Untapped Potential of Marine-Associated Cladosporium Species: An Overview on Secondary Metabolites, Biotechnological Relevance, and Biological Activities
Abstract
1. Introduction
2. Importance of Marine Associated Cladosporium Species
| Compound Name | Mol. Wt. | Mol. Formula | Fungal Source | Host (Sample, Family) | Place | Ref. |
|---|---|---|---|---|---|---|
| 1. Tetramic acid derivatives | ||||||
| Cladosin A (1) | 282 | C14H22N2O4 | C. sphaerospermum 2005-01-E3 | Deep-sea sludge, Pacific Ocean | Qingdao, China | [42] |
| Cladosin B (2) | 268 | C13H20N2O4 | C. sphaerospermum 2005-01-E3 | Deep-sea sludge, Pacific Ocean | Qingdao, China | [42] |
| C. sphaerospermum SW67 | Hydractinia echinata (Marine hydroid, Hydractiniidae) | South Korea | [53] | |||
| Cladosin C (3) | 250 | C13H18N2O3 | C. sphaerospermum 2005-01-E3 | Deep-sea sludge, Pacific Ocean | Qingdao, China | [42] |
| C. sphaerospermum SW67 | Hydractinia echinata (Marine hydroid, Hydractiniidae) | South Korea | [53] | |||
| Cladosin D (4) | 250 | C13H18N2O3 | C. sphaerospermum 2005-01-E3 | Deep-sea sludge, Pacific Ocean | Qingdao, China | [42] |
| Cladosin F (5) | 268 | C13H20N2O4 | C. sphaerospermum 2005-01-E3 | Deep-sea sludge, Pacific Ocean | Qingdao, China | [54] |
| C. sphaerospermum SW67 | Hydractinia echinata (Marine hydroid, Hydractiniidae) | South Korea | [53] | |||
| Cladosin G (6) | 282 | C14H22N2O4 | C. sphaerospermum 2005-01-E3 | Deep-sea sludge, Pacific Ocean | Qingdao, China | [54] |
| Cladosin H (7) | 358 | C20H26N2O4 | C. sphaerospermum L3P3 | Marine sediment | Mariana Trench, South Pacific Ocean, China | [55] |
| Cladosin I (8) | 358 | C20H26N2O4 | C. sphaerospermum L3P3 | Marine sediment | Mariana Trench, South Pacific Ocean, China | [55] |
| Cladosin J (9) | 419 | C25H29N3O3 | C. sphaerospermum L3P3 | Marine sediment | Mariana Trench, South Pacific Ocean, China | [55] |
| Cladosin K (10) | 419 | C25H29N3O3 | C. sphaerospermum L3P3 | Marine sediment | Mariana Trench, South Pacific Ocean, China | [55] |
| Cladosin L (11) | 270 | C13H22N2O4 | C. sphaerospermum SW67 | Hydractinia echinata (Marine hydroid, Hydractiniidae) | South Korea | [53] |
| Cladosporicin A (12) | 401 | C21H27N3O5 | C. sphaerospermum SW67 | Hydractinia echinata (Marine hydroid, Hydractiniidae) | South Korea | [38] |
| Cladodionen (13) | 233 | C13H15NO3 | Cladosporium sp. OUCMDZ-1635 | Unidentified sponge | Xisha Islands, China | [56] |
| C. sphaerospermum EIODSF 008. | Deep sea sediment | East Indian Ocean, China | [57] | |||
| C. sphaerospermum L3P3 | Marine sediment | Mariana Trench, South Pacific Ocean, China | [55] | |||
| Cladosporiumin A (14) | 349 | C19H27NO5 | Cladosporium sp. SCSIO z0025 | Deep sea sediment | Okinawa, Japan | [58] |
| Cladosporiumin B (15) | 349 | C19H27NO5 | Cladosporium sp. SCSIO z0025 | Deep sea sediment | Okinawa, Japan | [58] |
| Cladosporiumin C (16) | 349 | C19H27NO5 | Cladosporium sp. SCSIO z0025 | Deep sea sediment | Okinawa, Japan | [58] |
| Cladosporiumin D (17) | 253 | C13H19NO4 | Cladosporium sp. SCSIO z0025 | Deep sea sediment | Okinawa, Japan | [58] |
| Cladosporiumin E (18) | 251 | C13H17NO4 | Cladosporium sp. SCSIO z0025 | Deep sea sediment | Okinawa, Japan | [58] |
| Cladosporiumin F (19) | 269 | C13H19NO5 | Cladosporium sp. SCSIO z0025 | Deep sea sediment | Okinawa, Japan | [58] |
| Cladosporiumin G (20) | 253 | C13H19NO4 | Cladosporium sp. SCSIO z0025 | Deep sea sediment | Okinawa, Japan | [58] |
| Cladosporiumin H (21) | 285 | C14H23NO5 | Cladosporium sp. SCSIO z0025 | Deep sea sediment | Okinawa, Japan | [58] |
| Cladosporiumin I (22) | 235 | C13H17NO3 | C. sphaerospermum EIODSF 008. | Deep sea sediment | East Indian Ocean, China | [57] |
| Cladosporiumin J (23) | 251 | C13H17NO4 | C. sphaerospermum EIODSF 008. | Deep sea sediment | East Indian Ocean, China | [57] |
| Cladosporiumin K (24) | 251 | C13H17NO4 | C. sphaerospermum EIODSF 008. | Deep sea sediment | East Indian Ocean. China | [57] |
| Cladosporiumin L (25) | 887 | C41H65N3O15Mg2 | C. sphaerospermum EIODSF 008. | Deep sea sediment | East Indian Ocean, China | [57] |
| Cladosporiumin M (26) | 233 | C13H15NO3 | C. sphaerospermum EIODSF 008. | Deep sea sediment | East Indian Ocean, China | [57] |
| Cladosporiumin N (27) | 253 | C13H19NO4 | C. sphaerospermum EIODSF 008. | Deep sea sediment | East Indian Ocean. China | [57] |
| Cladosporiumin O (28) | 251 | C13H17NO4 | C. sphaerospermum EIODSF 008. | Deep sea sediment | East Indian Ocean, China | [57] |
| Cladosporiumin I (29) | 349 | C19H27NO5 | C. sphaerospermum SW67 | Hydractinia echinata (Marine hydroid, Hydractiniidae) | South Korea | [38] |
| Cladosporiumin J (30) | 349 | C19H27NO5 | C. sphaerospermum SW67 | Hydractinia echinata (Marine hydroid, Hydractiniidae) | South Korea | [38] |
| 2. Diketopiperazines | ||||||
| Cyclo-(Pro, Trp) (31) | 283 | C16H17N3O2 | Cladosporium sp. EF424419 | Porphyra yezoensis (Red alga, Bangiaceae) | Lianyungang, Jiangsu, China | [59] |
| Cyclo-(Val-Pro) (32) | 196 | C10H16N2O2 | Cladosporium sp. EF424419 | Porphyra yezoensis (Red alga, Bangiaceae) | Lianyungang, Jiangsu, China | [59] |
| Cyclo-(Phe-Pro) (33) | 244 | C14H16N2O2 | Cladosporium sp. F14 | Seawater from mangrove stand | Kei Ling Ha Lo Wai, Sai Kung, China | [60] |
| Cyclo-(Phe-Val) (34) | 246 | C14H18N2O2 | Cladosporium sp. F14 | Seawater from mangrove stand | Kei Ling Ha Lo Wai, Sai Kung, China | [60] |
| Cyclo-(Gly-Leu) (35) | 170 | C8H14N2O2 | Cladosporium sp. SCSIO41007 | Callyspongia sp. (Sponge, Callyspongiidae) | Xuwen, Guangdong, China | [61] |
| Cladosporin A (36) | 460 | C22H24N2O5S2 | Cladosporium sp. | Marine sediment | Yangshashan Bay, Ningbo, Zhejiang, China | [62] |
| Cladosporin B (37) | 442 | C22H22N2O4S2 | Cladosporium sp. | Marine sediment | Yangshashan Bay, Ningbo, Zhejiang, China | [62] |
| Haematocin (38) | 502 | C24H26N2O6S2 | Cladosporium sp. | Marine sediment | Yangshashan Bay, Ningbo, Zhejiang, China | [62] |
| 3. Alkaloids | ||||||
| 3.1. Indole alkaloids | ||||||
| 3.1.1 Simple indole alkaloids | ||||||
