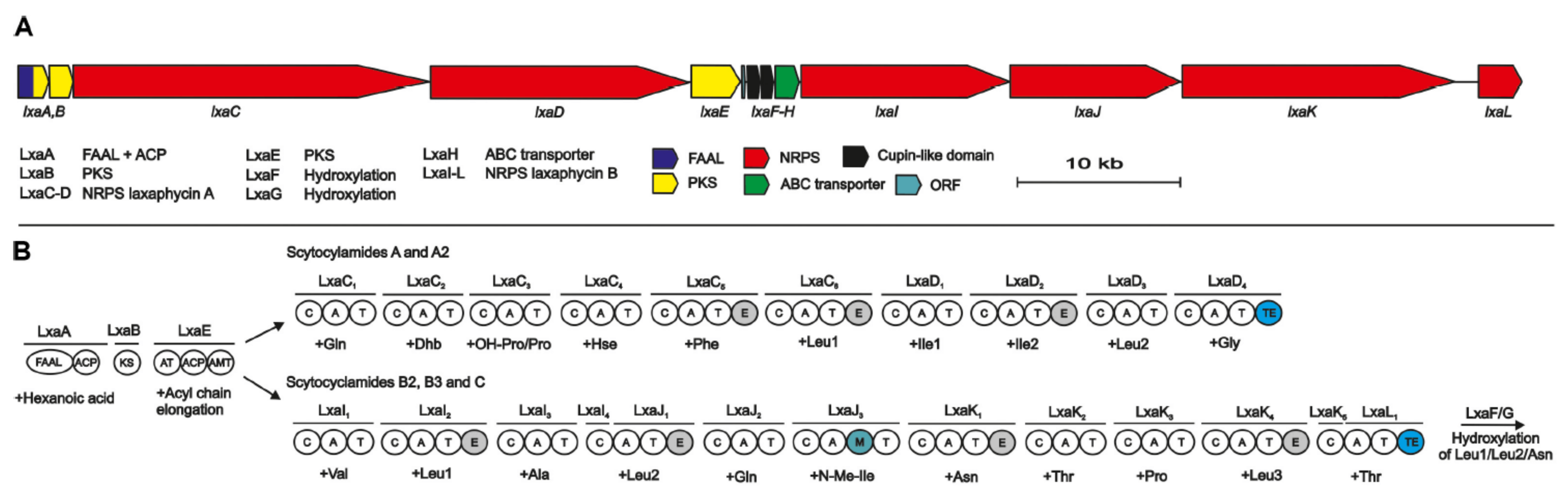
2.4. Scytocyclamides
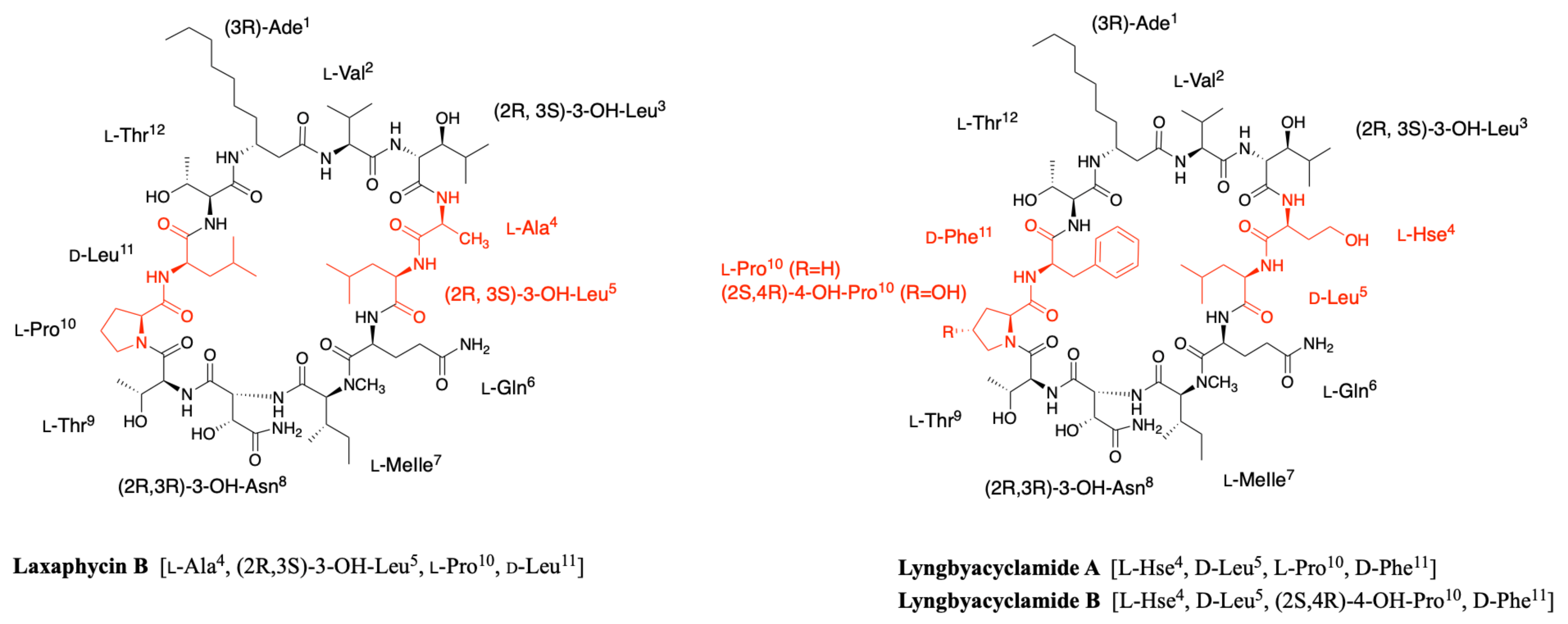
2.5. Lyngbyacyclamides
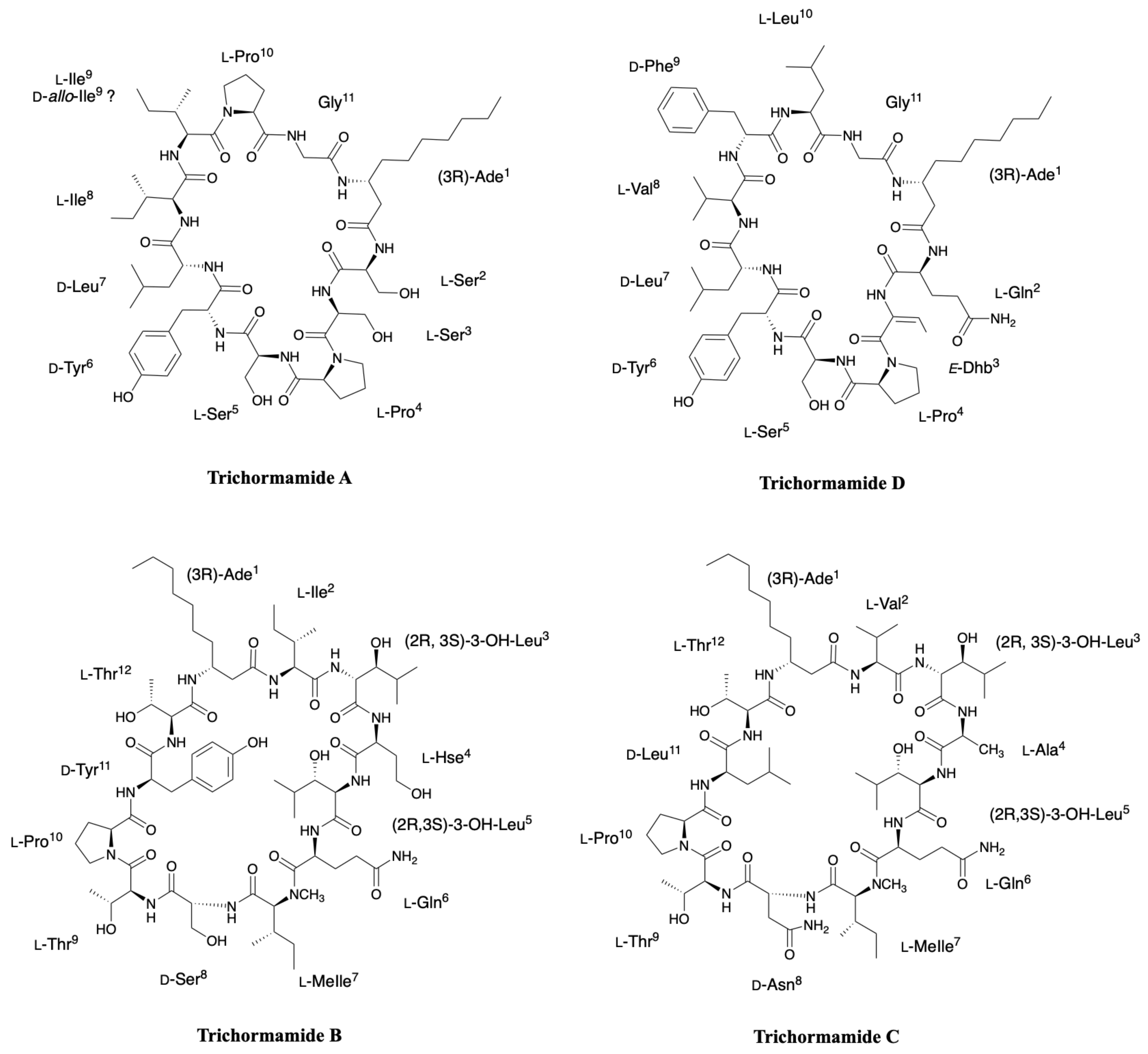
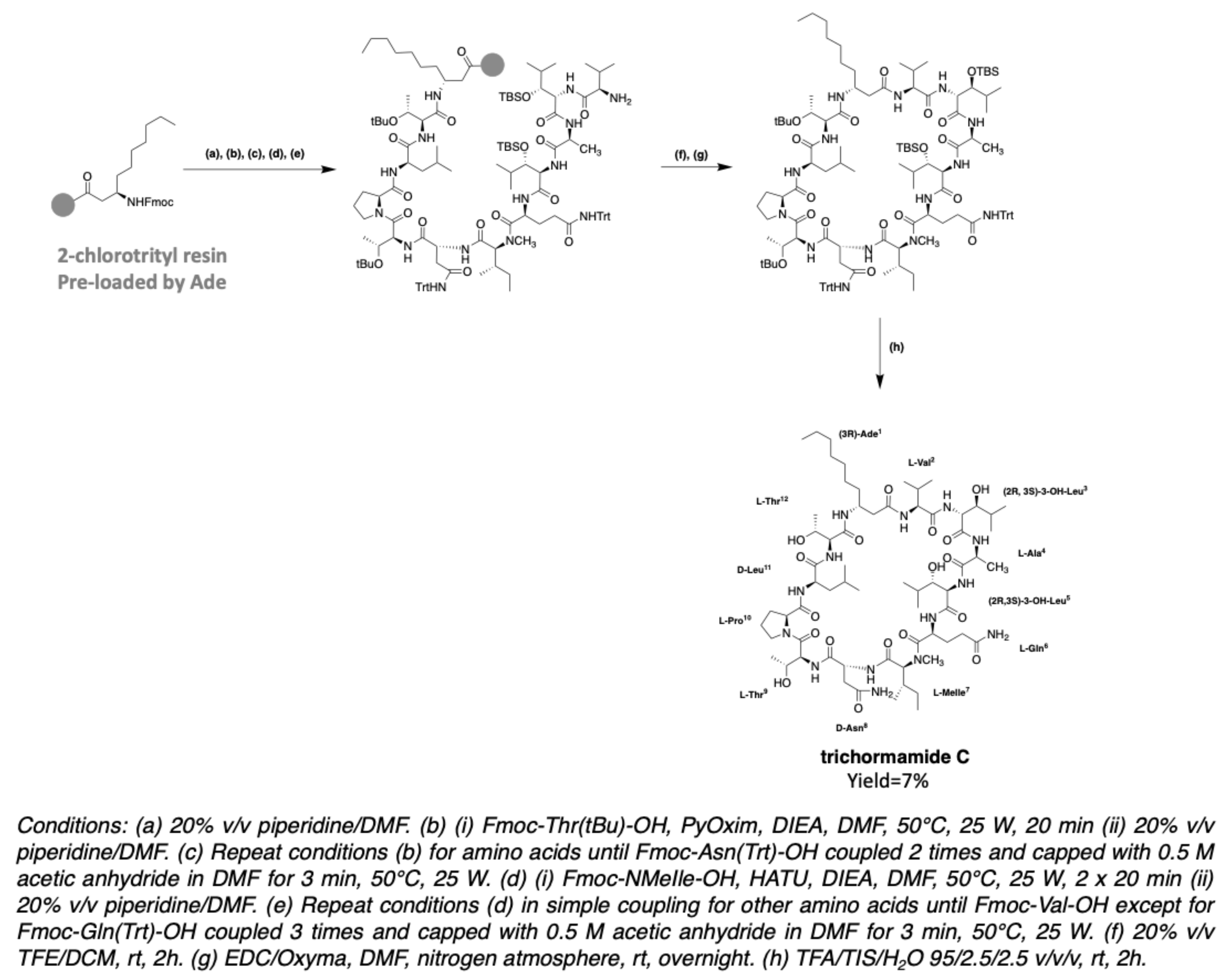
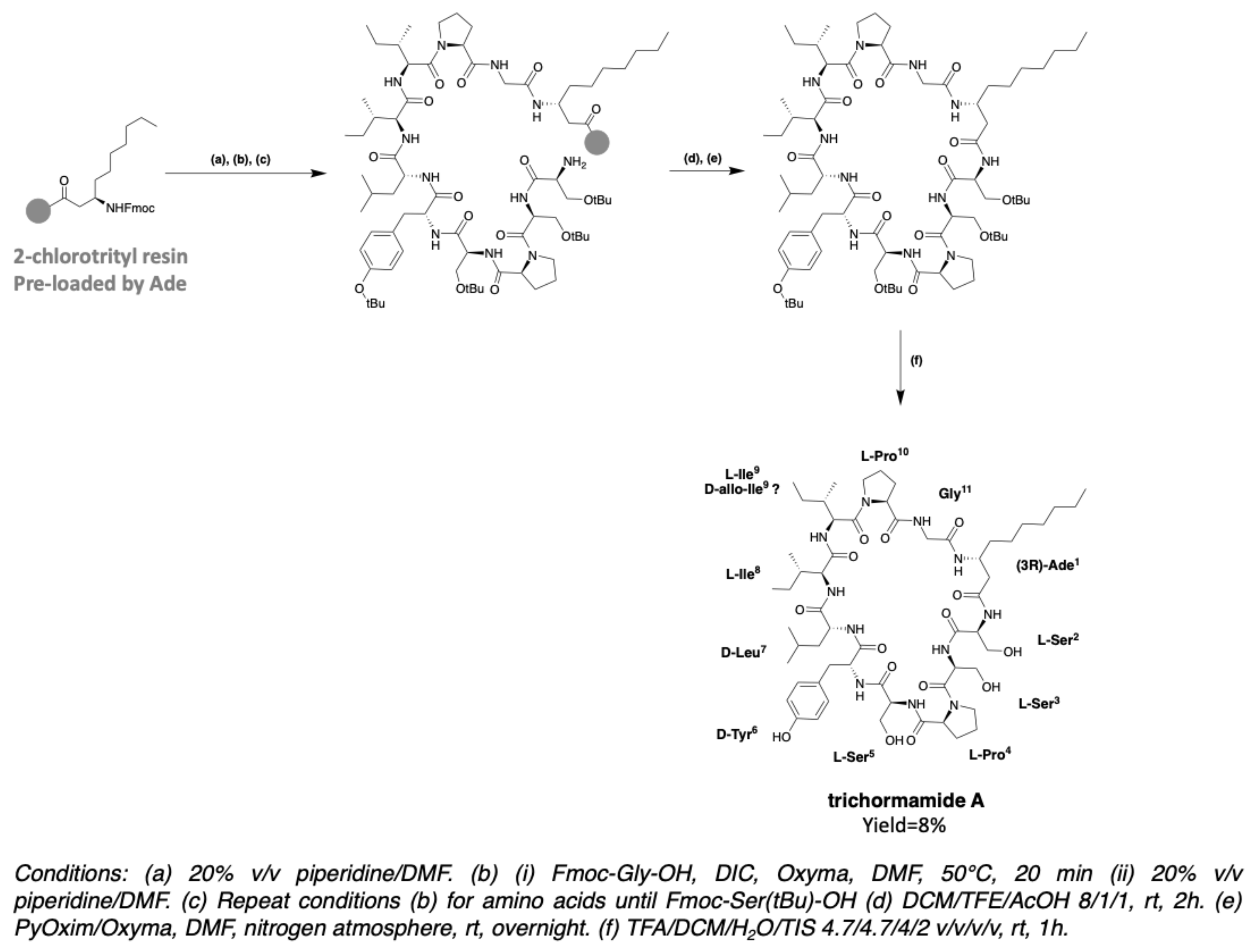
2.6. Trichormamides
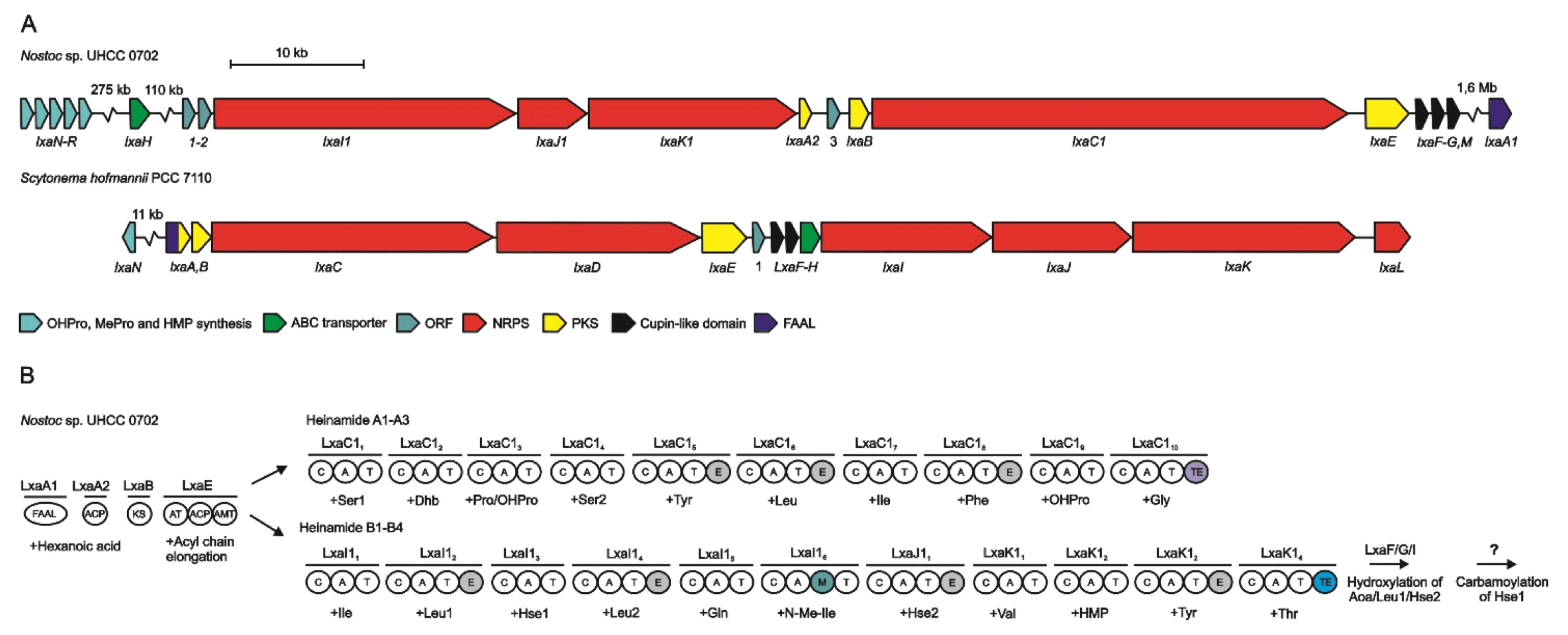
2.7. Heinamides
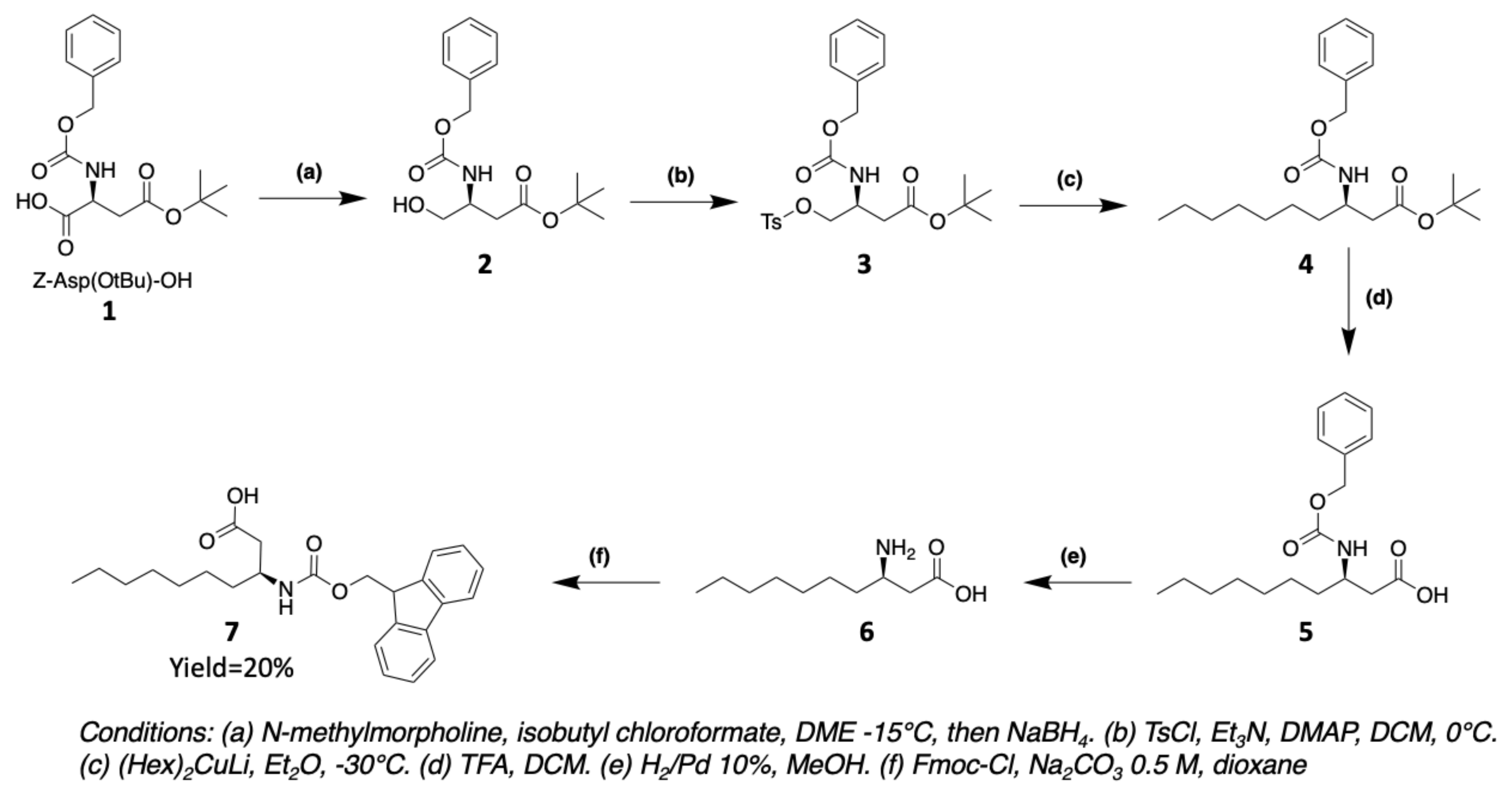
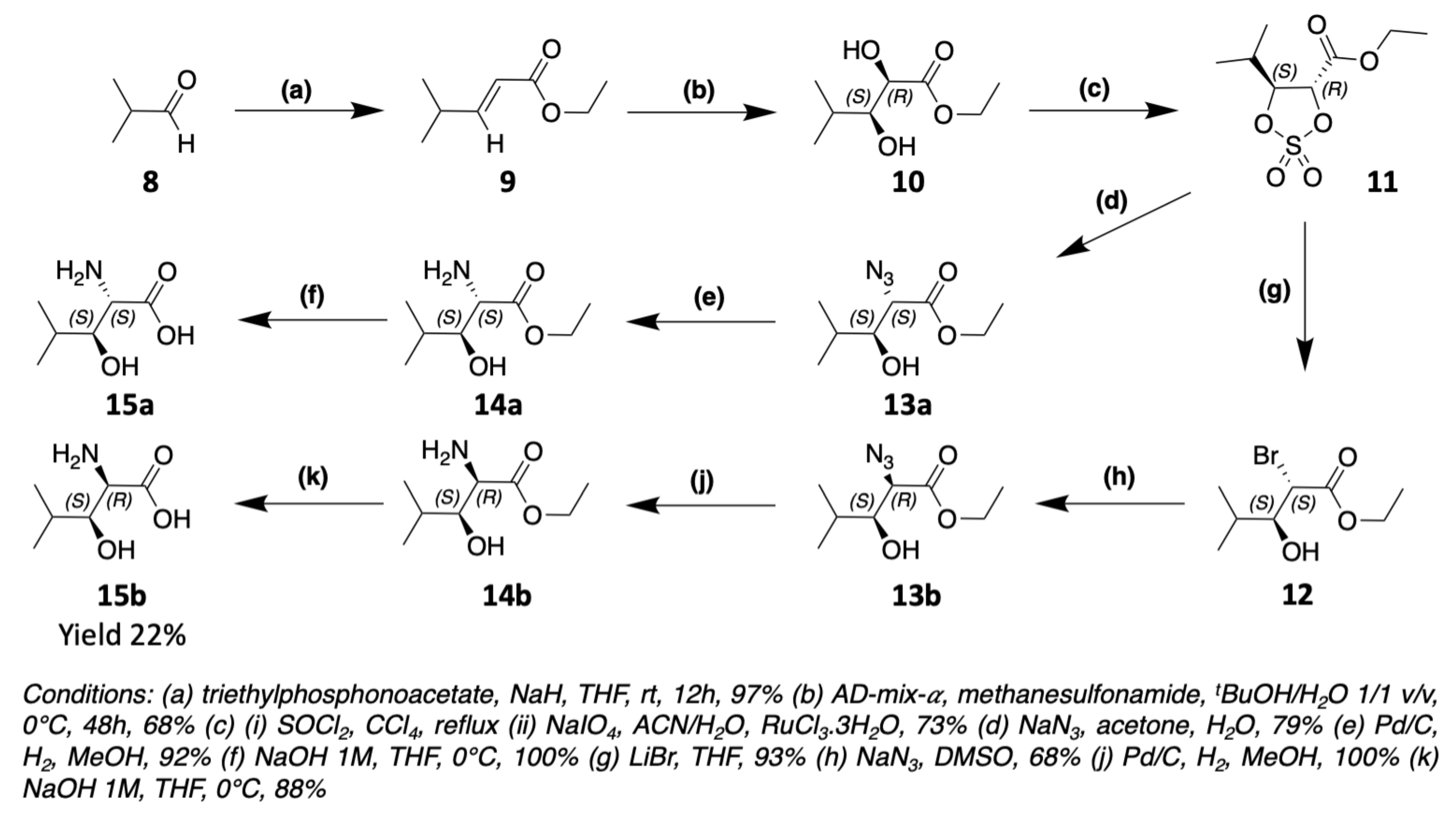
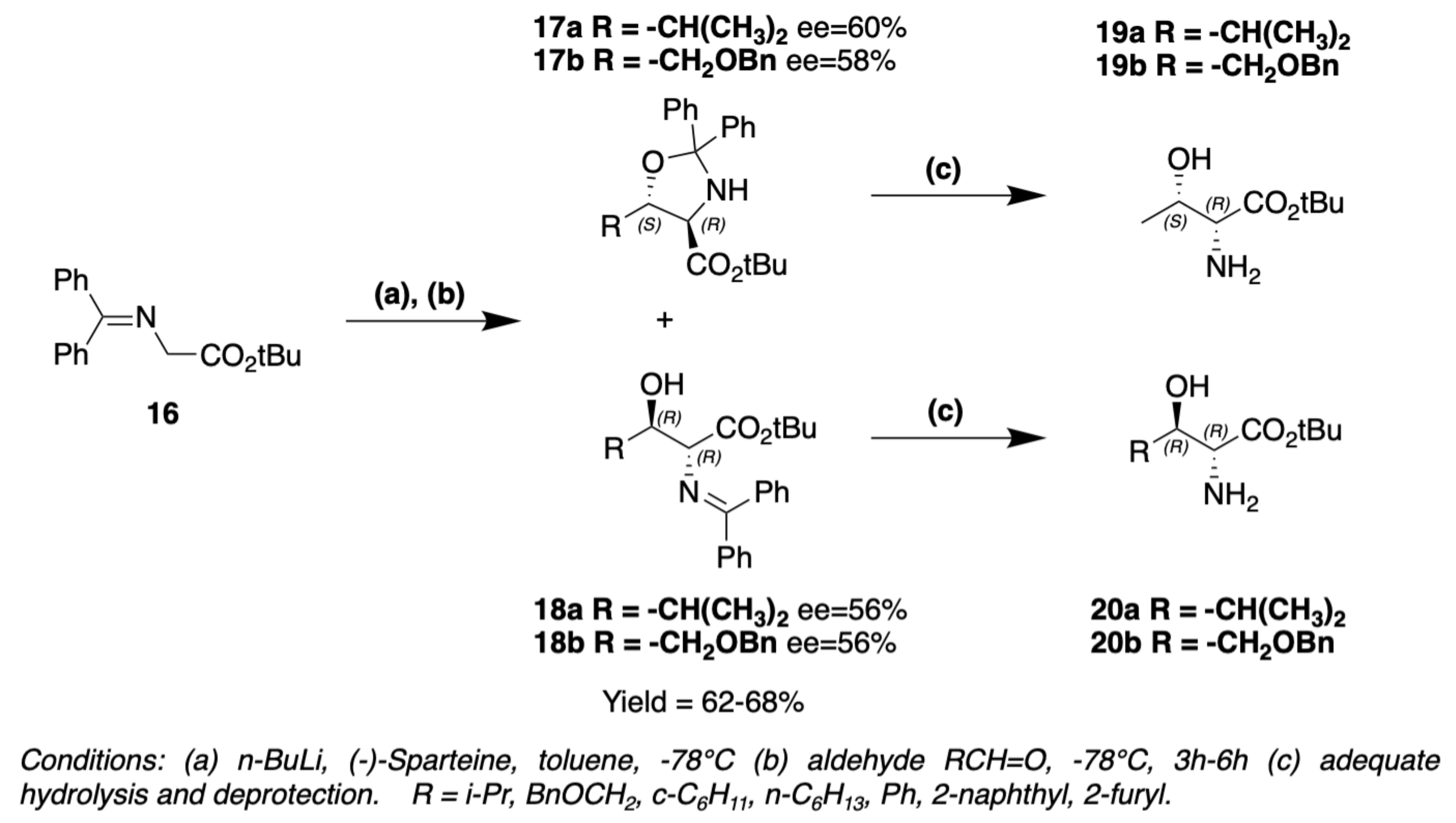
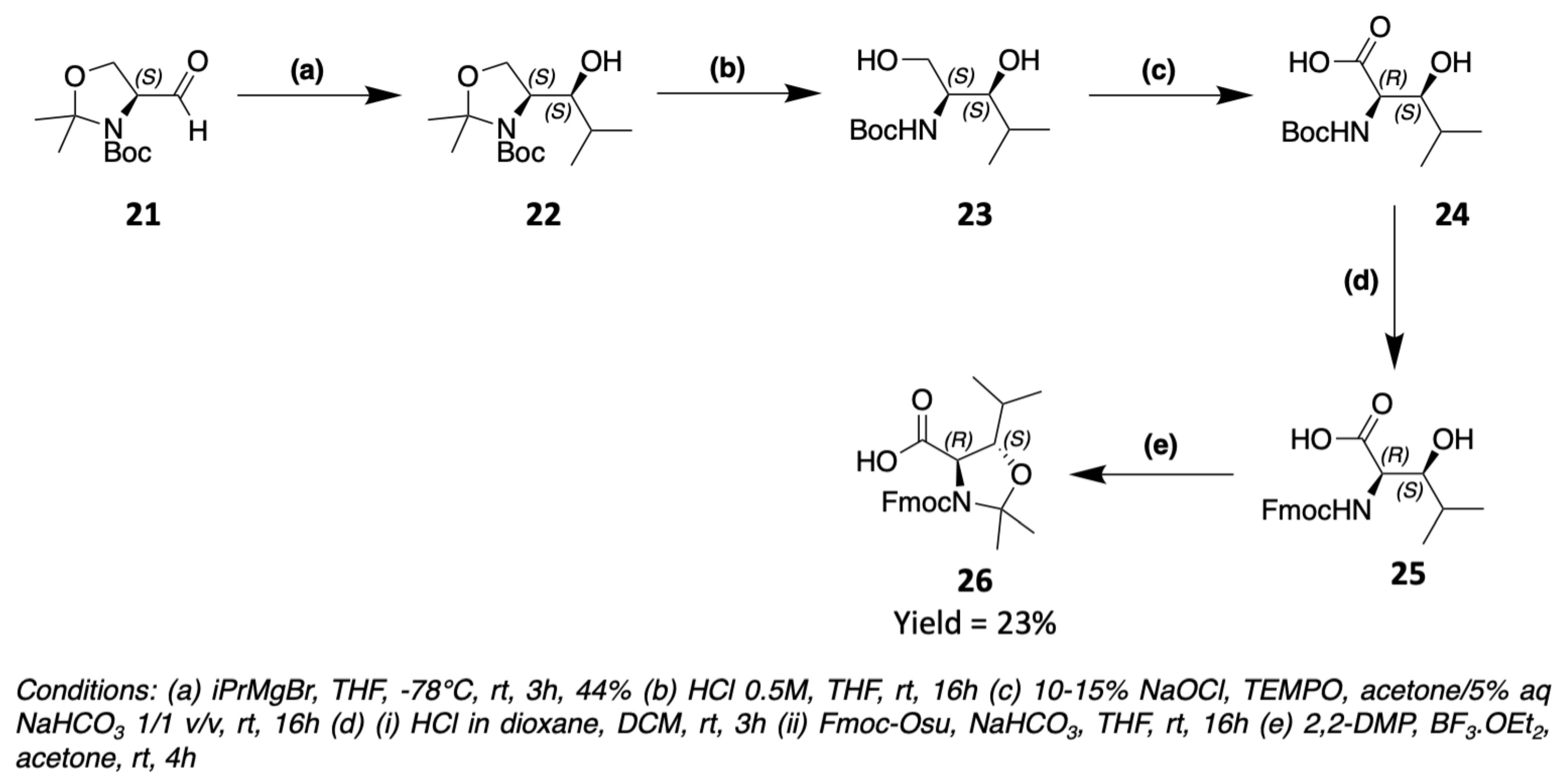
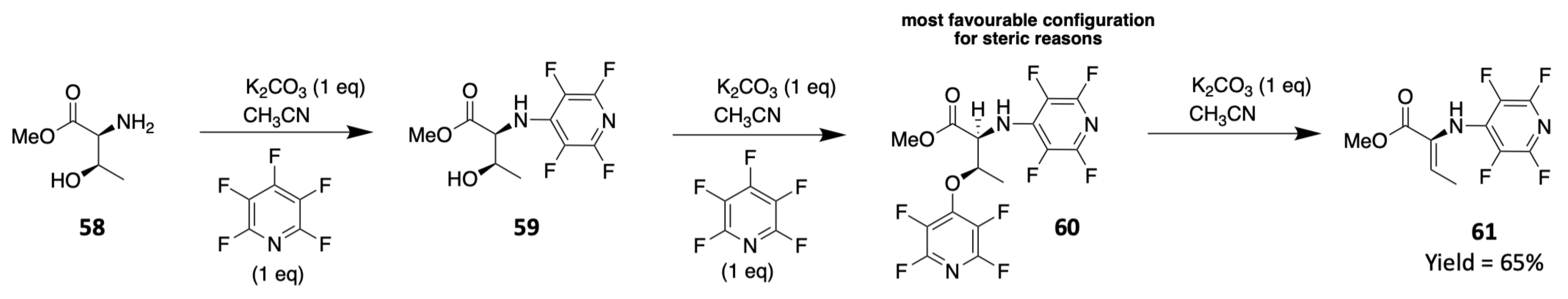
3. Chemical Synthesis
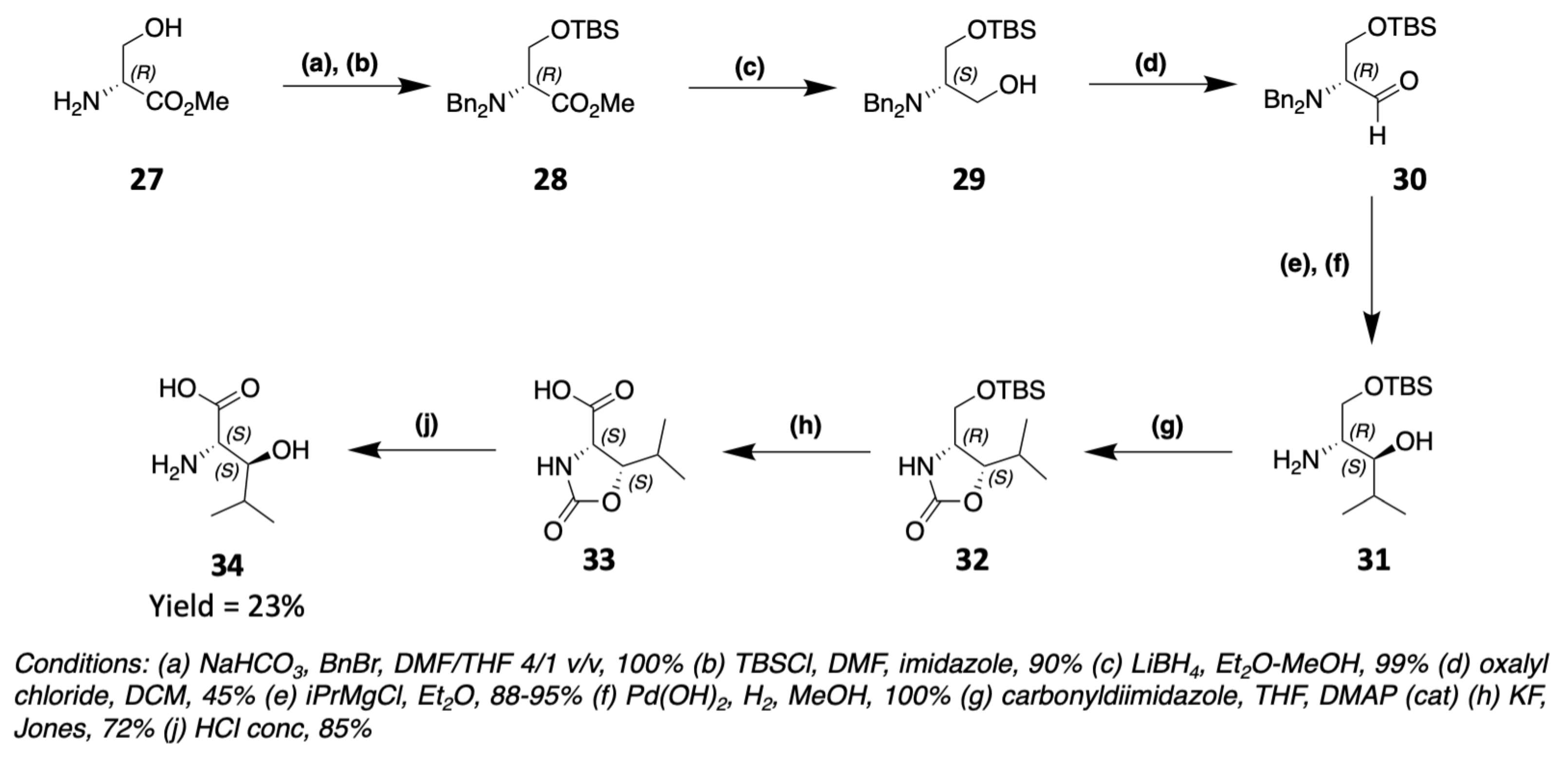
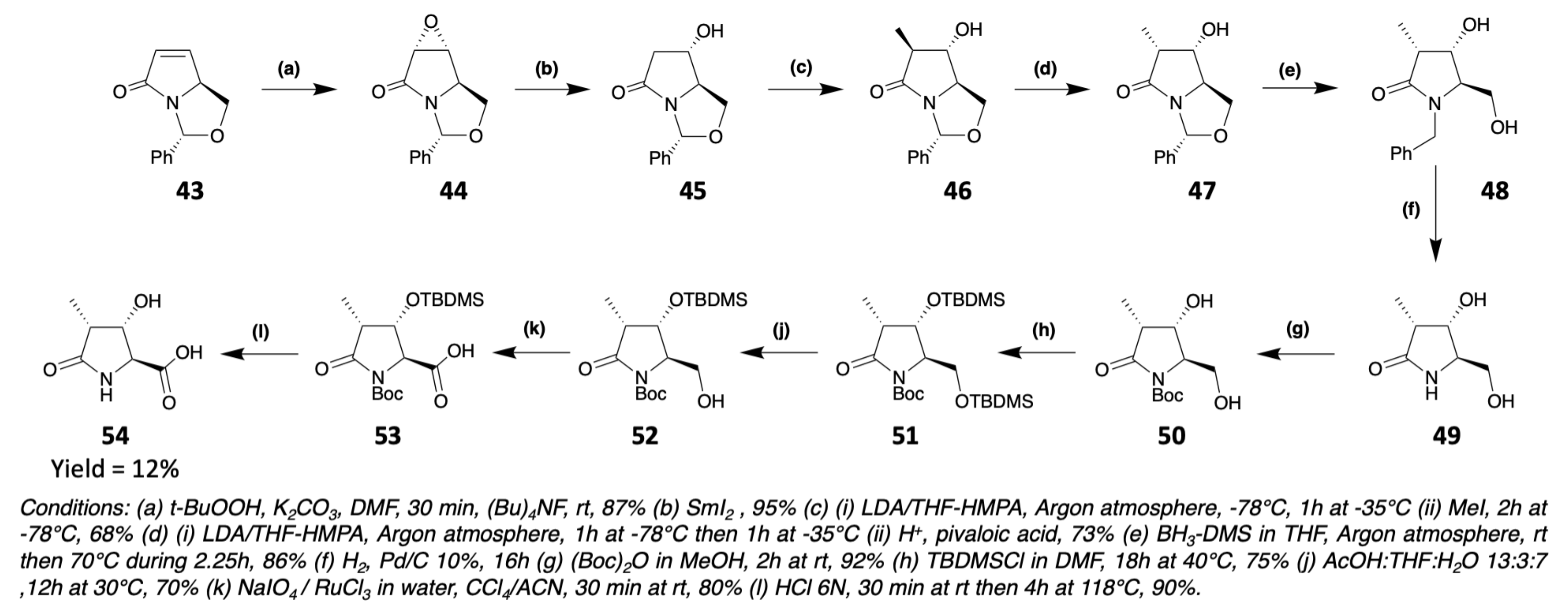
3.1. Synthesis of Characteristic Amino Acids in Laxaphycins
3.1.1. Synthesis of 3-Aminodecanoic acid and 3-Aminooctanoic acid
3.1.2. Synthesis of Hydroxylated Amino Acids
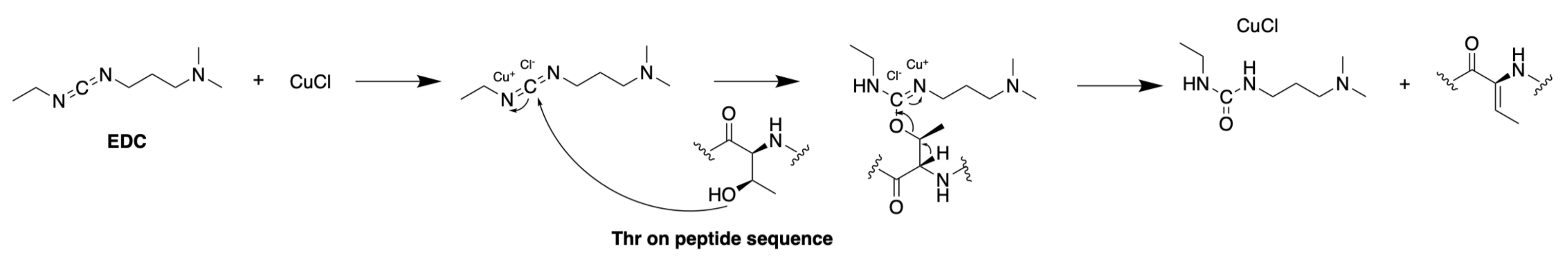
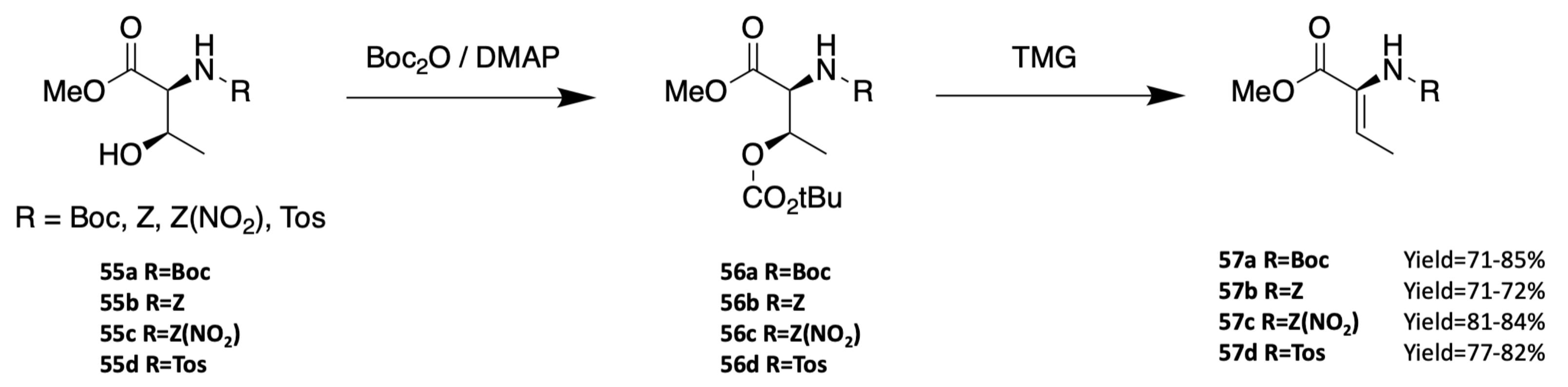
3.1.3. Synthesis of Dehydro Amino Acids
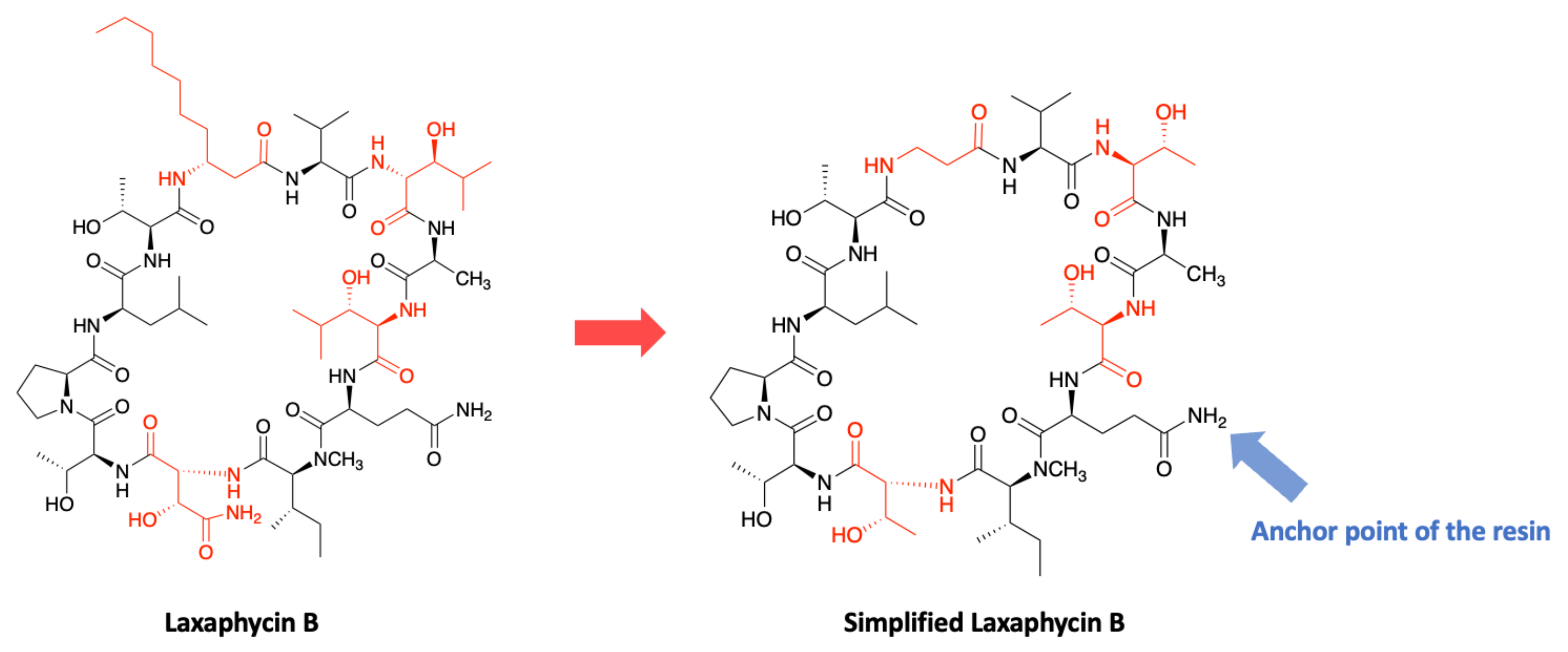
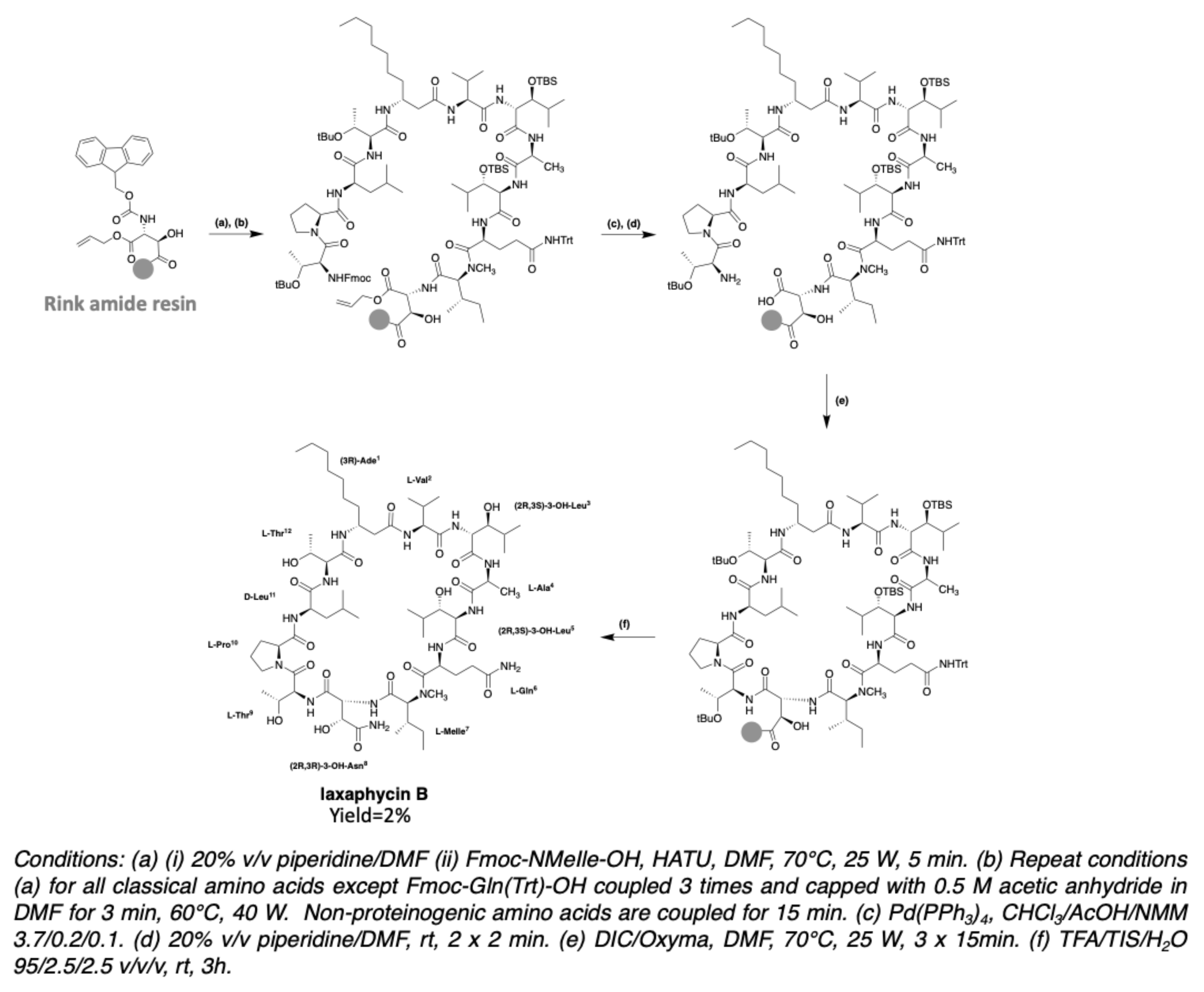
3.2. Synthesis of Laxaphycin B-Type Peptides
3.3. Synthesis of Laxaphycin A-Type Peptides
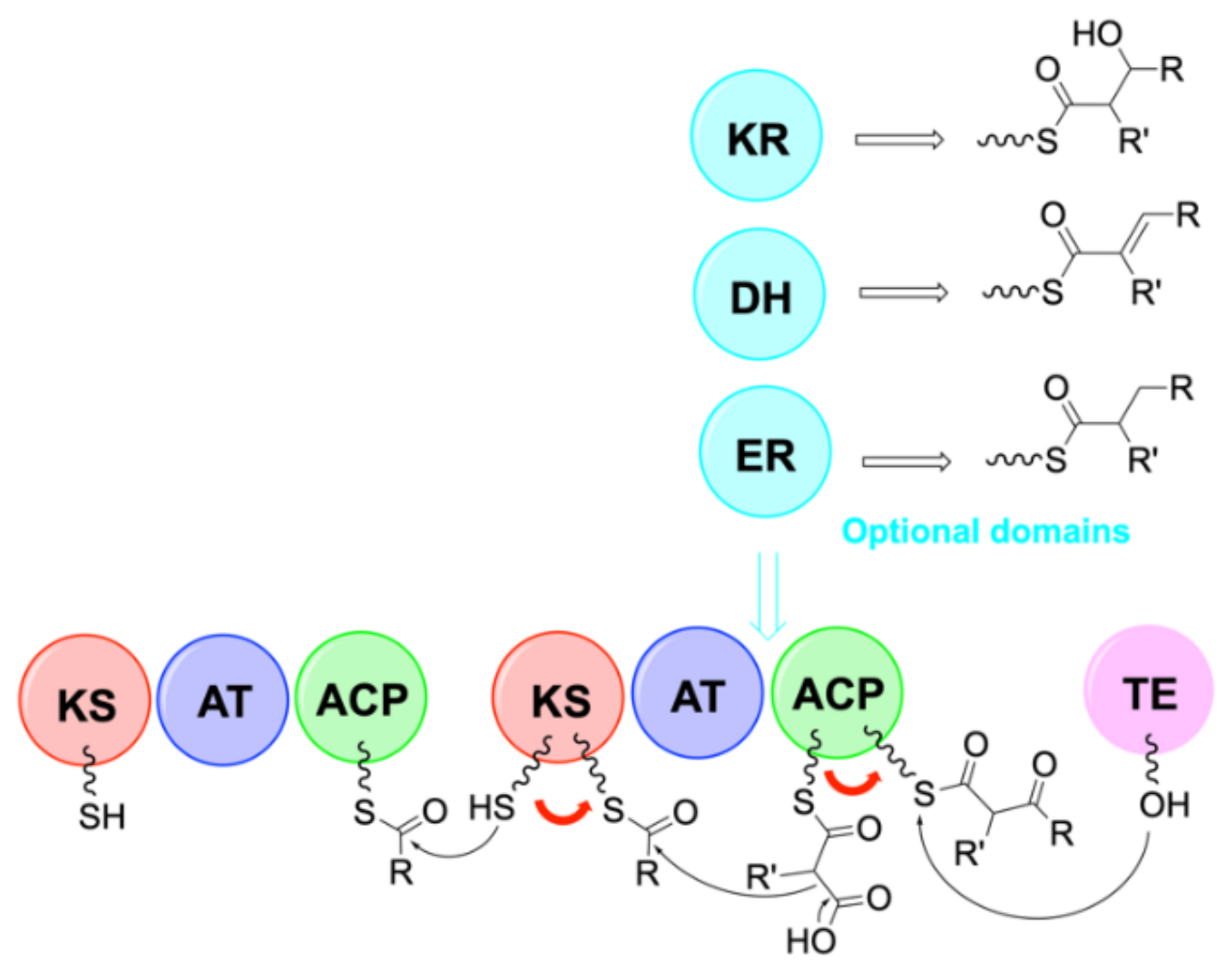
4. Biosynthesis
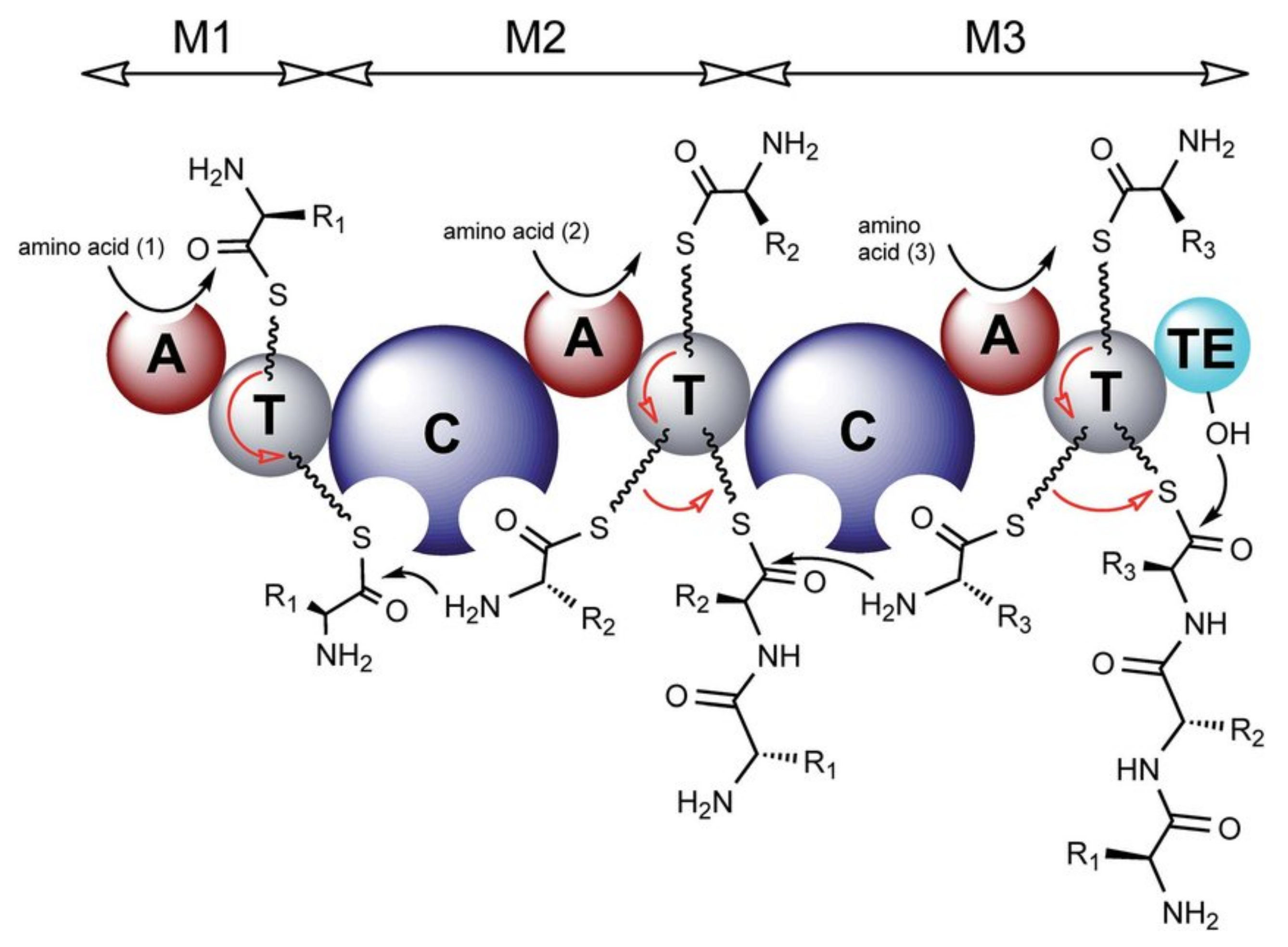
4.1. Biosynthesis of Characteristic Amino Acids in Laxaphycins
4.1.1. Biosynthesis of Hydroxy-Amino Acids
4.1.2. Biosynthesis of Dehydroamino Acids
4.2. Biosynthesis of Laxaphycins
5. In Situ and Ex Situ Biological Activities
5.1. Biological Properties
Laxaphycins
5.2. Feeding Deterrence as an Example of Ecological Role
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Montaser, R.; Luesch, H. Marine natural products: A new wave of drugs? Future Med. Chem. 2011, 3, 1475–1489. [Google Scholar] [CrossRef] [PubMed]
- Martins, A.; Vieira, H.; Gaspar, H.; Santos, S. Marketed Marine natural products in the pharmaceutical and cosmeceutical industries: Tips for success. Mar. Drugs 2014, 12, 1066–1101. [Google Scholar] [CrossRef] [PubMed]
- Zuo, W.; Kwok, H.F. Development of marine-derived compounds for cancer therapy. Mar. Drugs 2021, 19, 342. [Google Scholar] [CrossRef] [PubMed]
- Sotelo, C.G.; Blanco, M.; Ramos, P.; Vázquez, J.A.; Perez-Martin, R.I. Sustainable Sources from aquatic organisms for cosmeceuticals ingredients. Cosmetics 2021, 8, 48. [Google Scholar] [CrossRef]
- Singh, R.K.; Tiwari, S.P.; Rai, A.K.; Mohapatra, T.M. Cyanobacteria: An emerging source for drug discovery. J. Antibiot. 2011, 64, 401–412. [Google Scholar] [CrossRef]
- Tan, L.T. Pharmaceutical agents from filamentous marine cyanobacteria. Drug Discov. Today 2013, 18, 863–871. [Google Scholar] [CrossRef]
- Khalifa, S.A.M.; Shedid, E.S.; Saied, E.M.; Jassbi, A.R.; Jamebozorgi, F.H.; Rateb, M.E.; Du, M.; Abdel-Daim, M.M.; Kai, G.-Y.; Al-Hammady, M.A.M.; et al. Cyanobacteria—From the oceans to the potential biotechnological and biomedical applications. Mar. Drugs 2021, 19, 241. [Google Scholar] [CrossRef]
- Dahiya, S.; Dahiya, R. A Comprehensive review of chemistry and pharmacological aspects of natural cyanobacterial azoline-based circular and linear oligopeptides. Eur. J. Med. Chem. 2021, 218, 113406. [Google Scholar] [CrossRef]
- Banaigs, B.; Bonnard, I.; Witczak, A.; Inguimbert, N. Marine peptide secondary metabolites. In Outstanding Marine Molecules; La Barre, S., Kornprobst, J.-M., Eds.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2014; pp. 285–318. ISBN 978-3-527-68150-1. [Google Scholar]
- Kehr, J.-C.; Gatte Picchi, D.; Dittmann, E. Natural product biosyntheses in cyanobacteria: A treasure trove of unique enzymes. Beilstein J. Org. Chem. 2011, 7, 1622–1635. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, T.; Yao, J.; Lu, J.; Liu, Z.; Ding, L. Recent advances in chemistry and bioactivity of marine Cyanobacteria moorea species. Eur. J. Med. Chem. 2020, 201, 112473. [Google Scholar] [CrossRef]
- Kleigrewe, K.; Gerwick, L.; Sherman, D.H.; Gerwick, W.H. Unique Marine derived cyanobacterial biosynthetic genes for chemical diversity. Nat. Prod. Rep. 2016, 33, 348–364. [Google Scholar] [CrossRef] [PubMed]
- Clark, R.J.; Fischer, H.; Dempster, L.; Daly, N.L.; Rosengren, K.J.; Nevin, S.T.; Meunier, F.A.; Adams, D.J.; Craik, D.J. Engineering stable peptide toxins by means of backbone cyclization: Stabilization of the -Conotoxin MII. Proc. Natl. Acad. Sci. USA 2005, 102, 13767–13772. [Google Scholar] [CrossRef] [PubMed]
- Cardillo, G.; Gentilucci, L.; Tolomelli, A. Unusual amino acids: Synthesis and introduction into naturally occurring peptides and biologically active analogues. Med. Chem. 2006, 6, 293–304. [Google Scholar]
- Gentilucci, L.; De Marco, R.; Cerisoli, L. Chemical Modifications Designed to improve peptide stability: Incorporation of non-natural Amino acids, Pseudo-Peptide bonds, and cyclization. Curr. Pharm. Des. 2010, 16, 3185–3203. [Google Scholar] [CrossRef]
