Distribution of the Water-Soluble Astaxanthin Binding Carotenoprotein (AstaP) in Scenedesmaceae
Abstract
:1. Introduction
2. Results
2.1. Effect of Photooxidative Stresses on the Tested Strains
2.2. Effect of Photooxidative Stress Response on Model Microalgae
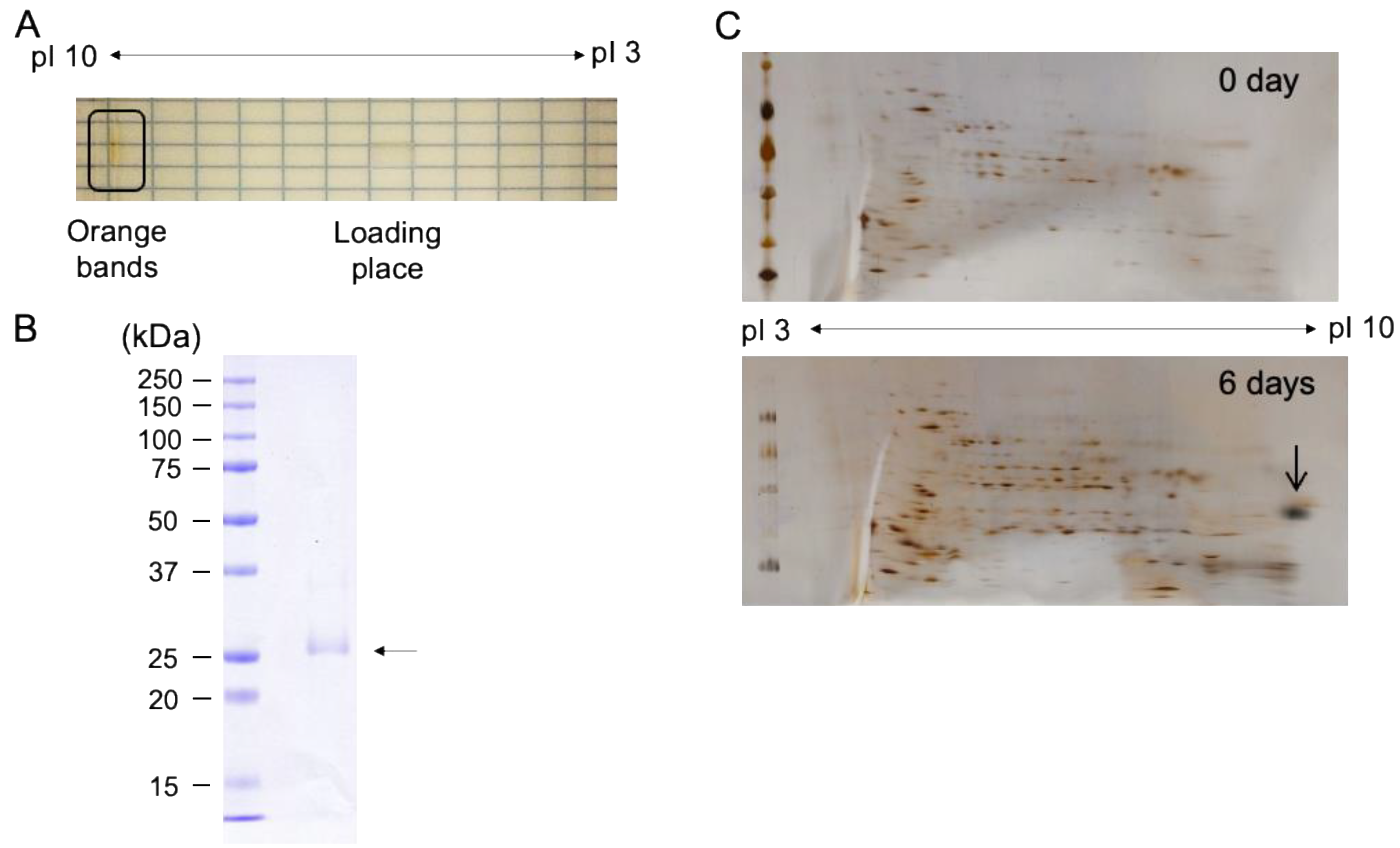
2.3. Purification and Characterization of Water-Soluble Astaxanthin-Binding Protein from S. costatus SAG 46.88
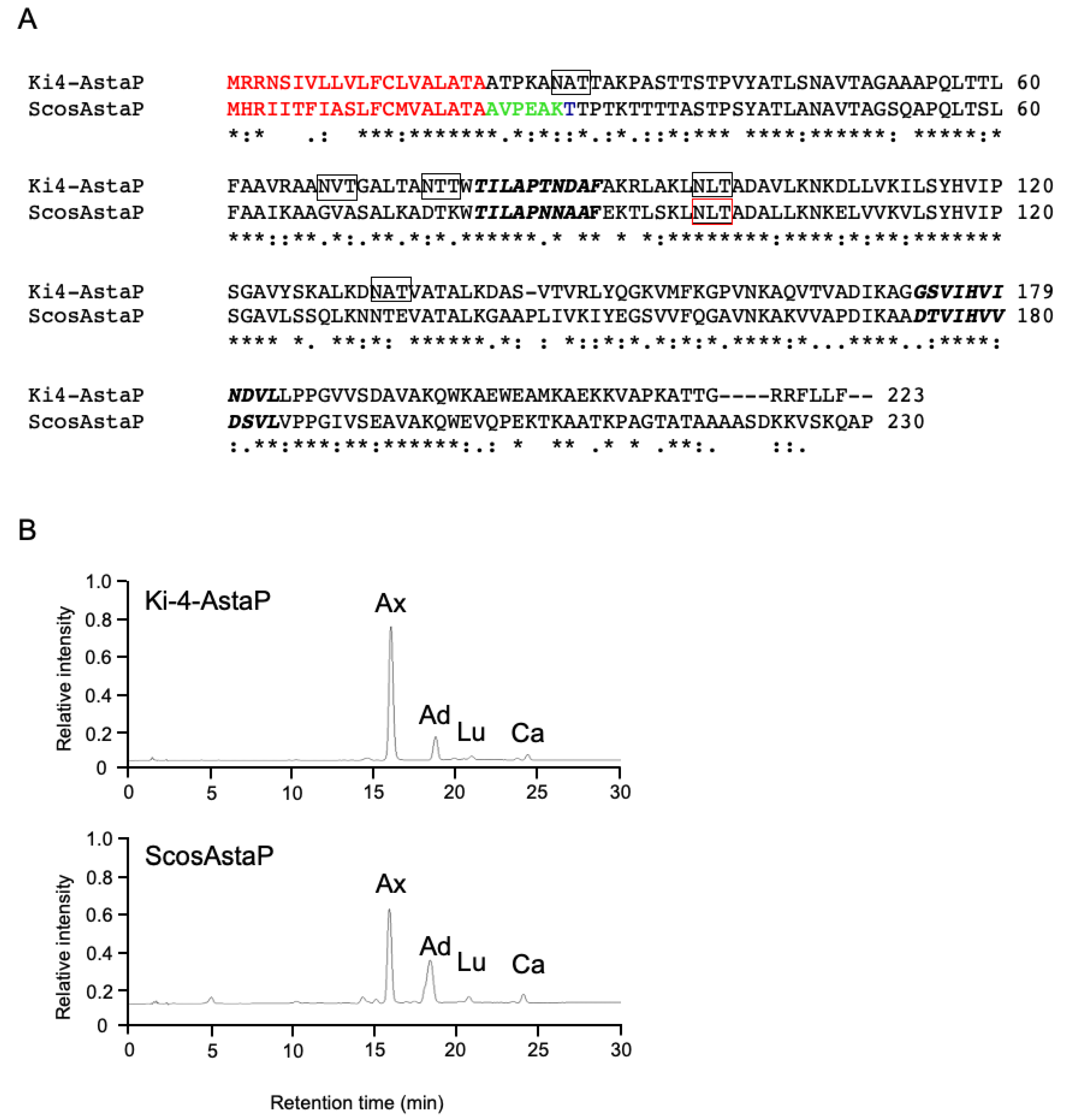
2.4. AstaP Ortholog in S. costatus Is a Fasciclin-Like Glycosylated Protein
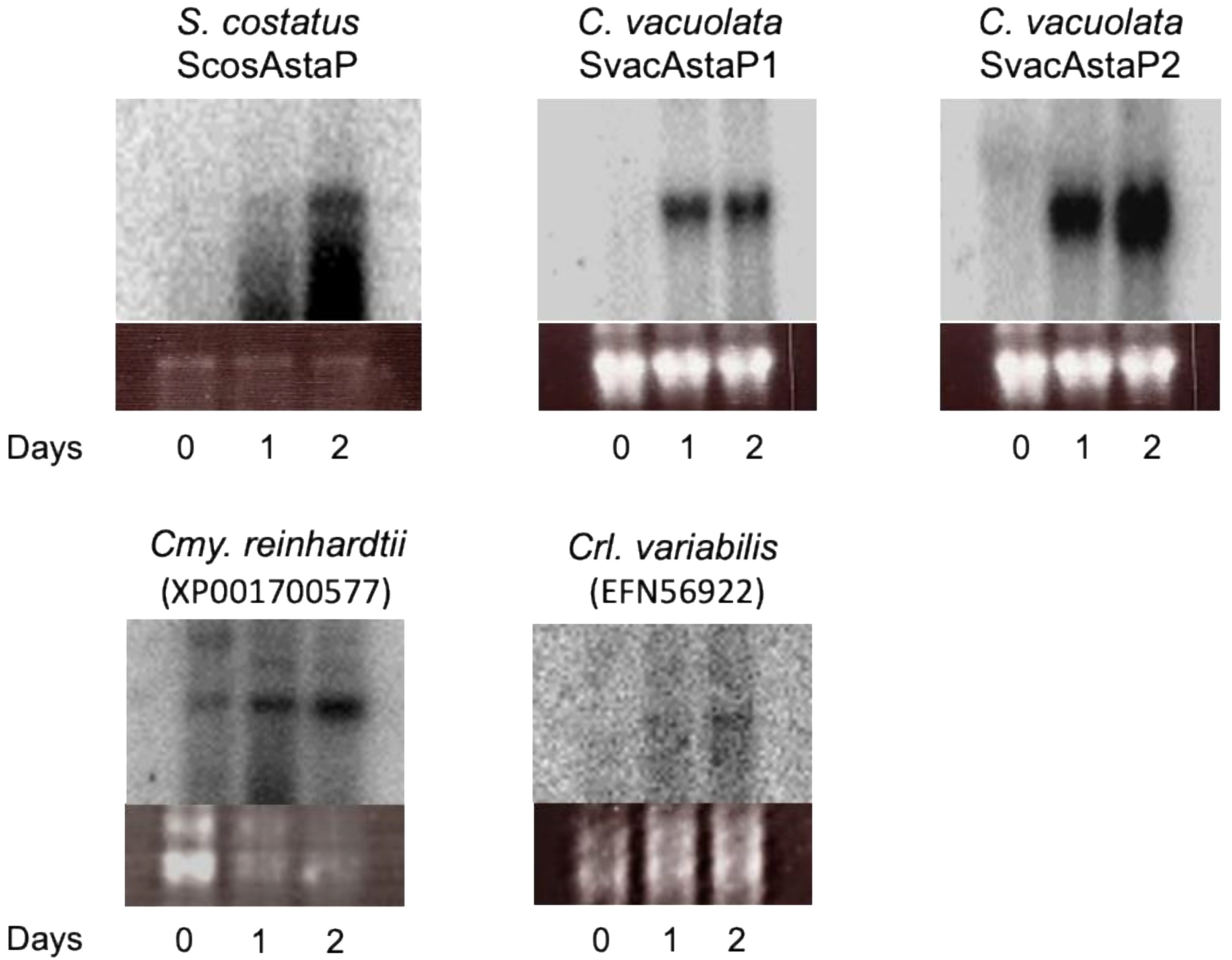
2.5. Identification of AstaP Orthologs in C. vacuolata and Two Model Microalgae
3. Discussion
4. Materials and Methods
4.1. Microalgal Strains and Growth Conditions
4.2. Gel Filtration Column Chromatography
4.3. Pigment Extraction and Identification
4.4. Purification of the Water Soluble Astaxanthin Binding Protein
4.5. Determination of Peptide Sequence
4.6. Isolation of RNA and Construction of a cDNA Library