| N-Acetyltryptamine (39) | 202 | C12H14N2O | Cladosporium sp. EF424419 | Porphyra yezoensis (Red alga, Bangiaceae) | Lianyungang, Jiangsu, China | [59] |
| N-methyl-1H-indole-2-carboxamide (40) | 174 | C10H10N2O | C. cladosporioides | Cliona sp. (Sponge, Clionaidae) | Los Molles, Chile | [63] |
| Indole-3-carboxylic acid (41) | 161 | C9H7NO2 | Cladosporium sp. SCSIO41007 | Callyspongia sp. (Sponge, Callyspongiidae) | Xuwen, Guangdong, China | [61] |
| 3.1.2 Glyantrypine derivatives | ||||||
| Glyantrypine (42) | 344 | C20H16N4O2 | Cladosporium sp. PJX-41 | Soil around a mangrove | Guangzhou, China | [64] |
| 3-Hydroxyglyantrypine (43) | 360 | C20H16N4O3 | Cladosporium sp. PJX-41 | Soil around a mangrove | Guangzhou, China | [64] |
| 14R-Oxoglyantrypine (44) | 358 | C20H14N4O3 | Cladosporium sp. PJX-41 | Soil around a mangrove | Guangzhou, China | [64] |
| 14S-Oxoglyantrypine (45) | 358 | C20H14N4O3 | Cladosporium sp. PJX-41 | Soil around a mangrove | Guangzhou, China | [64] |
| Prelapatin B (46) | 344 | C20H16N4O2 | Cladosporium sp. PJX-41 | Soil around a mangrove | Guangzhou, China | [64] |
| Cladoquinazoline (47) | 418 | C23H22N4O4 | Cladosporium sp. PJX-41 | Soil around a mangrove | Guangzhou, China | [64] |
| Epi-Cladoquinazoline (48) | 418 | C23H22N4O4 | Cladosporium sp. PJX-41 | Soil around a mangrove | Guangzhou, China | [64] |
| 3.2. Quinazoline alkaloids | ||||||
| Norquinadoline A (49) | 471 | C26H25N5O4 | Cladosporium sp. PJX-41 | Soil around a mangrove | Guangzhou, China | [64] |
| Quinadoline A (50) | 485 | C27H27N5O5 | Cladosporium sp. PJX-41 | Soil around a mangrove | Guangzhou, China | [64] |
| Deoxynortryptoquivaline (51) | 516 | C28H28N4O6 | Cladosporium sp. PJX-41 | Soil around a mangrove | Guangzhou, China | [64] |
| Deoxytryptoquivaline (52) | 530 | C29H30N4O6 | Cladosporium sp. PJX-41 | Soil around a mangrove | Guangzhou, China | [64] |
| Tryptoquivaline (53) | 546 | C29H30N4O7 | Cladosporium sp. PJX-41 | Soil around a mangrove | Guangzhou, China | [64] |
| CS-C (54) | 546 | C29H30N4O7 | Cladosporium sp. PJX-41 | Soil around a mangrove | Guangzhou, China | [64] |
| Quinadoline B (55) | 439 | C25H21N5O3 | Cladosporium sp. PJX-41 | Soil around a mangrove | Guangzhou, China | [64] |
| Circumdatin A (56) | 391 | C22H21N3O4 | Cladosporium sp. MFC353-b | Chondria crassicualis (Red alga, Rhodomelaceae) | Yokji Island, Kyeongnam, Korea | [65] |
| 3.3. Quinolone alkaloids | ||||||
| Quinolactacin A1 (57) | 270 | C16H18N2O2 | C. oxysporum BRS2A-AR2F | Conocarpus erectus (Mangrove plant, Combretaceae) Laguncularia racemosa (Mangrove plant, Combretaceae) Rhizophora racemosa (Mangrove plant, Rhizophoraceae) | Banks of the River Butre, Western Region of Ghana | [66] |
| Quinolactacin A2 (58) | 270 | C16H18N2O2 | C. oxysporum BRS2A-AR2F | Conocarpus erectus (Mangrove plant, Combretaceae) Laguncularia racemosa (Mangrove plant, Combretaceae) Rhizophora racemosa (Mangrove plant, Rhizophoraceae) | Banks of the River Butre, Western Region of Ghana | [66] |
| Quinolactacin B1 (59) | 256 | C15H16N2O2 | C. oxysporum BRS2A-AR2F | Conocarpus erectus (Mangrove plant, Combretaceae) Laguncularia racemosa (Mangrove plant, Combretaceae) Rhizophora racemosa (Mangrove plant, Rhizophoraceae) | Banks of the River Butre, Western Region of Ghana | [66] |
| Quinolactacin B2 (60) | 256 | C15H16N2O2 | C. oxysporum BRS2A-AR2F | Conocarpus erectus (Mangrove plant, Combretaceae) Laguncularia racemosa (Mangrove plant, Combretaceae) Rhizophora racemosa (Mangrove plant, Rhizophoraceae) | Banks of the River Butre, Western Region of Ghana | [66] |
| Quinolactacin C1 (61) | 286 | C16H18N2O3 | C. oxysporum BRS2A-AR2F | Conocarpus erectus (Mangrove plant, Combretaceae) Laguncularia racemosa (Mangrove plant, Combretaceae) Rhizophora racemosa (Mangrove plant, Rhizophoraceae) | Banks of the River Butre, Western Region of Ghana | [66] |
| Quinolactacin C2 (62) | 286 | C16H18N2O3 | C. oxysporum BRS2A-AR2F | Conocarpus erectus (Mangrove plant, Combretaceae) Laguncularia racemosa (Mangrove plant, Combretaceae) Rhizophora racemosa (Mangrove plant, Rhizophoraceae) | Banks of the River Butre, Western Region of Ghana | [66] |
| Quinolactacin D1 (63) | 286 | C16H18N2O3 | C. oxysporum BRS2A-AR2F | Conocarpus erectus (Mangrove plant, Combretaceae) Laguncularia racemosa (Mangrove plant, Combretaceae) Rhizophora racemosa (Mangrove plant, Rhizophoraceae) | Banks of the River Butre, Western Region of Ghana | [66] |
| Quinolactacin D2 (64) | 286 | C16H18N2O3 | C. oxysporum BRS2A-AR2F | Conocarpus erectus (Mangrove plant, Combretaceae) Laguncularia racemosa (Mangrove plant, Combretaceae) Rhizophora racemosa (Mangrove plant, Rhizophoraceae) | Banks of the River Butre, Western Region of Ghana | [66] |
| Quinocitrinine A (65) | 272 | C16H19N2O2 | C. oxysporum BRS2A-AR2F | Conocarpus erectus (Mangrove plant, Combretaceae) Laguncularia racemosa (Mangrove plant, Combretaceae) Rhizophora racemosa (Mangrove plant, Rhizophoraceae) | Banks of the River Butre, Western Region of Ghana | [66] |
| Quinocitrinine B (66) | 272 | C16H19N2O2 | C. oxysporum BRS2A-AR2F | Conocarpus erectus (Mangrove plant, Combretaceae) Laguncularia racemosa (Mangrove plant, Combretaceae) Rhizophora racemosa (Mangrove plant, Rhizophoraceae) | Banks of the River Butre, Western Region of Ghana | [66] |
| Quinolactacide (67) | 236 | C14H8N2O2 | C. oxysporum BRS2A-AR2F | Conocarpus erectus (Mangrove plant, Combretaceae) Laguncularia racemosa (Mangrove plant, Combretaceae) Rhizophora racemosa (Mangrove plant, Rhizophoraceae) | Banks of the River Butre, Western Region of Ghana | [66] |
| 3.4. Citrinadin derivatives | ||||||
| Citrinadin A (68) | 624 | C35H52N4O6 | C. oxysporum BRS2A-AR2F | Conocarpus erectus (Mangrove plant, Combretaceae) Laguncularia racemosa (Mangrove plant, Combretaceae) Rhizophora racemosa (Mangrove plant, Rhizophoraceae) | Banks of the River Butre, Western Region of Ghana | [66] |
| Citrinadin B (69) | 481 | C28H39N3O4 | C. oxysporum BRS2A-AR2F | Conocarpus erectus (Mangrove plant, Combretaceae) Laguncularia racemosa (Mangrove plant, Combretaceae) Rhizophora racemosa (Mangrove plant, Rhizophoraceae) | Banks of the River Butre, Western Region of Ghana | [66] |