- Dennison, S.; Wallace, J.; Harris, F.; Phoenix, D. Amphiphilic Alpha-helical antimicrobial peptides and their structure/function relationships. Protein Pept. Lett. 2005, 12, 31–39. [Google Scholar] [CrossRef]
- Molinski, T.F. NMR of Natural products at the ‘Nanomole-scale’. Nat. Prod. Rep. 2010, 27, 321–329. [Google Scholar] [CrossRef]
- Fujii, K.; Ikai, Y.; Mayumi, T.; Oka, H.; Suzuki, M.; Harada, K. A Nonempirical method using LC/MS for determination of the absolute configuration of constituent amino acids in a peptide: Elucidation of limitations of Marfey’s method and of its separation mechanism. Anal. Chem. 1997, 69, 3346–3352. [Google Scholar] [CrossRef]
- Frankmolle, W.P.; Larsen, L.K.; Caplan, F.R.; Patterson, G.M.L.; Knubel, G.; Levine, I.A.; Moore, R.E. Antifungal cyclic peptides from the terrestrial blue-green alga Anabaena Laxa. I. Isolation and biological properties. J. Antibiot. 1992, 45, 1451–1457. [Google Scholar] [CrossRef] [PubMed]
- Frankmolle, W.P.; Knubel, G.; Moore, E.; Patterson, G.M.L. Antifungal cyclic peptides from the terrestrial blue-green alga Anabaena Laxa. II. Structures of Laxaphycins A, B, C, D and E. J. Antibiot. 1992, 45, 1458–1466. [Google Scholar] [CrossRef]
- Bonnard, I.; Rolland, M.; Francisco, C.; Banaigs, B. Total structure and biological properties of Laxaphycins A and B, cyclic lipopeptides from the marine cyanobacterium Lyngbya majuscula. Lett. Pept. Sci. 1997, 4, 289–292. [Google Scholar] [CrossRef]
- Bonnard, I.; Rolland, M.; Salmon, J.-M.; Debiton, E.; Barthomeuf, C.; Banaigs, B. Total Structure and inhibition of tumor cell proliferation of laxaphycins. J. Med. Chem. 2007, 50, 1266–1279. [Google Scholar] [CrossRef]
- Boyaud, F.; Mahiout, Z.; Lenoir, C.; Tang, S.; Wdzieczak-Bakala, J.; Witczak, A.; Bonnard, I.; Banaigs, B.; Ye, T.; Inguimbert, N. first total synthesis and stereochemical revision of Laxaphycin B and its extension to lyngbyacyclamide A. Org. Lett. 2013, 15, 3898–3901. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.-D.; Nam, S.-J.; Chin, Y.-W.; Kim, M.-S. Misassigned natural products and their revised structures. Arch. Pharm. Res. 2016, 39, 143–153. [Google Scholar] [CrossRef]
- Cai, W.; Matthew, S.; Chen, Q.-Y.; Paul, V.J.; Luesch, H. Discovery of new A- and B-Type Laxaphycins with synergistic anticancer activity. Bioorg. Med. Chem. 2018, 26, 2310–2319. [Google Scholar] [CrossRef]
- Sullivan, P.; Krunic, A.; Burdette, J.E.; Orjala, J. Laxaphycins B5 and B6 from the cultured Cyanobacterium UIC 10484. J. Antibiot. 2020, 73, 526–533. [Google Scholar] [CrossRef] [PubMed]
- Bornancin, L.; Alonso, E.; Alvariño, R.; Inguimbert, N.; Bonnard, I.; Botana, L.M.; Banaigs, B. Structure and biological evaluation of new cyclic and acyclic Laxaphycin-A type peptides. Bioorg. Med. Chem. 2019, 27, 1966–1980. [Google Scholar] [CrossRef]
- Bonnard, I.; Bornancin, L.; Dalle, K.; Chinain, M.; Zubia, M.; Banaigs, B.; Roué, M. Assessment of the chemical diversity and potential toxicity of benthic cyanobacterial blooms in the lagoon of Moorea Island (French Polynesia). J. Mar. Sci. Eng. 2020, 8, 406. [Google Scholar] [CrossRef]
- Gerwick, W.H.; Mrozek, C.; Moghaddam, M.F.; Agarwal, S.K. Novel cytotoxic peptides from the tropical marine cyanobacterium Hormothamnion enteromorphoides 1. Discovery, isolation and initial chemical and biological characterization of the hormothamnins from wild and cultured material. Experientia 1989, 45, 115–121. [Google Scholar] [CrossRef]
- MacMillan, J.B.; Ernst-Russell, M.A.; de Ropp, J.S.; Molinski, T.F. Lobocyclamides A−C, lipopeptides from a cryptic cyanobacterial mat containing Lyngbya confervoides. J. Org. Chem. 2002, 67, 8210–8215. [Google Scholar] [CrossRef]
- Zhaxybayeva, O. Phylogenetic Analyses of cyanobacterial genomes: Quantification of horizontal gene transfer events. Genome Res. 2006, 16, 1099–1108. [Google Scholar] [CrossRef]
- Bornancin, L.; Boyaud, F.; Mahiout, Z.; Bonnard, I.; Mills, S.C.; Banaigs, B.; Inguimbert, N. Isolation and synthesis of laxaphycin B-Type peptides: A case study and clues to their biosynthesis. Mar. Drugs 2015, 13, 7285–7300. [Google Scholar] [CrossRef]
- Alvariño, R.; Alonso, E.; Bornancin, L.; Bonnard, I.; Inguimbert, N.; Banaigs, B.; Botana, L.M. Biological activities of cyclic and acyclic B-Type laxaphycins in SH-SY5Y human neuroblastoma cells. Mar. Drugs 2020, 18, 364. [Google Scholar] [CrossRef] [PubMed]
- Bornancin, L. Lipopeptides from Cyanobacteria: Structure and Role in a Trophic Cascade. Ph.D. Dissertation, University of Montpellier, Montpellier, France, 2016. [Google Scholar]
- Gerwick, W.H.; Jiang, Z.D.; Agarwal, S.K.; Farmer, B.T. Total structure of Hormothamnin A, a toxic cyclic undecapeptide from the tropical marine cyanobacterium Hormothamnion enteromorphoides. Tetrahedron 1992, 48, 2313–2324. [Google Scholar] [CrossRef]