4.7. Bioinformatics
4.8. Northern Hybridization
4.9. Two-Dimensional Electrophoresis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Cmy | Chlamydomonas |
| Crl | Chlorella |
| WSCP | Water-Soluble Carotenoprotein |
| w/HL | with High Light |
References
- Asada, K. The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 601–639. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Shigeoka, S. Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol. 2011, 155, 93–100. [Google Scholar] [CrossRef] [Green Version]
- Britton, G.; Liaaen-Jensen, S.; Pfander, H. (Eds.) Carotenoids Handbook; Birkhauseer: Basel, Switzerland, 2004. [Google Scholar]
- Keen, J.N.; Caceres, I.; Eliopoulos, E.E.; Zagalsky, P.F.; Findlay, J.B.C. Complete sequence and model for the A2 subunit of the carotenoid pigment complex, crustacyanin. Eur. J. Biochem. 1991, 197, 407–417. [Google Scholar] [CrossRef]
- Keen, J.N.; Caceres, I.; Eliopoulos, E.E.; Zagalsky, P.F.; Findlay, J.B.C. Complete sequence and model for the C1 subunit of the carotenoprotein crustacyanin, and model for the dimer, β-crustacyanin, formed from the C1 and A2 subunits with astaxanthin. Eur. J. Biochem. 1991, 202, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Bhosale, P.; Larson, A.J.; Frederick, J.M.; Southwick, K.; Thulin, C.D.; Bernstein, P.S. Identification and characterization of a Pi isoform of glutathione S-transferase (GSTP1) as a zeaxanthin-binding protein in the macula of the human eye. J. Biol. Chem. 2004, 279, 49447–49454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabunoki, H.; Sugiyama, H.; Tanaka, Y.; Fujii, H.; Banno, Y.; Jouni, Z.E.; Kobayashi, M.; Sato, R.; Maekawa, H.; Tsuchida, K. Isolation, characterization, and cDNA sequence of a carotenoid binding protein from the silk gland of Bombyx mori larvae. J. Biol. Chem. 2002, 277, 32133–32140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kay Holt, T.; Krogmann, D.W. A carotenoid-protein from cyanobacteria. BBA Bioenerg. 1981, 637, 408–414. [Google Scholar] [CrossRef]