| Butrecitrinadin (70) | 682 | C38H57N4O7 | C. oxysporum BRS2A-AR2F | Conocarpus erectus (Mangrove plant, Combretaceae) Laguncularia racemosa (Mangrove plant, Combretaceae) Rhizophora racemosa (Mangrove plant, Rhizophoraceae) | Banks of the River Butre, Western Region of Ghana | [66] |
| PF1270 A (71) | 566 | C32H43N3O6 | C. oxysporum BRS2A-AR2F | Conocarpus erectus (Mangrove plant, Combretaceae) Laguncularia racemosa (Mangrove plant, Combretaceae) Rhizophora racemosa (Mangrove plant, Rhizophoraceae) | Banks of the River Butre, Western Region of Ghana | [66] |
| PF1270 B (72) | 552 | C31H41N3O6 | C. oxysporum BRS2A-AR2F | Conocarpus erectus (Mangrove plant, Combretaceae) Laguncularia racemosa (Mangrove plant, Combretaceae) Rhizophora racemosa (Mangrove plant, Rhizophoraceae) | Banks of the River Butre, Western Region of Ghana | [66] |
| PF1270 C (73) | 538 | C30H39N3O6 | C. oxysporum BRS2A-AR2F | Conocarpus erectus (Mangrove plant, Combretaceae) Laguncularia racemosa (Mangrove plant, Combretaceae) Rhizophora racemosa (Mangrove plant, Rhizophoraceae) | Banks of the River Butre, Western Region of Ghana | [66] |
| 3.5. Pyrrolidine derivatives | ||||||
| Cladosporitin A (74) | 505 | C32H43NO4 | Cladosporium sp. HNWSW-1 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | Dong Zhai Gang, Hainan, China | [67] |
| Cladosporitin B (75) | 505 | C32H43NO4 | Cladosporium sp. HNWSW-1 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | Dong Zhai Gang, Hainan, China | [67] |
| Talaroconvolutin A (76) | 487 | C32H41NO3 | Cladosporium sp. HNWSW-1 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | Dong Zhai Gang, Hainan, China | [67] |
| Cladosporamide A (77) | 273 | C14H11NO5 | Cladosporium sp. TPU1507 | Unidentified marine sponge | Manado, Indonesia | [68] |
| 3.6. Other class of alkaloids | ||||||
| Cladosporilactam A (78) | 181 | C10H15NO2 | Cladosporium sp. RA07-1 | Anthogorgia ochracea (Gorgonian, Acanthogorgiidae) | Weizhou coral reef, South China Sea | [69] |
| Cladospamide A (79) | 268 | C13H20N2O4 | Cladosporium sp. SCNU-F0001 | Mangrove plant | Zhuhai Mangrove Nature, Guangdong, China | [70] |
| Cladosporin A (80) | 233 | C13H15NO3 | C. cladosporioides SCSIO z015 | Deep sea sediment | Okinawa, Japan | [36] |
| Cladosporin B (81) | 233 | C13H15NO3 | C. cladosporioides SCSIO z015 | Deep sea sediment | Okinawa, Japan | [36] |
| 2′-Deoxythymidine (82) | 241 | C11H15NO5 | Cladosporium sp. SCSIO41007 | Callyspongia sp. (Sponge, Callyspongiidae) | Xuwen, Guangdong, China | [61] |
| Nicotinic acid (83) | 123 | C6H5NO2 | Cladosporium sp. EF424419 | Porphyra yezoensis (Red alga, Bangiaceae) | Lianyungang, Jiangsu, China | [59] |
| 2-Methylacetate-3,5,6-trimethylpyrazine (84) | 194 | C10H14N2O2 | Cladosporium sp. JS1-2 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | Dongzhaigang, Hainan, China | [71] |
| Cytochalasin D (85) | 507 | C30H37NO6 | Cladosporium sp. JS1-2 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | Dongzhaigang, Hainan, China | [71] |
| Cladosin E (86) | 251 | C13H17NO4 | C. sphaerospermum 2005-01-E3 | Deep-sea sludge, Pacific Ocean | Qingdao, China | [42] |
| N-Acetyltyramine (87) | 179 | C10H13NO2 | Cladosporium sp. EF424419 | Porphyra yezoensis (Red alga, Bangiaceae) | Lianyungang, Jiangsu, China | [59] |
| 4. Macrolides | ||||||
| Cladospolide A (88) | 228 | C12H20O4 | Cladosporium sp. FT-0012 | Sponge | Pohnpei island, Federated State of Micronesia | [72] |
| Cladosporium sp. IFB3lp-2 | Rhizophora stylosa (Mangrove plant, Rhizophoraceae) | Mangrove forest, Hainan, China | [73] | |||
| Cladospolide B (89) | 228 | C12H20O4 | Cladosporium sp. FT-0012 | Sponge | Pohnpei island, Federated State of Micronesia | [72] |
| C. herbarum (Pers.) | Callyspongia aerizusa (Sponge, Callyspongiidae) | Bali Bata National Park, Indonesia, | [74] | |||
| Cladosporium sp. RA07-1 | Anthogorgia ochracea (Gorgonian, Acanthogorgiidae) | Weizhou coral reef, South China Sea | [69] | |||
| Cladosporium sp. SCNU-F0001 | Mangrove plant | Zhuhai Mangrove Nature, Guangdong, China | [70] | |||
| Cladospolide C (90) | 228 | C12H20O4 | C. cladosporioides MCCC 3A00182 | Marine sediment | Southwest Pacific Ocean | [75] |
| Cladospolide D (91) | 226 | C12H18O4 | Cladosporium sp. FT-0012 | Sponge | Pohnpei island, Federated State of Micronesia | [72] |
| Cladospolide E (92) | 188 | C8H12O5 | Cladosporium sp. F14. | Seawater nearby mangrove stand | Kei Ling Ha Lo Wai, Sai Kung, Hong Kong, China | [76] |
| Pandangolide 1 (93) | 244 | C12H20O5 | Cladosporium sp. | Niphates rowi (Sponge, Niphatidae) | Gulf of Aqaba, Israel | [77] |
| Cladosporium sp. F14 | Seawater from mangrove stand | Kei Ling Ha Lo Wai, Sai Kung, China | [60] | |||
| Cladosporium sp. IFB3lp-2 | Rhizophora stylosa (Mangrove plant, Rhizophoraceae) | Mangrove forest, Hainan, China | [73] | |||
| C. cladosporioides MA-299 | Bruguiera gymnorrhiza (Mangrove plant, Rhizophoraceae) | Hainan Island, China | [40] | |||
| Pandangolide 1a (94) | 244 | C12H20O5 | Cladosporium sp. | Niphates rowi (Sponge, Niphatidae) | Gulf of Aqaba, Israel | [77] |
| Cladosporium sp. IFB3lp-2 | Rhizophora stylosa (Mangrove plant, Rhizophoraceae) | Mangrove forest, Hainan, China | [73] | |||
| Pandangolide 2 (95) | 318 | C14H22O6S | C. herbarum (Pers.) | Callyspongia aerizusa (Sponge, Callyspongiidae) | Bali Bata National Park, Indonesia | [74] |
| Cladosporium sp. IFB3lp-2 | Rhizophora stylosa (Mangrove plant, Rhizophoraceae) | Mangrove forest, Hainan, China | [73] | |||
| Pandangolide 3 (96) | 362 | C16H26O7S | C. herbarum (Pers.) | Callyspongia aerizusa (Sponge, Callyspongiidae) | Bali Bata National Park, Indonesia, | [74] |
| Cladosporium sp. IFB3lp-2 | Rhizophora stylosa (Mangrove plant, Rhizophoraceae) | Mangrove forest, Hainan, China | [73] | |||
| C. cladosporioides MA-299 | Bruguiera gymnorrhiza (Mangrove plant, Rhizophoraceae) | Hainan Island, China | [39] | |||
| C. oxysporum HDN13-314 | Avicennia marina (Mangrove plant, Acanthaceae) | Hainan, China | [78] | |||