- Gerwick, W.H.; Jiang, Z.D.; Agarwal, S.K.; Farmer, B.T. Total structure of hormothamnin A, a toxic cyclic undecapeptide from the tropical marine cyanobacterium Hormothamnion enteromorphoides. Tetrahedron 1992, 48, 5755. [Google Scholar] [CrossRef]
- Grewe, J.C. Cyanopeptoline und Scytocyclamide: Zyklische Peptide aus Scytonema hofmanni PCC 7110 Struktur und Biologische Aktivität. Ph.D. Dissertation, Universität Freiburg im Breisgau, Breisgau, Germany, 2005. [Google Scholar]
- Heinilä, L.M.P.; Fewer, D.P.; Jokela, J.K.; Wahlsten, M.; Jortikka, A.; Sivonen, K. Shared PKS module in biosynthesis of synergistic laxaphycins. Front. Microbiol. 2020, 11, 578878. [Google Scholar] [CrossRef]
- Maru, N.; Ohno, O.; Uemura, D. Lyngbyacyclamides A and B, novel cytotoxic peptides from marine cyanobacteria Lyngbya sp. Tetrahedron Lett. 2010, 51, 6384–6387. [Google Scholar] [CrossRef]
- Luo, S.; Krunic, A.; Kang, H.-S.; Chen, W.-L.; Woodard, J.L.; Fuchs, J.R.; Swanson, S.M.; Orjala, J. Trichormamides A and B with antiproliferative activity from the cultured freshwater cyanobacterium Trichormus sp. UIC 10339. J. Nat. Prod. 2014, 77, 1871–1880. [Google Scholar] [CrossRef]
- Luo, S.; Kang, H.-S.; Krunic, A.; Chen, W.-L.; Yang, J.; Woodard, J.L.; Fuchs, J.R.; Hyun Cho, S.; Franzblau, S.G.; Swanson, S.M.; et al. Trichormamides C and D, antiproliferative cyclic lipopeptides from the cultured freshwater cyanobacterium Cf. Oscillatoria sp. UIC 10045. Bioorg. Med. Chem. 2015, 23, 3153–3162. [Google Scholar] [CrossRef] [PubMed]
- Gaillard, M.; Das, S.; Djibo, M.; Raviglione, D.; Roumestand, C.; Legrand, B.; Inguimbert, N. Towards the total synthesis of trichormamide A, a cyclic undecapeptide. Tetrahedron Lett. 2018, 59, 3713–3718. [Google Scholar] [CrossRef]
- Darcel, L.; Djibo, M.; Gaillard, M.; Raviglione, D.; Bonnard, I.; Banaigs, B.; Inguimbert, N. Trichormamide C structural confirmation through total synthesis and extension to analogs. Org. Lett. 2020, 22, 145–149. [Google Scholar] [CrossRef]
- Heinilä, L.M.P.; Fewer, D.P.; Jokela, J.K.; Wahlsten, M.; Ouyang, X.; Permi, P.; Jortikka, A.; Sivonen, K. The Structure and biosynthesis of heinamides A1–A3 and B1–B5, antifungal members of the Laxaphycin lipopeptide family. Org. Biomol. Chem. 2021, 19, 5577–5588. [Google Scholar] [CrossRef]
- Yue, Q.; Chen, L.; Zhang, X.; Li, K.; Sun, J.; Liu, X.; An, Z.; Bills, G.F. Evolution of chemical diversity in Echinocandin lipopeptide antifungal metabolites. Eukaryot. Cell 2015, 14, 698–718. [Google Scholar] [CrossRef]
- Gang, D.; Kim, D.; Park, H.-S. Cyclic peptides: Promising scaffolds for biopharmaceuticals. Genes 2018, 9, 557. [Google Scholar] [CrossRef]
- Rezai, T.; Yu, B.; Millhauser, G.L.; Jacobson, M.P.; Lokey, R.S. Testing the conformational hypothesis of passive membrane permeability using synthetic cyclic peptide diastereomers. J. Am. Chem. Soc. 2006, 128, 2510–2511. [Google Scholar] [CrossRef]
- Xue, Y.; Zhao, P.; Quan, C.; Zhao, Z.; Gao, W.; Li, J.; Zu, X.; Fu, D.; Feng, S.; Bai, X.; et al. Cyanobacteria-derived peptide antibiotics discovered since 2000. Peptides 2018, 107, 17–24. [Google Scholar] [CrossRef]
- Ghareeb, M.A.; Tammam, M.A.; El-Demerdash, A.; Atanasov, A.G. Insights about clinically approved and preclinically investigated marine natural products. Curr. Res. Biotechnol. 2020, 2, 88–102. [Google Scholar] [CrossRef]
- Buron, F.; Turck, A.; Plé, N.; Bischoff, L.; Marsais, F. Towards a biomimetic synthesis of Barrenazine A. Tetrahedron Lett. 2007, 48, 4327–4330. [Google Scholar] [CrossRef]
- Boyaud, F. Synthèse Totale de la Laxaphycine B: Un Lipopeptide Cyclique d’origine Marine: Extension à D’autres PEPTIDES Apparentés. Ph.D. Dissertation, Montpellier University, Montpellier, France, 2013. [Google Scholar]
- Hale, K.J.; Manaviazar, S.; Delisser, V.M. A practical new asymmetric synthesis of (2S,3S)- and (2R,3R)-3-Hydroxyleucine. Tetrahedron 1994, 50, 9181–9188. [Google Scholar] [CrossRef]
- MacMillan, J.B.; Molinski, T.F. Lobocyclamide B from Lyngbya Confervoides. Configuration and asymmetric synthesis of β-Hydroxy-α-Amino acids by (−)-Sparteine-Mediated aldol addition. Org. Lett. 2002, 4, 1883–1886. [Google Scholar] [CrossRef] [PubMed]
- Giltrap, A.M.; Haeckl, F.P.J.; Kurita, K.L.; Linington, R.G.; Payne, R.J. Total synthesis of skyllamycins A-C. Chem.Eur. J. 2017, 23, 15046–15049. [Google Scholar] [CrossRef] [PubMed]
- Ries, O.; Büschleb, M.; Granitzka, M.; Stalke, D.; Ducho, C. Amino Acid Motifs in Natural Products: Synthesis of O-Acylated Derivatives of (2S,3S)-3-Hydroxyleucine. Beilstein J. Org. Chem. 2014, 10, 1135–1142. [Google Scholar] [CrossRef]
- Laib, T.; Chastanet, J.; Zhu, J. A Highly stereoselective synthesis of (2S,3S)-Beta-hydroxyleucine. Tetrahedron Lett. 1997, 38, 1771–1772. [Google Scholar] [CrossRef]