- Kawasaki, S.; Mizuguchi, K.; Sato, M.; Kono, T.; Shimizu, H. A novel astaxanthin-binding photooxidative stress-inducible aqueous carotenoprotein from a eukaryotic microalga isolated from asphalt in midsummer. Plant Cell Physiol. 2013, 54, 1027–1040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawasaki, S.; Yamazaki, K.; Nishikata, T.; Ishige, T.; Toyoshima, H.; Miyata, A. Photooxidative stress-inducible orange and pink water-soluble astaxanthin-binding proteins in eukaryotic microalga. Commun. Biol. 2020, 3, 490. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.; Kinney, J.N.; Zwart, P.H.; Punginelli, C.; D’Haene, S.; Perreau, F.; Klein, M.G.; Kirilovsky, D.; Kerfeld, C.A. Structural determinants underlying photoprotection in the photoactive orange carotenoid protein of cyanobacteria. J. Biol. Chem. 2010, 285, 18364–18375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerfeld, C.A.; Sawaya, M.R.; Brahmandam, V.; Cascio, D.; Ho, K.K.; Trevithick-Sutton, C.C.; Krogmann, D.W.; Yeates, T.O. The crystal structure of a cyanobacterial water-soluble carotenoid binding protein. Structure 2003, 11, 55–65. [Google Scholar] [CrossRef] [Green Version]
- Kawasaki, S.; Yoshida, R.; Kiichi, O.A.; Toyoshima, H. Coelastrella astaxanthina sp. nov. (Sphaeropleales, Chlorophyceae), a novel microalga isolated from an asphalt surface in midsummer in Japan. Phycol. Res. 2019, 68, 107–114. [Google Scholar] [CrossRef]
- Torelli, A.; Marieschi, M.; Castagnoli, B.; Zanni, C.; Gorbi, G.; Corradi, M.G. Identification of S2-T A63: A cDNA fragment corresponding to a gene differentially expressed in a Cr-tolerant strain of the unicellular green alga Scenedesmus acutus. Aquat. Toxicol. 2008, 86, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Schmidle, W. Beiträge zur alpinen Algenflora. Österreichische Bot. Z. 1895, 45, 305–311. [Google Scholar] [CrossRef]