| Pandangolide 4 (97) | 486 | C24H38O8S | C. herbarum (Pers.) | Callyspongia aerizusa (Sponge, Callyspongiidae) | Bali Bata National Park, Indonesia | [74] |
| 5R-Hydroxyrecifeiolide (98) | 212 | C12H20O3 | C. cladosporioides MA-299 | Bruguiera gymnorrhiza (Mangrove plant, Rhizophoraceae) | Hainan Island, China | [40] |
| 5S-Hydroxyrecifeiolide (99) | 212 | C12H20O3 | C. cladosporioides MA-299 | Bruguiera gymnorrhiza (Mangrove plant, Rhizophoraceae) | Hainan Island, China | [40] |
| Methyl 2-(((4R,6R,12R)-6-hydroxy-12-methyl-2,5-dioxooxacyclodo decan-4-yl)thio)acetate (100) | 332 | C15H24O6S | Cladosporium sp. IFB3lp-2 | Rhizophora stylosa (Mangrove plant, Rhizophoraceae) | Mangrove forest, Hainan, China | [73] |
| Thiocladospolide A (101) | 346 | C16H26O6S | C. cladosporioides MA-299 | Bruguiera gymnorrhiza (Mangrove plant, Rhizophoraceae) | Hainan Island, China | [39] |
| C. oxysporum HDN13-314 | Avicennia marina (Mangrove plant, Acanthaceae) | Hainan, China | [78] | |||
| Thiocladospolide B (102) | 360 | C16H24O7S | C. cladosporioides MA-299 | Bruguiera gymnorrhiza (Mangrove plant, Rhizophoraceae) | Hainan Island, China | [39] |
| Thiocladospolide C (103) | 330 | C15H22O6S | C. cladosporioides MA-299 | Bruguiera gymnorrhiza (Mangrove plant, Rhizophoraceae) | Hainan Island, China | [39] |
| Thiocladospolide D (104) | 364 | C16H28O7S | C. cladosporioides MA-299 | Bruguiera gymnorrhiza (Mangrove plant, Rhizophoraceae) | Hainan Island, China | [39] |
| Thiocladospolide E (105) | 306 | C14H26O5S | Cladosporium sp. SCNU-F0001 | Mangrove plant | Zhuhai Mangrove Nature, Guangdong, China | [70] |
| Thiocladospolide F (106) | 332 | C16H28O5S | C. cladosporioides MA-299 | Bruguiera gymnorrhiza (Mangrove plant, Rhizophoraceae) | Hainan Island, China | [79] |
| Thiocladospolide F (107) | 386 | C24H38O8S | C. oxysporum HDN13-314 | Avicennia marina (Mangrove plant, Acanthaceae) | Hainan, China | [78] |
| Thiocladospolide G (108) | 348 | C16H28O6S | C. cladosporioides MA-299 | Bruguiera gymnorrhiza (Mangrove plant, Rhizophoraceae) | Hainan Island, China | [79] |
| Thiocladospolide G (109) | 348 | C15H24O7S | C. oxysporum HDN13-314 | Avicennia marina (Mangrove plant, Acanthaceae) | Hainan, China | [78] |
| Thiocladospolide H (110) | 332 | C15H24O6S | C. oxysporum HDN13-314 | Avicennia marina (Mangrove plant, Acanthaceae) | Hainan, China | [78] |
| Thiocladospolide I (111) | 560 | C27H44O10S | C. oxysporum HDN13-314 | Avicennia marina (Mangrove plant, Acanthaceae) | Hainan, China | [78] |
| Thiocladospolide J (112) | 558 | C27H42O10S | C. oxysporum HDN13-314 | Avicennia marina (Mangrove plant, Acanthaceae) | Hainan, China | [78] |
| Sporiolide A (113) | 348 | C19H24O6 | Cladosporium sp. L037 | Actinotrichia fragilis (Red alga, Galaxauraceae) | Seragaki Beach, Okinawa Island, Japan | [80] |
| Sporiolide B (114) | 258 | C13H22O5 | Cladosporium sp. L037 | Actinotrichia fragilis (Red alga, Galaxauraceae) | Seragaki Beach, Okinawa Island, Japan | [80] |
| (6R,12S)-6-Hydroxy-12-methyl-1-oxacyclododecane-2,5-dione (115) | 228 | C12H20O4 | Cladosporium sp. F14 | Seawater from the mangrove stand | Kei Ling Ha Lo Wai, Sai Kung, China | [60] |
| (3R,6S)-6-Hydroxy-12-methyl-2,5-dioxooxacyclododecan-3-yl (E)-4,11-dihydroxydodec-2-enoate (116) | 456 | C24H40O8 | Cladosporium sp. IFB3lp-2 | Rhizophora stylosa (Mangrove plant, Rhizophoraceae) | Mangrove forest, Hainan, China | [73] |
| Dendrodolide A (117) | 256 | C13H20O5 | Cladosporium sp. RA07-1 | Anthogorgia ochracea (Gorgonian, Acanthogorgiidae) | Weizhou coral reef, South China Sea | [69] |
| Dendrodolide C (118) | 242 | C12H18O5 | Cladosporium sp. RA07-1 | Anthogorgia ochracea (Gorgonian, Acanthogorgiidae) | Weizhou coral reef, South China Sea | [69] |
| Dendrodolide L (119) | 228 | C12H20O4 | Cladosporium sp. RA07-1 | Anthogorgia ochracea (Gorgonian, Acanthogorgiidae) | Weizhou coral reef, South China Sea | [69] |
| Dendrodolide M (120) | 256 | C13H20O5 | Cladosporium sp. RA07-1 | Anthogorgia ochracea (Gorgonian, Acanthogorgiidae) | Weizhou coral reef, South China Sea | [69] |
| Cladocladosin A (121) | 224 | C12H16O4 | C. cladosporioides MA-299 | Bruguiera gymnorrhiza (Mangrove plant, Rhizophoraceae) | Hainan Island, China | [79] |
| 5. Butenolides and butanolides | ||||||
| Cladospolide F (122) | 230 | C12H22O4 | Cladosporium sp. TZP29 | Unidentified soft coral | Guangzhou, China | [41] |
| Ent-cladospolide F (123) | 230 | C14H24O5 | C. cladosporioides MA-299 | Bruguiera gymnorrhiza (Mangrove plant, Rhizophoraceae) | Hainan Island, China | [40] |
| Cladospolide G (124) | 272 | C14H24O5 | C. cladosporioides MA-299 | Bruguiera gymnorrhiza (Mangrove plant, Rhizophoraceae) | Hainan Island, China | [40] |
| 11-Hydroxy-γ-dodecalactone (125) | 214 | C12H22O3 | Cladosporium sp. TZP29 | Unidentified soft coral | Guangzhou, China | [41] |
| Iso-Cladospolide B (126) | 228 | C12H20O4 | C. herbarum (Pers.) | Callyspongia aerizusa (Sponge, Callyspongiidae) | Bali Bata National Park, Indonesia, | [74] |
| Cladosporium sp. | Niphates rowi (Sponge, Niphatidae) | Gulf of Aqaba, Israel | [77] | |||
| Cladosporium sp. F14 | Seawater from the mangrove stand | Kei Ling Ha Lo Wai, Sai Kung, China | [60] | |||
| Cladosporium sp. IFB3lp-2 | Rhizophora stylosa (Mangrove plant, Rhizophoraceae) | Mangrove forest, Hainan, China | [73] | |||
| Cladosporium sp. RA07-1 | Anthogorgia ochracea (Gorgonian, Acanthogorgiidae) | Weizhou coral reef, South China Sea | [70] | |||
| Cladosporium sp. TZP29 | Unidentified soft coral | Guangzhou, China | [41] | |||
| C. cladosporioides MA-299 | Bruguiera gymnorrhiza (Mangrove plant, Rhizophoraceae) | Hainan Island, China | [40] | |||
| C. oxysporum HDN13-314 | Avicennia marina (Mangrove plant, Acanthaceae) | Hainan, China | [78] | |||
| Cladospolide H (127) | 210 | C12H18O3 | C. cladosporioides MA-299 | Bruguiera gymnorrhiza (Mangrove plant, Rhizophoraceae) | Hainan Island, China | [40] |
| 6. Seco-acids | ||||||
| Cladospolide A II (128) | Cladosporium sp. IFB3lp-2 | Rhizophora stylosa (Mangrove plant, Rhizophoraceae) | Mangrove forest, Hainan, China | [73] | ||