- Davis, F.A.; Srirajan, V.; Fanelli, D.L.; Portonovo, P. Concise Asymmetric synthesis of β-Hydroxy α-Amino Acids using the sulfinimine-mediated asymmetric strecker synthesis: Phenylserine and β-Hydroxyleucine. J. Org. Chem. 2000, 65, 7663–7666. [Google Scholar] [CrossRef]
- Nagamitsu, T.; Sunazuka, T.; Tanaka, H.; Ōmura, S.; Sprengeler, P.A.; Smith, A.B. Total synthesis of (+)-Lactacystin. J. Am. Chem. Soc. 1996, 118, 3484–3590. [Google Scholar] [CrossRef]
- Kaur, H.; Harris, P.W.R.; Little, P.J.; Brimble, M.A. Total synthesis of the cyclic depsipeptide YM-280193, a platelet aggregation inhibitor. Org. Lett. 2015, 17, 492–495. [Google Scholar] [CrossRef]
- Makino, K.; Goto, T.; Hiroki, Y.; Hamada, Y. Stereoselective synthesis of Anti—β—Hydroxy—α—Amino acids through dynamic kinetic resolution. Angew. Chem. Int. Ed. 2004, 43, 882–884. [Google Scholar] [CrossRef]
- Boyaud, F.; Viguier, B.; Inguimbert, N. Synthesis of a protected derivative of (2R,3R)-β-Hydroxyaspartic acid suitable for fmoc-based solid phase synthesis. Tetrahedron Lett. 2013, 54, 158–161. [Google Scholar] [CrossRef]
- Charvillon, F.B.; Amouroux, R. Synthesis of 3-Hydroxylated analogues of D-Aspartic acid β-Hydroxamate. Synth. Commun. 1997, 27, 395–403. [Google Scholar] [CrossRef]
- Spengler, J.; Pelay, M.; Tulla-Puche, J.; Albericio, F. Synthesis of orthogonally protected L-Threo-β-Ethoxyasparagine. Amino Acids 2010, 39, 161–165. [Google Scholar] [CrossRef]
- Inoue, M.; Shinohara, N.; Tanabe, S.; Takahashi, T.; Okura, K.; Itoh, H.; Mizoguchi, Y.; Iida, M.; Lee, N.; Matsuoka, S. Total synthesis of the large non-ribosomal peptide Polytheonamide B. Nat. Chem. 2010, 2, 280–285. [Google Scholar] [CrossRef]
- Boger, D.L.; Lee, R.J.; Bounaud, P.-Y.; Meier, P. Asymmetric synthesis of orthogonally protected l—Threo -β-Hydroxyasparagine. J. Org. Chem. 2000, 65, 6770–6772. [Google Scholar] [CrossRef]
- Cardillo, G.; Gentilucci, L.; Tolomelli, A.; Tomasini, C. A practical method for the synthesis of B-Amino a-Hydroxy acids. Synthesis of enantiomerically pure hydroxyaspartic acid and isoserine. Synlett 1999, 11, 1727–1730. [Google Scholar] [CrossRef]
- Kurokawa, N.; Ohfune, Y. Synthetic studies on antifungal cyclic peptides, echinocandins. Stereoselective total synthesis of Echinocandin D via a novel peptide coupling. Tetrahedron 1993, 49, 6195–6222. [Google Scholar] [CrossRef]
- Langlois, N.; Rakotondradany, F. Diastereoselective synthesis of (2S,3S,4S)-3-Hydroxy-4-Methylproline, a Common constituent of several antifungal cyclopeptides. Tetrahedron 2000, 56, 2437–2448. [Google Scholar] [CrossRef]
- Carlsen, P.H.J.; Katsuki, T.; Martin, V.S.; Sharpless, K.B. A Greatly improved procedure for ruthenium tetroxide catalyzed oxidations of organic compounds. J. Org. Chem. 1981, 46, 3936–3938. [Google Scholar] [CrossRef]
- Siodłak, D. α,β-Dehydroamino acids in naturally occurring peptides. Amino Acids 2015, 47, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Patel, H.C.; Singh, T.P.; Chauhan, V.S.; Kaur, P. Synthesis, crystal structure, and molecular conformation of peptide N-Boc-L-Pro-Dehydro-Phe-L-Gly-OH. Biopolymers 1990, 29, 509–515. [Google Scholar] [CrossRef]
- Jain, R.; Chauhan, V.S. Conformational Characteristics of Peptides Containing α,β-Dehydroamino Acid Residues. Biopolym. Pept. Sci. 1996, 40, 105–119. [Google Scholar] [CrossRef]
- Bogart, J.W.; Bowers, A.A. Dehydroamino acids: Chemical multi-tools for late-stage diversification. Org. Biomol. Chem. 2019, 17, 3653–3669. [Google Scholar] [CrossRef]
- Goodall, K.; Parsons, A.F. A new and efficient preparation of α,β-Dehydroamino acids. Tetrahedron Lett. 1995, 36, 3259–3260. [Google Scholar] [CrossRef]
- Moyá, D.A.; Lee, M.A.; Chanthakhoun, J.C.; LeSueur, A.K.; Joaquin, D.; Barfuss, J.D.; Castle, S.L. Towards a streamlined synthesis of peptides containing α,β-Dehydroamino acids. Tetrahedron Lett. 2021, 74, 153175. [Google Scholar] [CrossRef]
- Jiménez, J.C.; Nicolas, E.; Giralt, E.; Albericio, F. Synthesis of peptides containing α, β-Didehydroamino acids. Scope and limitations. Lett. Pept. Sci. 2002, 9, 135–141. [Google Scholar] [CrossRef]
- López-Macià, À.; Jiménez, J.C.; Royo, M.; Giralt, E.; Albericio, F. Synthesis and structure determination of Kahalalide F. J. Am. Chem. Soc. 2001, 123, 11398–11401. [Google Scholar] [CrossRef]
- Das, S. Methodological Development in Peptide Chemistry for Synthesis of Antimicrobial and Antifungal Derivatives of Marine Natural Peptides. Ph.D. Dissertation, University of Perpignan Via Domitia, Perpignan, France, 2018. [Google Scholar]
- Ferreira, P.M.T.; Maia, H.L.S.; Monteiro, L.S.; Sacramento, J. High yielding synthesis of dehydroamino acid and dehydropeptide derivatives. J. Chem. Soc. Perkin 1 1999, 3697–3703. [Google Scholar] [CrossRef]