- Hegewald, E.; Hanagata, N. Phylogenetic studies on Scenedesmaceae (Chlorophyta). Algol. Stud. 2000, 100, 29–49. [Google Scholar] [CrossRef]
- Chodat, R. Matériaux pour l’histoire des algues de la Suisse. Bull. Soc. Bot Geneve Ser. 1921, 2, 66–114. [Google Scholar]
- Guiry, M.D.; Guiry, G.M. AlgaeBase; World-Wide Electronic Publication, National University of Ireland: Galway, Ireland, 2021. [Google Scholar]
- Toyoshima, H.; Takaichi, S.; Kawasaki, S. Water-soluble astaxanthin-binding protein (AstaP) from Coelastrella astaxanthina Ki-4 (Scenedesmaceae) involving in photo-oxidative stress tolerance. Algal Res. 2020, 50, 101988. [Google Scholar] [CrossRef]
- Kaufnerová, V.; Eliáš, M. The demise of the genus Scotiellopsis Vinatzer (Chlorophyta). Nov. Hedwigia 2013, 97, 415–428. [Google Scholar] [CrossRef] [Green Version]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]

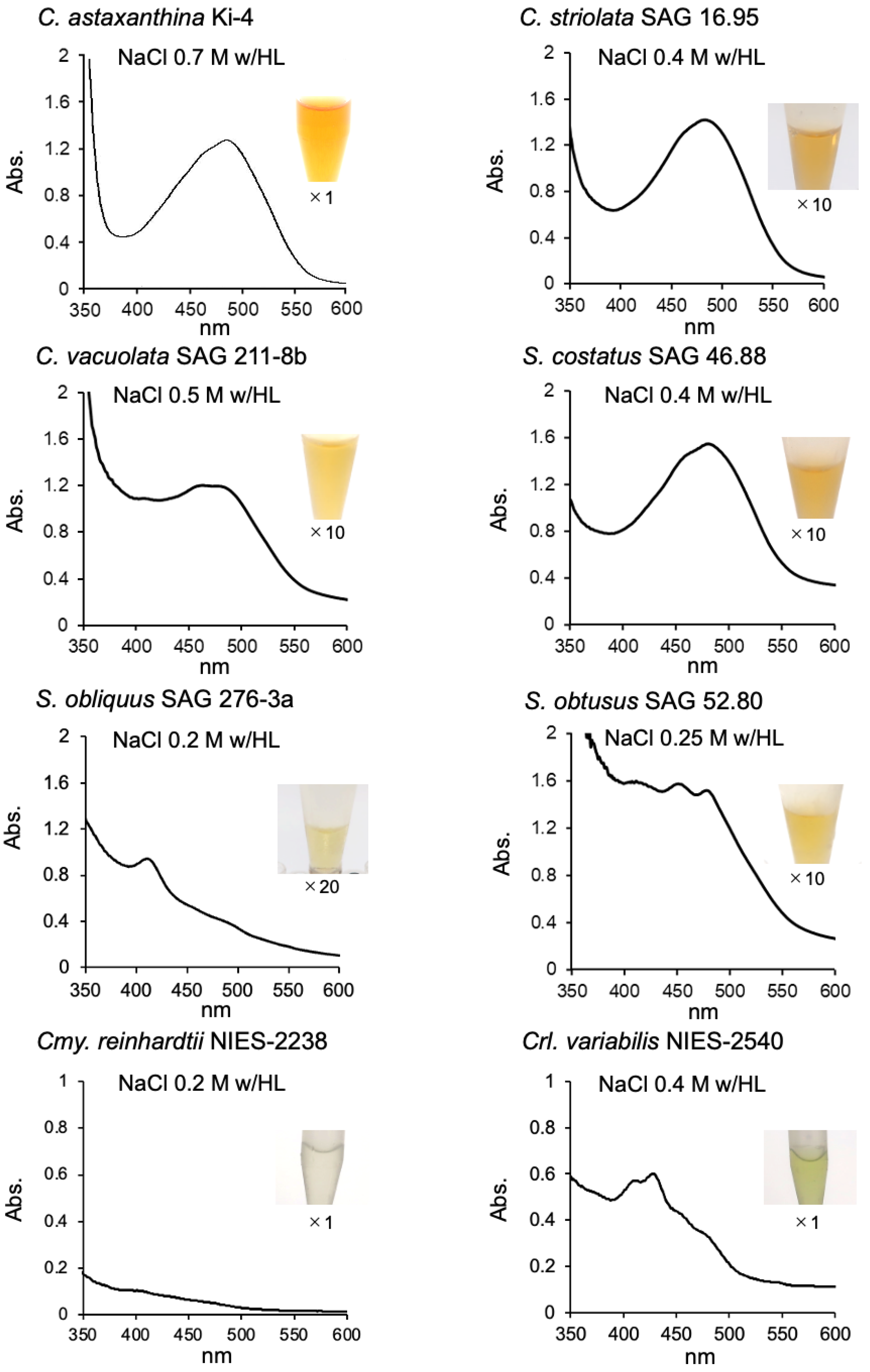
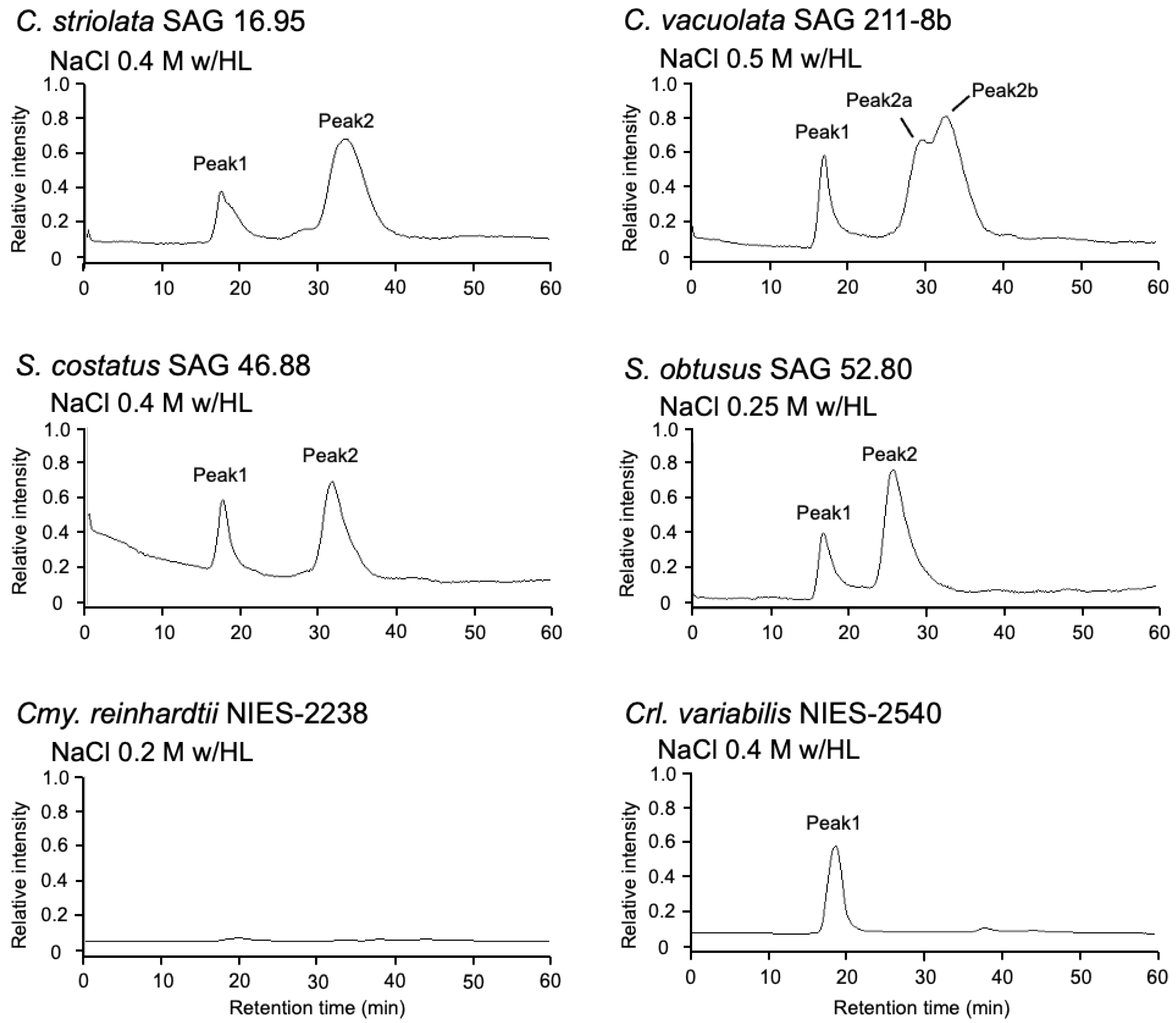
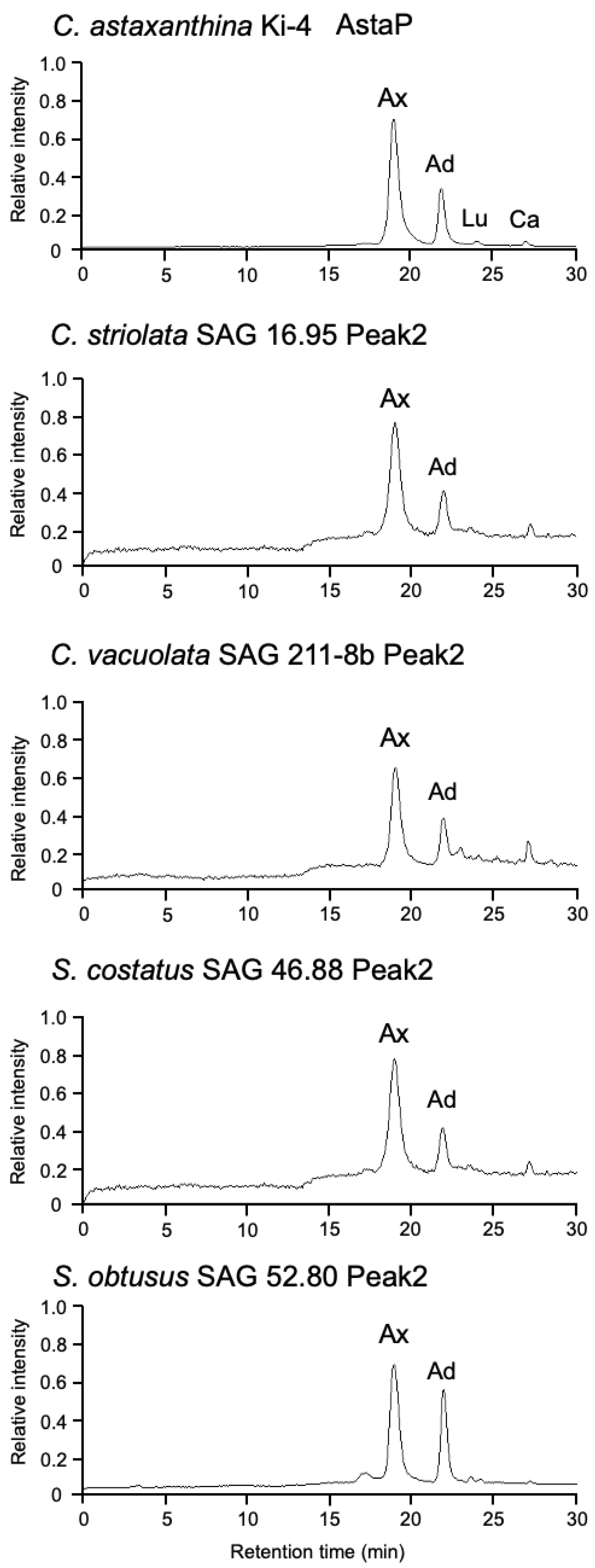

| Species and Strain | Stress Conditions (with/ HL) | OD480 (1 g Wet Cell/10 mL) | Reference |
|---|---|---|---|
| Coelastrella astaxanthina Ki-4 | 0.7 M NaCl | 1.3 | [9] |
| Scenedesmus obtusus Oki-4N | 0.5 M NaCl | 1.0 | [10] |
| Coelastrella striolata SAG 16.95 | 0.4 M NaCl | 0.14 | This study |
| Coelastrella vacuolata SAG 211-8b | 0.5M NaCl | 0.12 | This study |
| Scenedesmus costatus SAG 46.88 | 0.4 M NaCl | 0.16 | This study |
| Scenedesmus obliquus SAG 276-3a | 0.2 M NaCl | 0.02 | This study |
| Scenedesmus obtusus SAG 52.80 | 0.25 M NaCl | 0.15 | This study |
| Chlamydomonas reinhardtii NIES-2238 | 0.2 M NaCl | 0.04 | This study |
| Chlorella variabilis NIES-2540 | 0.4 M NaCl | 0.33 | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toyoshima, H.; Miyata, A.; Yoshida, R.; Ishige, T.; Takaichi, S.; Kawasaki, S. Distribution of the Water-Soluble Astaxanthin Binding Carotenoprotein (AstaP) in Scenedesmaceae. Mar. Drugs 2021, 19, 349. https://doi.org/10.3390/md19060349
Toyoshima H, Miyata A, Yoshida R, Ishige T, Takaichi S, Kawasaki S. Distribution of the Water-Soluble Astaxanthin Binding Carotenoprotein (AstaP) in Scenedesmaceae. Marine Drugs. 2021; 19(6):349. https://doi.org/10.3390/md19060349
Chicago/Turabian StyleToyoshima, Hiroki, Ami Miyata, Risako Yoshida, Taichiro Ishige, Shinichi Takaichi, and Shinji Kawasaki. 2021. "Distribution of the Water-Soluble Astaxanthin Binding Carotenoprotein (AstaP) in Scenedesmaceae" Marine Drugs 19, no. 6: 349. https://doi.org/10.3390/md19060349
APA StyleToyoshima, H., Miyata, A., Yoshida, R., Ishige, T., Takaichi, S., & Kawasaki, S. (2021). Distribution of the Water-Soluble Astaxanthin Binding Carotenoprotein (AstaP) in Scenedesmaceae. Marine Drugs, 19(6), 349. https://doi.org/10.3390/md19060349