| Cladospolide E (129) | 228 | C12H20O4 | Cladosporium sp. TZP29 | Unidentified soft coral | Guangzhou, China | [41] |
| Seco-Patulolide A (130) | 228 | C12H20O4 | Cladosporium sp. TZP29 | Unidentified soft coral | Guangzhou, China | [41] |
| Seco-Patulolide C (131) | 230 | C12H22O4 | Cladosporium sp. F14 | Seawater from the Mangrove stand | Kei Ling Ha Lo Wai, Sai Kung, China | [60] |
| Cladosporium sp. TZP29 | Unidentified soft coral | Guangzhou, China | [41] | |||
| C. cladosporioides MA-299 | Bruguiera gymnorrhiza (Mangrove plant, Rhizophoraceae) | Hainan Island, China | [39] | |||
| Seco-Secopatulolide C (132) | 230 | C12H22O4 | C. oxysporum HDN13-314 | Avicennia marina (Mangrove plant, Acanthaceae) | Hainan, China | [78] |
| Cladosporester A (133) | 244 | C13H24O4 | C. cladosporioides OUCMDZ-187 | Rhizophora stylosa (Mangrove plant, Rhizophoraceae) | Shankou, Guangxi, China | [81] |
| Cladosporester B (134) | 244 | C13H24O4 | C. cladosporioides OUCMDZ-187 | Rhizophora stylosa (Mangrove plant, Rhizophoraceae) | Shankou, Guangxi, China | [81] |
| Cladosporacid A (135) | 230 | C12H22O4 | C. cladosporioides OUCMDZ-187 | Rhizophora stylosa (Mangrove plant, Rhizophoraceae) | Shankou, Guangxi, China | [81] |
| Cladosporacid B (136) | 230 | C12H22O4 | C. cladosporioides OUCMDZ-187 | Rhizophora stylosa (Mangrove plant, Rhizophoraceae) | Shankou, Guangxi, China | [81] |
| Cladosporacid D (137) | 228 | C12H20O4 | C. cladosporioides OUCMDZ-187 | Rhizophora stylosa (Mangrove plant, Rhizophoraceae) | Shankou, Guangxi, China | [81] |
| Cladosporester C (138) | 288 | C14H24O6 | C. cladosporioides OUCMDZ-187 | Rhizophora stylosa (Mangrove plant, Rhizophoraceae) | Shankou, Guangxi, China | [81] |
| Cladosporacid C (139) | 230 | C12H22O4 | C. cladosporioides OUCMDZ-187 | Rhizophora stylosa (Mangrove plant, Rhizophoraceae) | Shankou, Guangxi, China | [81] |
| Cladosporacid E (140) | 200 | C10H16O4 | C. cladosporioides OUCMDZ-187 | Rhizophora stylosa (Mangrove plant, Rhizophoraceae) | Shankou, Guangxi, China | [81] |
| 11-Hydroxy-4,5-dioxododecanoic acid (141) | 244 | C10H16O4 | Cladosporium sp. IFB3lp-2 | Rhizophora stylosa (Mangrove plant, Rhizophoraceae) | Mangrove forest, Hainan, China | [73] |
| 7. Tetralones (napthalenones) | ||||||
| Cladosporol/Cladosporol A (142) | 352 | C20H16O6 | Cladosporium sp. KcFL6′ | Kandelia candel (Mangrove plant, Rhizophoraceae) | Daya Bay, Shenzhen city, Guangdong, China | [82] |
| Cladosporol C (143) | 338 | C20H18O5 | Cladosporium sp. KcFL6′ | Kandelia candel (Mangrove plant, Rhizophoraceae) | Daya Bay, Shenzhen city, Guangdong, China | [82] |
| C. cladosporioides HDN14-342 | Marine sediment | Indian Ocean, Qingdao, China | [83] | |||
| C. cladosporioides EN-399 | Laurencia okamurai (Red alga, Rhodomelaceae) | Qingdao, China | [84] | |||
| Cladosporium sp. JS1-2 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | Dongzhaigang, Hainan, China | [71] | |||
| C. cladosporioides MCCC 3A00182 | Marine sediment | Southwest Pacific Ocean | [75] | |||
| Cladosporol D (144) | 354 | C20H18O6 | Cladosporium sp. KcFL6′ | Kandelia candel (Mangrove plant, Rhizophoraceae) | Daya Bay, Shenzhen city, Guangdong, China | [82] |
| Cladosporol E (145) | 370 | C20H18O7 | C. cladosporioides HDN14-342 | Marine sediment | Indian Ocean, Qingdao, China | [83] |
| Cladosporium sp. JS1-2 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | Dongzhaigang, Hainan, China | [71] | |||
| Cladosporol F (146) | 352 | C21H20O5 | C. cladosporioides HDN14-342 | Marine sediment | Indian Ocean, Qingdao, China | [83] |
| C. cladosporioides EN-399 | Laurencia okamurai (Red alga, Rhodomelaceae) | Qingdao, China | [84] | |||
| Cladosporol G (147) | 388 | C20H17ClO6 | C. cladosporioides HDN14-342 | Marine sediment | Indian Ocean, Qingdao, China | [83] |
| Cladosporol G (148) | 352 | C21H20O5 | C. cladosporioides EN-399 | Laurencia okamurai (Red alga, Rhodomelaceae) | Qingdao, China | [84] |
| Cladosporol H (149) | 336 | C20H16O5 | C. cladosporioides EN-399 | Laurencia okamurai (Red alga, Rhodomelaceae) | Qingdao, China | [84] |
| Cladosporol I = Cladosperanol A (150) | 338 | C20H18O5 | C. cladosporioides EN-399 | Laurencia okamurai (Rhodomelaceae) | Qingdao, China | [84] |
| Cladosporium sp. KFD33 | Blood cockle (Bivalve mollusk, Cardiidae) | Haikou Bay, China | [85] | |||
| 338 | C20H18O5 | C. perangustum FS62 | - | China | [86] | |
| Cladosporol J (151) | 338 | C20H18O5 | C. cladosporioides EN-399 | Laurencia okamurai (Red alga, Rhodomelaceae) | Qingdao, China | [84] |
| Cladosporone A (152) | 352 | C20H16O6 | Cladosporium sp. KcFL6′ | Kandelia candel (Mangrove plant, Rhizophoraceae) | Daya Bay, Shenzhen city, Guangdong, China | [82] |
| Altertoxin XII (153) | 322 | C20H18O4 | Cladosporium sp. KFD33 | Blood cockle (Bivalve mollusk, Cardiidae) | Haikou Bay, China | [85] |
| Clindanone A (154) | 394 | C22H18O7 | C. cladosporioides HDN14-342 | Marine sediment | Indian Ocean, Qingdao, China | [83] |
| Clindanone B (155) | 394 | C22H18O7 | C. cladosporioides HDN14-342 | Marine sediment | Indian Ocean, Qingdao, China | [83] |
| Isosclerone = (-)-(4R)-Regiolone (156) | 178 | C10H10O3 | C. perangustm FS62 | Marine sediment | South China Sea, china | [87] |
| 178 | C10H10O3 | C. cladosporioides HDN14-342 | Marine sediment | Indian Ocean, Qingdao, China | [83] | |
| 178 | C10H10O3 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | South China Sea, Dongzhaigang, Hainan, China | [88] | |
| (-)-trans-(3R,4R)-3,4,8-Trihydroxy-6,7-dimethyl-3,4- dihydronaphthalen-1(2H)-one (157) | 222 | C12H14O4 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | South China Sea, Dongzhaigang, Hainan, China | [88] |
| (3S)-3,8-Dihydroxy-6,7-dimethyl-α-tetralone (158) | 206 | C12H14O3 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | South China Sea, Dongzhaigang, Hainan, China | [88] |
| (3R,4R)-3,4-Dihydro-3,4,8-trihydroxy-1(2H)-napthalenone (159) | 194 | C10H10O4 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | South China Sea, Dongzhaigang, Hainan, China | [88] |
| Cladosporium sp. HDN17-58 | Deep-sea sediment | Western Pacific Ocean, China | [89] | |||