- Ferreira, P.M.T.; Monteiro, L.S.; Pereira, G.; Ribeiro, L.; Sacramento, J.; Silva, L. Reactivity of dehydroamino acids and dehydrodipeptides towardsN-Bromosuccinimide: Synthesis of β-Bromo- and β,β-Dibromodehydroamino acid derivatives and of substituted 4-Imidazolidinones. Eur. J. Org. Chem. 2007, 2007, 5934–5949. [Google Scholar] [CrossRef]
- Tian, X.; Li, L.; Han, J.; Zhen, X.; Liu, S. Stereoselectively Synthesis and structural confirmation of dehydrodipeptides with dehydrobutyrine. SpringerPlus 2016, 5. [Google Scholar] [CrossRef][Green Version]
- Webster, A.M.; Coxon, C.R.; Kenwright, A.M.; Sandford, G.; Cobb, S.L. A mild method for the synthesis of a novel dehydrobutyrine-containing amino acid. Tetrahedron 2014, 70, 4661–4667. [Google Scholar] [CrossRef]
- Nagano, T.; Kinoshita, H. A new and convenient method for the synthesis of dehydroamino acids starting from ethyl N-Boc- and N-Z-α-tosylglycinates and various nitro compounds. Bull. Chem. Soc. Jpn. 2000, 73, 1605–1613. [Google Scholar] [CrossRef]
- Bonauer, C.; Walenzyk, T.; König, B. α,β-Dehydroamino Acids. Synthesis 2006, 1, 1–20. [Google Scholar] [CrossRef]
- Gude, M.; Ryf, J.; White, P.D. An Accurate Method for the Quantitation of Fmoc-Derivatized Solid Phase Supports. Lett. Pept. Sci. 2002, 9, 203–206. [Google Scholar] [CrossRef]
- Echalier, C.; Al-Halifa, S.; Kreiter, A.; Enjalbal, C.; Sanchez, P.; Ronga, L.; Puget, K.; Verdié, P.; Amblard, M.; Martinez, J.; et al. Heating and microwave assisted SPPS of C-Terminal acid peptides on trityl resin: The truth behind the yield. Amino Acids 2013, 45, 1395–1403. [Google Scholar] [CrossRef] [PubMed]
- Malesevic, M.; Strijowski, U.; Bächle, D.; Sewald, N. An improved method for the solution cyclization of peptides under pseudo-high dilution conditions. J. Biotechnol. 2004, 112, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Darcel, L.; Bornancin, L.; Raviglione, D.; Bonnard, I.; Mills, S.C.; Sáez-Vásquez, J.; Banaigs, B.; Inguimbert, N. D-Peptidase activity in a marine mollusk detoxifies a nonribosomal cyclic lipopeptide: An ecological model to study antibiotic resistance. J. Med. Chem. 2021, 64, 6198–6208. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Moore, B.S.; Yoon, Y.J. Reinvigorating natural product combinatorial biosynthesis with synthetic biology. Nat. Chem. Biol. 2015, 11, 649–659. [Google Scholar] [CrossRef]
- Süssmuth, R.D.; Mainz, A. Nonribosomal Peptide Synthesis-Principles and Prospects. Angew. Chem. Int. Ed. 2017, 56, 3770–3821. [Google Scholar] [CrossRef]
- Walsh, C.T.; O’Brien, R.V.; Khosla, C. Nonproteinogenic Amino Acid Building Blocks for Nonribosomal Peptide and Hybrid Polyketide Scaffolds. Angew. Chem. Int. Ed. 2013, 52, 7098–7124. [Google Scholar] [CrossRef]
- Kim, W.E.; Patel, A.; Hur, G.H.; Tufar, P.; Wuo, M.G.; McCammon, J.A.; Burkart, M.D. Mechanistic Probes for the epimerization domain of nonribosomal peptide synthetases. Chembiochem Eur. J. Chem. Biol. 2019, 20, 147–152. [Google Scholar] [CrossRef]
- Cryle, M.J.; Brieke, C.; Haslinger, K. Oxidative transformations of amino acids and peptides catalysed by Cytochromes P450. In Amino Acids, Peptides and Proteins; Farkas, E., Ryadnov, M., Eds.; Royal Society of Chemistry: Cambridge, UK, 2013; Volume 38, pp. 1–36. ISBN 978-1-84973-585-8. [Google Scholar]
- Miyanaga, A.; Kudo, F.; Eguchi, T. Protein–Protein interactions in polyketide Synthase–Nonribosomal Peptide synthetase hybrid assembly lines. Nat. Prod. Rep. 2018, 35, 1185–1209. [Google Scholar] [CrossRef]
- Winn, M.; Fyans, J.K.; Zhuo, Y.; Micklefield, J. Recent advances in engineering nonribosomal peptide assembly lines. Nat. Prod. Rep. 2016, 33, 317–347. [Google Scholar] [CrossRef]
- Wang, H.; Fewer, D.P.; Holm, L.; Rouhiainen, L.; Sivonen, K. Atlas of Nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc. Natl. Acad. Sci. USA 2014, 111, 9259–9264. [Google Scholar] [CrossRef]
- Hou, J.; Robbel, L.; Marahiel, M.A. Identification and Characterization of the lysobactin biosynthetic gene cluster reveals mechanistic insights into an unusual termination module architecture. Chem. Biol. 2011, 18, 655–664. [Google Scholar] [CrossRef]
- Strieker, M.; Kopp, F.; Mahlert, C.; Essen, L.-O.; Marahiel, M.A. Mechanistic and structural basis of stereospecific Cβ-Hydroxylation in calcium-dependent antibiotic, a daptomycin-type lipopeptide. ACS Chem. Biol. 2007, 2, 187–196. [Google Scholar] [CrossRef]
- Strieker, M.; Nolan, E.M.; Walsh, C.T.; Marahiel, M.A. Stereospecific synthesis of Threo- and Erythro -β-Hydroxyglutamic acid during kutzneride biosynthesis. J. Am. Chem. Soc. 2009, 131, 13523–13530. [Google Scholar] [CrossRef]