| Aladothalen (160) | 194 | C10H10O4 | Cladosporium sp. HDN17-58 | Deep-sea sediment | Western Pacific Ocean, China | [89] |
| 8. Perylenequinones | ||||||
| Altertoxin VIII (161) | 304 | C20H16O3 | Cladosporium sp. KFD33 | Blood cockle (Bivalve mollusk, Cardiidae) | Haikou Bay, Hainan, China | [85] |
| Altertoxin IX (162) | 290 | C20H18O2 | Cladosporium sp. KFD33 | Blood cockle (Bivalve mollusk, Cardiidae) | Haikou Bay, China | [85] |
| Altertoxin X (163) | 290 | C20H18O2 | Cladosporium sp. KFD33 | Blood cockle (Bivalve mollusk, Cardiidae) | Haikou Bay, China | [85] |
| Altertoxin XI (164) | 304 | C21H20O2 | Cladosporium sp. KFD33 | Blood cockle (Bivalve mollusk, Cardiidae) | Haikou Bay, China | [85] |
| 9. Naphthalene derivatives | ||||||
| 8-Methoxynaphthalen-1-ol (165) | 174 | C11H10O2 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | South China Sea, China | [90] |
| 1,8-Dimethoxynaphthalene (166) | 188 | C12H12O2 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | South China Sea, Dongzhaigang, Hainan, China | [88] |
| Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | South China Sea, China | [90] | |||
| Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | South China Sea, China | [91] | |||
| 4-Methoxynaphthalene-1,5-diol (167) | 190 | C11H10O3 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | South China Sea, China | [91] |
| 8-Methoxynaphthalene-1,7-diol (168) | 190 | C11H10O3 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | South China Sea, China | [91] |
| Cladonaphchrom A (169) | 350 | C22H22O4 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | South China Sea, China | [90] |
| Cladonaphchrom B (170) | 350 | C22H22O4 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | South China Sea, China | [90] |
| 10. Xanthones | ||||||
| 8-Hydroxy-6-methylxanthone-1-carboxylic acid (171) | 270 | C15H10O5 | C. halotolerans GXIMD 02502 | Porites lutea (Stony coral, Poritidae) | Weizhou Islands coral reef, Guangxi Zhuang autonomous region, China | [92] |
| Methyl 8-hydroxy-6-methyl-9- oxo-9H-xanthene-1-carboxylate (172) | 284 | C16H12O5 | C. halotolerans GXIMD 02502 | Porites lutea (Stony coral, Poritidae) | Weizhou Islands coral reef, Guangxi Zhuang autonomous region, China | [92] |
| Methyl 8-hydroxy-6- (hydroxymethyl)-9-oxo-9H-xanthene-1-carboxylate (173) | 300 | C16H12O6 | C. halotolerans GXIMD 02502 | Porites lutea (Stony coral, Poritidae) | Weizhou Islands coral reef, Guangxi Zhuang autonomous region, China | [92] |
| Vertixanthone (174) | 270 | C15H10O5 | C. halotolerans GXIMD 02502 | Porites lutea (Stony coral, Poritidae) | Weizhou Islands coral reef, Guangxi Zhuang autonomous region, China | [92] |
| 8-(Methoxycarbonyl)-1-hydroxy-9-oxo-9H-xanthene-3-carboxylic acid (175) | 314 | C16H10O7 | C. halotolerans GXIMD 02502 | Porites lutea (Stony coral, Poritidae) | Weizhou Islands coral reef, Guangxi Zhuang autonomous region, China | [92] |
| 3,8-Dihydroxy-6-methyl-9-oxo-9H-xanthene-1-Carboxylate (176) | 300 | C16H12O6 | C. halotolerans GXIMD 02502 | Porites lutea (Stony coral, Poritidae) | Weizhou Islands coral reef, Guangxi Zhuang autonomous region, China | [92] |
| Conioxanthone A (177) | 316 | C16H12O7 | C. halotolerans GXIMD 02502 | Porites lutea (Stony coral, Poritidae) | Weizhou Islands coral reef, Guangxi Zhuang autonomous region, China | [92] |
| 11. Tropolones | ||||||
| Malettinin A (178) | 288 | C16H16O5 | Cladosporium sp. KF501 | Water sample | German Wadden Sea | [93] |
| Malettinin B (179) | 292 | C16H20O5 | Cladosporium sp. KF501 | Water sample | German Wadden Sea | [93] |
| Malettinin C (180) | 292 | C16H20O5 | Cladosporium sp. KF501 | Water sample | German Wadden Sea | [93] |
| Malettinin E (181) | 292 | C16H20O5 | Cladosporium sp. KF501 | Water samples | German Wadden Sea | [93] |
| 12. Binaphthopyrones | ||||||
| Cladosporinone (182) | 650 | C33H30O14 | C. cladosporioides | Sediment of a hypersaline lake El Hamra | Wadi el Natrun, Egypt | [94] |
| Viriditoxin (183) | 662 | C34H30O14 | C. cladosporioides | Sediment of a hypersaline lake El Hamra | Wadi el Natrun, Egypt | [94] |
| Viriditoxin derivative 1 (184) | 646 | C34H30O13 | C. cladosporioides | Sediment of a hypersaline lake El Hamra | Wadi el Natrun, Egypt | [94] |
| Viriditoxin derivative 2 (185) | 646 | C34H30O13 | C. cladosporioides | Sediment of a hypersaline lake El Hamra | Wadi el Natrun, Egypt | [94] |
| 13. Benzopyranes, benzopyrones, and pyrones | ||||||
| (2S)-5-Hydroxy-2-methyl-chroman-4-one (186) | 178 | C10H10O3 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | South China Sea, Dongzhaigang, Hainan, China | [88] |
| (R)-5-Hydroxy-2-methylchroman-4-one (187) | 178 | C10H10O3 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | South China Sea, China | [90] |
| Cladosporium sp. OUCMDZ-302 | Excoecaria agallocha (Mangrove plant, Euphorbiaceae) | Wenchang, Hainan, China | [95] | |||
| (2R)-7-O-α-D-Ribofuranosyl-5-hydroxy-2-methyl chroman-4-one (188) | 326 | C15H18O8 | Cladosporium sp. OUCMDZ-302 | Excoecaria agallocha (Mangrove plant, Euphorbiaceae) | Wenchang, Hainan, China | [95] |
| Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | South China Sea, China | [91] | |||
| (2S)-7-O-α-D-Ribofuranosyl-5-hydroxy-2-methylchroman-4-one (189) | 326 | C15H108O8 | Cladosporium sp. OUCMDZ-302 | Excoecaria agallocha (Mangrove plant, Euphorbiaceae) | Wenchang, Hainan, China | [95] |
| (±)-5,7-Dihydroxy-2-methyl chroman-4-one (190) | 194 | C10H10O4 | Cladosporium sp. OUCMDZ-302 | Excoecaria agallocha (Mangrove plant, Euphorbiaceae) | Wenchang, Hainan, China | [95] |
| 5-Hydroxy-2-methyl-4H-chromen-4-one (191) | 176 | C10H8O3 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | South China Sea, China | [90] |
| Clapone (192) | 216 | C13H12O3 | Cladosporium sp. HNWSW-1 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | Dong Zhai Gang Mangrove, Hainan, China | [67] |
| 7-O-α-D-Ribosyl-5-hydroxy-2-propylchromone (193) | 352 | C17H20O8 | Cladosporium sp. OUCMDZ-302 | Excoecaria agallocha (Mangrove plant, Euphorbiaceae) | Wenchang, Hainan, China | [95] |
| Coniochaetone A (194) | 230 | C13H10O4 | C. halotolerans GXIMD 02502 | Porites lutea (Stony coral, Poritidae) | Weizhou Islands coral reef, Guangxi Zhuang autonomous region, China | [92] |
| Coniochaetone B (195) | 232 | C13H12O4 | C. halotolerans GXIMD 02502 | Porites lutea (Stony coral, Poritidae) | Weizhou Islands coral reef, Guangxi Zhuang autonomous region, China | [92] |
| Coniochaetone K (196) | 262 | C13H10O6 | C. halotolerans GXIMD 02502 | Porites lutea (Stony coral, Poritidae) | Weizhou Islands coral reef, Guangxi Zhuang autonomous region, China | [92] |
| α-Diversonolic ester (197) | 320 | C16H16O7 | C. halotolerans GXIMD 02502 | Porites lutea (Poritidae) | Weizhou Islands coral reef, Guangxi Zhuang autonomous region, China | [92] |
| β-Diversonolic ester (198) | 320 | C16H16O7 | C. halotolerans GXIMD 02502 | Porites lutea (Stony coral, Poritidae) | Weizhou Islands coral reef, Guangxi Zhuang autonomous region, China | [92] |
| Secalonic acid D (199) | 638 | C32H30O14 | Cladosporium sp. JS1-2 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | Dongzhaigang, Hainan, China | [71] |
| (2S,3S,4R)-2-Methylchroman-3,4,5-triol (200) | 196 | C10H12O4 | Cladosporium sp. OUCMDZ-302 | Excoecaria agallocha (Mangrove plant, Euphorbiaceae) | Wenchang, Hainan, China | [95] |
| (2S,4S)-4-Methoxy-2-methylchroman-5-ol (201) | 194 | C11H14O3 | Cladosporium sp. OUCMDZ-302 | Excoecaria agallocha (Mangrove plant, Euphorbiaceae) | Wenchang, Hainan, China | [95] |
| (2R,4R)-3,4-Dihydro-4-methoxy-2-methyl-2H-1-benzopyran-5-ol (202) | 194 | C11H14O3 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | South China Sea, China | [91] |
| (2S,4S)-2-methylchroman-4,5-diol (203) | 180 | C10H12O3 | Cladosporium sp. OUCMDZ-302 | Excoecaria agallocha (Mangrove plant, Euphorbiaceae) | Wenchang, Hainan, China | [95] |
| (2R,4S)-2,3-Dihydro-2-methyl-benzopyran-4,5-diol (204) | 180 | C10H12O3 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | South China Sea, China | [91] |
| (2R*,4R*)-3,4-Dihydro-5-methoxy-2-methyl-1(2H)-benzopyran-4-ol (205) | 164 | C10H12O2 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | South China Sea, Dongzhaigang, Hainan, China | [88] |
| Citrinin H1 (206) | 428 | C24H28O7 | Cladosporium sp. JS1-2 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | Dongzhaigang, Hainan, China | [71] |
| Cladosporin C (207) | 248 | C14H16O4 | C. cladosporioides SCSIO z015 | Deep sea sediment | Okinawa, Japan | [36] |
| (S)-5-Hydroxy-4-methylchroman-2-one (208) | 178 | C10H10O3 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | South China Sea, China | [91] |
| (3R)-3-(2-Hydroxypropyl)-6,8-dihydroxy-3,4-dihydroiso-coumarin (209) | 238 | C12H14O5 | Cladosporium sp. CSIO41007 | Callyspongia sp. (Sponge, Callyspongiidae) | Xuwen, Guangdong, China | [61] |
| Phomasatin (210) | 208 | C10H8O5 | C. cladosporioides MCCC 3A00182 | Marine sediment | Southwest Pacific Ocean | [75] |
| 14. Pyrone derivatives | ||||||
| Herbarin A (211) | 236 | C12H12O5 | C. herbarum (Pers.) | Aplysina aerophoba (Sponge, Aplysinidae) | Bali Bata National Park, Indonesia | [96] |
| Callyspongia aerizusa (Sponge, Callyspongiidae) | Bali Bata National Park, Indonesia | [96] | ||||
| Herbarin B (212) | 210 | C10H10O5 | C. herbarum (Pers.) | Aplysina aerophoba (Sponge, Aplysinidae) | Bali Bata National Park, Indonesia | [96] |
| Callyspongia aerizusa (Sponge, Callyspongiidae) | Bali Bata National Park, Indonesia | [96] | ||||
| Citreoviridin A (213) | 402 | C23H30O6 | C. herbarum (Pers.) | Aplysina aerophoba (Sponge, Aplysinidae) | Bali Bata National Park, Indonesia | [96] |
| Callyspongia aerizusa (Sponge, Callyspongiidae) | Bali Bata National Park, Indonesia | [96] | ||||
| Vermistatin (214) | 328 | C18H16O6 | Cladosporium sp. JS1-2 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | Dongzhaigang, Hainan, China | [71] |
| 15. Lactones, cyclohexene, and azaphilone derivatives | ||||||
| (R)-Mevalonolactone (215) | 130 | C8H10O3 | Cladosporium sp. EF424419 | Porphyra yezoensis (Red alga, Bangiaceae) | Lianyungang, Jiangsu, China | [59] |
| Cladosporactone A (216) | 196 | C10H12O4 | C. cladosporioides MCCC 3A00182 | Marine Sediment | Southwest Pacific Ocean | [75] |
| Helicascolide A (217) | 212 | C12H20O3 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | South China Sea, China | [91] |
| Cladoscyclitol A (218) | 244 | C12H20O5 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | Dongzhaigang of Hainan Province, China | [97] |
| Cladoscyclitol B (219) | 290 | C13H22O7 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | Dongzhaigang of Hainan Province, China | [97] |
| Cladoscyclitol C (220) | 230 | C12H22O4 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | Dongzhaigang of Hainan Province, China | [97] |
| Cladoscyclitol D (221) | 246 | C12H22O5 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | Dongzhaigang of Hainan Province, China | [97] |
| 2-Butyryl-3,5-dihydroxycyclohex-2-enone (222) | 198 | C10H14O4 | Cladosporium sp. OUCMDZ-302 | Excoecaria agallocha (Mangrove plant, Euphorbiaceae) | Wenchang, Hainan, China | [95] |
| Perangustol A (223) | 210 | C11H14O4 | C. perangustm FS62 | Marine sediment | South China Sea, China | [87] |
| Perangustol B (224) | 210 | C11H14O4 | C. perangustm FS62 | Marine sediment | South China Sea, China | [87] |
| Bicyclic diol (225) | 210 | C11H14O4 | C. perangustm FS62 | Marine sediment | South China Sea, China | [87] |
| 16. Phenolics and other aromatic compounds | ||||||
| 3-Phenyl-propionic acid (226) | 210 | C11H14O4 | Cladosporium sp. JJM22 | Ceriops tagal (Rhizophoraceae) | South China Sea, China | [91] |
| P-Toluic acid (227) | 136 | C8H8O2 | C. cladosporioides | Marine sponge | Argentina | [98] |
| L-β-Phenyllactic acid (228) | 166 | C9H10O3 | Cladosporium sp. EF424419 | Porphyra yezoensis (Red alga, Bangiaceae) | Lianyungang, Jiangsu, China | [59] |
| α-Resorcylic acid (229) | 154 | C7H6O4 | Cladosporium sp. EF424419 | Porphyra yezoensis (Red alga, Bangiaceae) | Lianyungang, Jiangsu, China | [59] |
| Phenylacetic acid (230) | 136 | C8H8O2 | Cladosporium sp. EF424419 | Porphyra yezoensis (Red alga, Bangiaceae) | Lianyungang, Jiangsu, China | [59] |