- Uhlmann, S.; Süssmuth, R.D.; Cryle, M.J. Cytochrome P450 sky interacts directly with the nonribosomal peptide synthetase to generate three amino acid precursors in skyllamycin biosynthesis. ACS Chem. Biol. 2013, 8, 2586–2596. [Google Scholar] [CrossRef]
- Fu, C.; Keller, L.; Bauer, A.; Brönstrup, M.; Froidbise, A.; Hammann, P.; Herrmann, J.; Mondesert, G.; Kurz, M.; Schiell, M.; et al. Biosynthetic studies of telomycin reveal new lipopeptides with enhanced activity. J. Am. Chem. Soc. 2015, 137, 7692–7705. [Google Scholar] [CrossRef]
- Parkinson, E.I.; Tryon, J.H.; Goering, A.W.; Ju, K.-S.; McClure, R.A.; Kemball, J.D.; Zhukovsky, S.; Labeda, D.P.; Thomson, R.J.; Kelleher, N.L.; et al. Discovery of the tyrobetaine natural products and their biosynthetic gene cluster via metabologenomics. ACS Chem. Biol. 2018, 13, 1029–1037. [Google Scholar] [CrossRef]
- Li, Y.; Lan, N.; Xu, L.; Yue, Q. Biosynthesis of pneumocandin lipopeptides and perspectives for its production and related echinocandins. Appl. Microbiol. Biotechnol. 2018, 102, 9881–9891. [Google Scholar] [CrossRef]
- Mattay, J.; Houwaart, S.; Hüttel, W. Cryptic production of Trans-3-hydroxyproline in Echinocandin B biosynthesis. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef]
- Gu, G.; Smith, L.; Liu, A.; Lu, S.-E. Genetic and Biochemical map for the biosynthesis of occidiofungin, an antifungal produced by Burkholderia contaminans strain MS14. Appl. Environ. Microbiol. 2011, 77, 6189–6198. [Google Scholar] [CrossRef]
- Freeman, M.F.; Gurgui, C.; Helf, M.J.; Morinaka, B.I.; Uria, A.R.; Oldham, N.J.; Sahl, H.-G.; Matsunaga, S.; Piel, J. Metagenome mining reveals polytheonamides as posttranslationally modified ribosomal peptides. Science 2012, 338, 387–390. [Google Scholar] [CrossRef]
- McAuliffe, O.; Ross, R.P.; Hill, C. Lantibiotics: Structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 2001, 25, 285–308. [Google Scholar] [CrossRef]
- Willey, J.M.; van der Donk, W.A. Lantibiotics: Peptides of diverse structure and function. Annu. Rev. Microbiol. 2007, 61, 477–501. [Google Scholar] [CrossRef] [PubMed]
- You, Y.O.; van der Donk, W.A. Mechanistic Investigations of the dehydration reaction of lacticin 481 synthetase using site-directed mutagenesis. Biochemistry 2007, 46, 5991–6000. [Google Scholar] [CrossRef] [PubMed]
- Wieland Brown, L.C.; Acker, M.G.; Clardy, J.; Walsh, C.T.; Fischbach, M.A. Thirteen Posttranslational modifications convert a 14-residue peptide into the antibiotic thiocillin. Proc. Natl. Acad. Sci. USA 2009, 106, 2549–2553. [Google Scholar] [CrossRef] [PubMed]
- Malcolmson, S.J.; Young, T.S.; Ruby, J.G.; Skewes-Cox, P.; Walsh, C.T. The Posttranslational modification cascade to the thiopeptide berninamycin generates linear forms and altered macrocyclic scaffolds. Proc. Natl. Acad. Sci.USA 2013, 110, 8483–8488. [Google Scholar] [CrossRef] [PubMed]
- Kelly, W.L.; Pan, L.; Li, C. Thiostrepton Biosynthesis: Prototype for a New Family of Bacteriocins. J. Am. Chem. Soc. 2009, 131, 4327–4334. [Google Scholar] [CrossRef]
- Liao, R.; Duan, L.; Lei, C.; Pan, H.; Ding, Y.; Zhang, Q.; Chen, D.; Shen, B.; Yu, Y.; Liu, W. Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem. Biol. 2009, 16, 141–147. [Google Scholar] [CrossRef]
- Yu, Y.; Duan, L.; Zhang, Q.; Liao, R.; Ding, Y.; Pan, H.; Wendt-Pienkowski, E.; Tang, G.; Shen, B.; Liu, W. Nosiheptide biosynthesis featuring a unique indole side ring formation on the characteristic thiopeptide framework. ACS Chem. Biol. 2009, 4, 855–864. [Google Scholar] [CrossRef]
- Moffitt, M.C.; Neilan, B.A. Characterization of the Nodularin Synthetase Gene Cluster and Proposed Theory of the Evolution of Cyanobacterial Hepatotoxins. Appl. Environ. Microbiol. 2004, 70, 6353–6362. [Google Scholar] [CrossRef]
- Scholz-Schroeder, B.K.; Soule, J.D.; Lu, S.-E.; Grgurina, I.; Gross, D.C. A physical map of the syringomycin and syringopeptin gene clusters localized to an approximately 145-Kb DNA region of Pseudomonas Syringae Pv. Syringae strain B301D. Mol. Plant Microbe Interact. 2001, 14, 1426–1435. [Google Scholar] [CrossRef]
- McIntosh, J.A.; Donia, M.S.; Schmidt, E.W. Ribosomal Peptide natural products: Bridging the ribosomal and nonribosomal worlds. Nat. Prod. Rep. 2009, 26, 537. [Google Scholar] [CrossRef]
- Tillett, D.; Dittmann, E.; Erhard, M.; von Döhren, H.; Börner, T.; Neilan, B.A. Structural Organization of microcystin biosynthesis in Microcystis Aeruginosa PCC7806: An integrated Peptide–Polyketide synthetase system. Chem. Biol. 2000, 7, 753–764. [Google Scholar] [CrossRef]
- Liu, W.-T.; Lamsa, A.; Wong, W.R.; Boudreau, P.D.; Kersten, R.; Peng, Y.; Moree, W.J.; Duggan, B.M.; Moore, B.S.; Gerwick, W.H.; et al. MS/MS-based networking and peptidogenomics guided genome mining revealed the stenothricin gene cluster in Streptomyces roseosporus. J. Antibiot. 2014, 67, 99–104. [Google Scholar] [CrossRef]
- Mareš, J.; Hájek, J.; Urajová, P.; Kopecky, J.; Hrouzek, P. A Hybrid non-ribosomal peptide/polyketide synthetase containing fatty-acyl ligase (FAAL) synthesizes the b- Amino fatty acid lipopeptides puwainaphycins in the cyanobacterium Cylindrospermum alatosporum. PLoS ONE 2014, 9, e111904. [Google Scholar] [CrossRef]
- Mareš, J.; Hájek, J.; Urajová, P.; Kust, A.; Jokela, J.; Saurav, K.; Galica, T.; Čapková, K.; Mattila, A.; Haapaniemi, E.; et al. Alternative biosynthetic starter units enhance the structural diversity of cyanobacterial lipopeptides. Appl. Environ. Microbiol. 2018, 85, e02675-18. [Google Scholar] [CrossRef]
- Nishizawa, T.; Asayama, M.; Fujii, K.; Harada, K.-I.; Shirai, M. Genetic analysis of the peptide synthetase genes for a cyclic heptapeptide microcystin in Microcystis spp. J. Biochem. 1999, 126, 520–529. [Google Scholar] [CrossRef]
- Duitman, E.H.; Hamoen, L.W.; Rembold, M.; Venema, G.; Seitz, H.; Saenger, W.; Bernhard, F.; Reinhardt, R.; Schmidt, M.; Ullrich, C.; et al. The mycosubtilin synthetase of Bacillus Subtilis ATCC6633: A Multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc. Natl. Acad. Sci. USA 1999, 96, 13294–13299. [Google Scholar] [CrossRef]
- Markolovic, S.; Leissing, T.M.; Chowdhury, R.; Wilkins, S.E.; Lu, X.; Schofield, C.J. Structure–Function relationships of human JmjC oxygenases—Demethylases versus hydroxylases. Curr. Opin. Struct. Biol. 2016, 41, 62–72. [Google Scholar] [CrossRef]
- Liu, L.; Budnjo, A.; Jokela, J.; Haug, B.E.; Fewer, D.P.; Wahlsten, M.; Rouhiainen, L.; Permi, P.; Fossen, T.; Sivonen, K. Pseudoaeruginosins, nonribosomal peptides in Nodularia spumigena. ACS Chem. Biol. 2015, 10, 725–733. [Google Scholar] [CrossRef]
- Leão, P.N.; Engene, N.; Antunes, A.; Gerwick, W.H.; Vasconcelos, V. The chemical ecology of cyanobacteria. Nat. Prod. Rep. 2012, 29, 372–391. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, B.; Liu, J.; Zhou, J.; Ma, J.; Huang, H.; Ju, J. Identification of the biosynthetic gene cluster and regulatory cascade for the synergistic antibacterial antibiotics griseoviridin and viridogrisein in Streptomyces griseoviridis. ChemBioChem 2012, 13, 2745–2757. [Google Scholar] [CrossRef]
- Gbankoto, A.; Vigo, J.; Dramane, K.; Banaigs, B.; Aina, E.; Salmon, J.-M. Cytotoxic effect of laxaphycins A and B on human lymphoblastic cells (CCRF-CEM) using digitised videomicrofluorometry. In Vivo 2005, 19, 577–582. [Google Scholar]
- Dussault, D.; Vu, K.D.; Vansach, T.; Horgen, F.D.; Lacroix, M. Antimicrobial effects of marine algal extracts and cyanobacterial pure compounds against five foodborne pathogens. Food Chem. 2016, 199, 114–118. [Google Scholar] [CrossRef]
- Cruz-Rivera, E.; Paul, V.J. Coral reef benthic cyanobacteria as food and refuge: Diversity, chemistry and complex interactions. In Proceedings of the 9th International Coral Reef Symposium, Bali, Indonesia, 23–27 October 2000; Volume 1. [Google Scholar]
- Pennings, S.C.; Pablo, S.R.; Paul, V.J. Chemical defenses of the tropical, benthic marine cyanobacterium Hormothamnion Enteromorphoides: Diverse consumers and synergisms. Limnol. Oceanogr. 1997, 42, 911–917. [Google Scholar] [CrossRef]
- Capper, A.; Tibbetts, I.R.; O’Neil, J.M.; Shaw, G.R. The Fate of Lyngbya Majuscula Toxins in Three Potential Consumers. J. Chem. Ecol. 2005, 31, 1595–1606. [Google Scholar] [CrossRef]
- Capper, A.; Cruz-Rivera, E.; Paul, V.J.; Tibbetts, I.R. Chemical deterrence of a marine cyanobacterium against sympatric and non-sympatric consumers. Hydrobiologia 2006, 553, 319–326. [Google Scholar] [CrossRef]
- Capper, A.; Erickson, A.A.; Ritson-Williams, R.; Becerro, M.A.; Arthur, K.A.; Paul, V.J. Palatability and Chemical defences of benthic cyanobacteria to a suite of herbivores. J. Exp. Mar. Biol. Ecol. 2016, 474, 100–108. [Google Scholar] [CrossRef]
- Cruz-Rivera, E.; Paul, V.J. Chemical deterrence of a cyanobacterial metabolite against generalized and specialized grazers. J. Chem. Ecol. 2006, 33, 213–217. [Google Scholar] [CrossRef]
- Li, Y.-X.; Zhong, Z.; Hou, P.; Zhang, W.-P.; Qian, P.-Y. Resistance to nonribosomal peptide antibiotics mediated by D-stereospecific peptidases. Nat. Chem. Biol. 2018, 14, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Shishido, T.K.; Jokela, J.; Fewer, D.P.; Wahlsten, M.; Fiore, M.F.; Sivonen, K. Simultaneous Production of anabaenopeptins and namalides by the cyanobacterium Nostoc sp. CENA543. ACS Chem. Biol. 2017, 12, 2746–2755. [Google Scholar] [CrossRef] [PubMed]
- Lazos, O.; Tosin, M.; Slusarczyk, A.L.; Boakes, S.; Cortés, J.; Sidebottom, P.J.; Leadlay, P.F. Biosynthesis of the putative siderophore erythrochelin requires unprecedented crosstalk between separate nonribosomal peptide gene clusters. Chem. Biol. 2010, 17, 160–173. [Google Scholar] [CrossRef]
- Hoffmann, D.; Hevel, J.M.; Moore, R.E.; Moore, B.S. Sequence Analysis and biochemical characterization of the nostopeptolide A biosynthetic gene cluster from Nostoc sp. GSV224. Gene 2003, 311, 171–180. [Google Scholar] [CrossRef]




| % AA Variation vs LaxA | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | Exact Mass | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 54.5% | Heinamide A3 | Ser | Pro | Ser | (2R)Tyr | (2R)Phe | Pro | 1186.65122 | |||||
| Heinamide A2 | Ser | Pro | Ser | (2R)Tyr | (2R)Phe | (2S,4R)Hyp | 1202.64614 | ||||||
| 45.5% | Heinamide A1 | Ser | Ser | (2R)Tyr | (2R)Phe | (2S,4R)Hyp | 1218.64105 | ||||||
| 18.2% or 27.3% | Lobocyclamide A | Ser | (2R)Tyr | (2R,3S)Ile or (2S,3R)Ile | 1197.70091 | ||||||||
| 18.2% | Scytocyclamide A2 | Gln | Pro | 1206.73763 | |||||||||
| [Des-(Leu10-Gly11)]AcyclolaxA | 1043,62669 | ||||||||||||
| 9.1% | Scytocyclamide A | Gln | 1222.73255 | ||||||||||
| Hormothamnin A | Z-Dhb | 1195.72165 | |||||||||||
| [D-Val9]LaxA | (2R)Val | 1181.70599 | |||||||||||
| LaxA2 | Val | 1181.70599 | |||||||||||
| [Des-Gly11]AcyclolaxA | 1156,71075 | ||||||||||||
| AcyclolaxA | 1213,73221 | ||||||||||||
| LaxA | Aoc | Hse | E-Dhb | (2S,4R)Hyp | Hse | (2R)Phe | (2R)Leu | Ile | (2R,3S)Ile | Leu | Gly | 1195.72165 | |
| 9.1% | LaxE | Ade | 1223.75295 | ||||||||||
| 63.6% | Trichormamide D | Ade | Gln | Pro | Ser | (2R)Tyr | Val | (2R)Phe | 1256.71989 | ||||
| 63.6% or 72.7% | Trichormamide A | Ade | Ser | Ser | Pro | Ser | (2R)Tyr | (2R,3S)Ile or Ile | Pro | 1183.68526 |
| % AA Variation vs LaxB | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | Exact Mass | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 41.7% | LaxB6 | Ile | Val | (2R)Leu | (2R)Asn | (2R)Tyr | 1454.87486 | |||||||
| 33.3% | LaxB5 | Ile | Val | (2R)Asn | (2R)Tyr | 1470.86977 | ||||||||
| Trichormamide B | Ile | Hse | (2R)Ser | (2R)Tyr | 1445.83814 | |||||||||
| Lyngbyacyclamide B | Hse | (2R)Leu | (2S,4R)Hyp | (2R)Phe | 1458.83338 | |||||||||
| 25% | Lyngbyacyclamide A | Hse | (2R)Leu | (2R)Phe | 1442.83847 | |||||||||
| [Des-(Ala4-Hle5)]AcyclolaxB1211 | Δ | 1211.70131 | ||||||||||||
| [Des-(Ala4-Hle5)]AcyclolaxB3 | (2S,4R)Hyp | 1228.72786 | ||||||||||||
| 16.7% | Lobocyclamide B | 3-OHHse* | (2S,4R)Hyp | 1397.83814 | ||||||||||
| LaxB4 | Hse | (2S,4R)Hyp | 1440.84395 | |||||||||||
| [Des-(Ala4-Hle5)]AcyclolaxB1195 | Δ | 1195.70639 | ||||||||||||
| [Des-(Ala4-Hle5)]AcyclolaxB | 1212.73294 | |||||||||||||
| 8.3% | AcyclolaxB3 | (2S,4R)Hyp | 1428.84395 | |||||||||||
| LaxB3 | (2S,4R)Hyp | 1410.83339 | ||||||||||||
| LaxB2 | (2R)Leu | 1378.84355 | ||||||||||||
| Trichormamide C | (2R)Asn | 1378.84355 | ||||||||||||
| AcyclolaxB | 1412.84904 | |||||||||||||
| LaxB | Ade | Val | (2R,3S)Hle | Ala | (2R,3S) Hle | Gln | (N)-MeIle | (2R,3R) Hasn | Thr | Pro | (2R)Leu | Thr | 1394.83847 | |
| 8.3% | LaxD or Scytocyclamide B | Aoc | 1366.80717 | |||||||||||
| 16.7% | Scytocyclamide B2 | Aoc | (2R)Asn | 1350.81226 | ||||||||||
| Scytocyclamide C | Aoc | (2R)Leu | 1350.81226 | |||||||||||
| 25% | Scytocyclamide B3 | Aoc | (2R)Leu | (2R)Asn | 1334.81734 | |||||||||
| Lobocyclamide C | Aoc | 3-OHHse* | (2S,4R)Hyp | 1369.80684 | ||||||||||
| 50.0% | Heinamide B3 | Aoc | Ile | O-carbamoyl-Hse | (2R)Leu | 3-OHHse | Val | (2R)Tyr | 1472.84903 | |||||
| 58.3% | Heinamide B2 | 5-OH-Aoc | Ile | O-carbamoyl-Hse | (2R)Leu | 3-OHHse | Val | (2R)Tyr | 1488.84395 | |||||
| Heinamide B4 | Aoc | Ile | O-carbamoyl-Hse | (2R)Leu | 3-OHHse | Val | 3-OH-4-MePro | (2R)Tyr | 1502.85960 | |||||
| 66.7% | Heinamide B1 | 5-OH-Aoc | Ile | O-carbamoyl-Hse | (2R)Leu | 3-OHHse | Val | 3-OH-4-MePro | (2R)Tyr | 1518.85451 | ||||
| Heinamide B5 | 5-OH-Aoc | Ile | O-carbamoyl-Hse | (2R)Leu | 3-OHHse | Val | 4-MePro | (2R)Tyr | 1502.85960 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Darcel, L.; Das, S.; Bonnard, I.; Banaigs, B.; Inguimbert, N. Thirtieth Anniversary of the Discovery of Laxaphycins. Intriguing Peptides Keeping a Part of Their Mystery. Mar. Drugs 2021, 19, 473. https://doi.org/10.3390/md19090473
Darcel L, Das S, Bonnard I, Banaigs B, Inguimbert N. Thirtieth Anniversary of the Discovery of Laxaphycins. Intriguing Peptides Keeping a Part of Their Mystery. Marine Drugs. 2021; 19(9):473. https://doi.org/10.3390/md19090473
Chicago/Turabian StyleDarcel, Laurine, Sanjit Das, Isabelle Bonnard, Bernard Banaigs, and Nicolas Inguimbert. 2021. "Thirtieth Anniversary of the Discovery of Laxaphycins. Intriguing Peptides Keeping a Part of Their Mystery" Marine Drugs 19, no. 9: 473. https://doi.org/10.3390/md19090473
APA StyleDarcel, L., Das, S., Bonnard, I., Banaigs, B., & Inguimbert, N. (2021). Thirtieth Anniversary of the Discovery of Laxaphycins. Intriguing Peptides Keeping a Part of Their Mystery. Marine Drugs, 19(9), 473. https://doi.org/10.3390/md19090473