| P-Hydroxyphenylacetic acid (231) | 152 | C8H8O3 | Cladosporium sp. EF424419 | Porphyra yezoensis (Red alga, Bangiaceae) | Lianyungang, Jiangsu, China | [59] |
| Cinnamic acid (3-Phenyl-2-propenoic acid) (232) | 148 | C9H8O2 | Cladosporium sp. F14 | Seawater from the mangrove stand | Kei Ling Ha Lo Wai, Sai Kung, China | [60] |
| 3-(2,3-Dihydroxy phenoxy) butanoic acid (233) | 212 | C10H12O5 | Cladosporium sp. OUCMDZ-302 | Excoecaria agallocha (Mangrove plant, Euphorbiaceae) | Wenchang, Hainan, China | [95] |
| P-Hydroxy benzoic acid methyl ester (234) | 152 | C8H8O3 | Cladosporium sp. EF424419 | Porphyra yezoensis (Red alga, Bangiaceae) | Lianyungang, Jiangsu, China | [59] |
| Methyl (3S)-3-(2,3-dihydroxy phenyloxy)butanoate (235) | 226 | C11H14O5 | Cladosporium sp. OUCMDZ-302 | Excoecaria agallocha (Mangrove plant, Euphorbiaceae) | Wenchang, Hainan, China | [95] |
| P-Hydroxyphenylethyl alcohol (236) | 138 | C8H10O2 | Cladosporium sp. EF424419 | Porphyra yezoensis (Red alga, Bangiaceae) | Lianyungang, Jiangsu Province, China | [59] |
| P-Hydroxybenzyl alcohol (237) | 142 | C7H8O2 | Cladosporium sp. EF424419 | Porphyra yezoensis (Red alga, Bangiaceae) | Lianyungang, Jiangsu Province, China | [59] |
| 2-Phenylethanol (238) | 122 | C8H10O | Cladosporium sp. F14 | Seawater from the mangrove stand | Kei Ling Ha Lo Wai, Sai Kung, China | [60] |
| 4-O-α-D-Ribofuranose-3-hydroxymethyl-2-pentyl- phenol (239) | 342 | C17H26O7 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | South China Sea, Dongzhaigang, Hainan, China | [88] |
| 4-O-α-D-Ribofuranose-2-pentyl-3-phemethylol (240) | 326 | C17H26O6 | Cladosporium sp. JJM22 | Ceriops tagal (Mangrove plant, Rhizophoraceae) | Dongzhaigang of Hainan Province, China | [97] |
| Clavatol (241) | 180 | C10H12O3 | Cladosporium sp.MFC353-b | Chondria crassicualis (Red alga, Rhodomelaceae) | Yokji Island, Kyeongnam, Korea | [65] |
| 1-(3,5-Dihydroxy-4-methylphenyl)propan-2-one (242) | 180 | C10H12O3 | C. perangustm FS62 | Marine sediment | South China Sea, china | [87] |
| α-Acetylorcinol (243) | 166 | C9H10O3 | C. perangustm FS62 | Marine sediment | South China Sea, china | [87] |
| 1-(2,6-Dihydroxyphenyl) ethanone (244) | 152 | C8H8O3 | Cladosporium sp. OUCMDZ-302 | Excoecaria agallocha (Mangrove plant, Euphorbiaceae) | Wenchang, Hainan, China | [95] |
| 1-(2,6-Hihydroxyphenyl)-1-butanone (245) | 180 | C10H12O3 | Cladosporium sp. OUCMDZ-302 | Excoecaria agallocha (Mangrove plant, Euphorbiaceae) | Wenchang, Hainan, China | [95] |
| (R)-3-Methoxyl-1-(2,6-dihydroxyphenyl)-butan-1-one (246) | 210 | C11H14O4 | Cladosporium sp. JJM22 | Ceriops tagal (Rhizophoraceae) | South China Sea, China | [91] |
| Cladosporin D (247) | 224 | C12H16O4 | C. cladosporioides SCSIO z015 | Deep sea sediment | Okinawa, Japan | [36] |
| (2S)-7,4′-dihydroxy-5-methoxy-8-(γ,γ-dimethylallyl)-flavanone (248) | 354 | C21H22O5 | Cladosporium sp. TPU1507 | Unidentified marine sponge | Manado, Indonesia | [68] |
| Bis(2-Ethylhexyl)phthalate (249) | 390 | C24H38O4 | Cladosporium sp. F14 | Seawater from the mangrove stand | Kei Ling Ha Lo Wai, Sai Kung, China | [60] |
| Herbaric acid (250) | 196 | C9H8O5 | C. herbarum (Pers.) | Callyspongia aerizusa (Sponge, Callyspongiidae) | Bali Bata National Park, Indonesia | [96] |
| Cladosacid (251) | 250 | C15H22O3 | Cladosporium sp. OUCMDZ-1635 | Unidentified sponge | Xisha Islands, China | [56] |
| 1,1′-Dioxine-2,2′-dipropionic acid (252) | 228 | C10H12O6 | Cladosporium sp. JS1-2 | Ceriops tagal (Mangrove, plant, Rhizophoraceae) | Dongzhaigang, Hainan, China | [71] |
| Sumiki’s acid (253) | 142 | C6H6O4 | C. herbarum (Pers.) | Callyspongia aerizusa (Sponge, Callyspongiidae) | Bali Bata National Park, Indonesia | [73] |
| Acetyl Sumiki’s acid (254) | 184 | C8H8O5 | C. herbarum (Pers.) | Callyspongia aerizusa (Sponge, Callyspongiidae) | Bali Bata National Park, Indonesia | [74] |
| 17. Sterols and terpenes | ||||||
| 5α,8α-Epidioxy-24(R)-methyl-cholesta-6,22-diene-3-β-ol (255) | 428 | C28H44O3 | C. sphaerospermum Penz | Ceramium condi (Red alga, Ceramiaceae) | Ussuriysk Bay, Japan | [99] |
| C.cladosporioides MCCC 3A00182 | Marine sediment | Southwest Pacific Ocean | [75] | |||
| 5α,8α-Epidioxy-ergosta-6,22E-dien-3β-ol (256) | 428 | C28H44O3 | Cladosporium sp. WZ-2008-0042 | Dichotella gemmacea (Gorgonian, Ellisellidae) | Weizhou Island coral reef, South China Sea | [100] |
| C. cladosporioides MCCC 3A00182 | Marine Sediment | Southwest Pacific Ocean | [75] | |||
| 5α,8α-Epidioxy-24(R)-methyl-cholesta-6,9(11),22-triene-3-β-ol (257) | 426 | C28H42O3 | C. sphaerospermum Penz | Ceramium condi (Red alga, Ceramiaceae) | Ussuriysk Bay, Japan | [99] |
| 5α,8α-Epidioxy-ergosta-6,9,22E-triene-3β-ol (258) | 426 | C28H42O3 | Cladosporium sp. WZ-2008-0042 | Dichotella gemmacea (Gorgonian, Ellisellidae) | Weizhou Island coral reef, South China Sea | [100] |
| 3β,5α,6β- Trihydroxyergosta-7,22-diene = Cerevisterol (259) | 430 | C28H46O3 | Cladosporium sp. SCSIO41007 | Callyspongia sp. (Sponge, Callyspongiidae) | Xuwen, Guangdong, China | [61] |
| Ergosta-7,22E-diene-3β,5α,6β-triol (260) | 430 | C28H46O3 | Cladosporium sp. WZ-2008-0042 | Dichotella gemmacea (Gorgonian, Ellisellidae) | Weizhou Island coral reef, South China Sea | [100] |
| 3β,5α,6α-Trihydroxy-(22E,24R) -ergosta-7,22-diene (261) | 430 | C28H46O3 | C. cladosporioides MCCC 3A00182 | Marine Sediment | Southwest Pacific Ocean | [75] |
| 3β,5α-Dihydroxy-6β-methoxyergosta-7,22-diene (262) | 444 | C29H48O3 | Cladosporium sp. WZ-2008-0042 | Dichotella gemmacea (Gorgonian, Ellisellidae) | Weizhou Island coral reef, South China Sea | [100] |
| Ergosterol (263) | 396 | C28H44O | Cladosporium sp. WZ-2008-0042 | Dichotella gemmacea (Gorgonian, Ellisellidae) | Weizhou Island coral reef, South China Sea | [100] |
| Cladosporisteroid A (264) | 460 | C28H44O5 | Cladosporium sp. SCSIO41007 | Callyspongia sp. (Sponge, Callyspongiidae) | Xuwen, Guangdong, China | [61] |
| 3β,5α,9α-Trihydroxy-(22E,24R)-ergosta-7,22-diene-6-one (265) | 444 | C28H44O4 | Cladosporium sp. SCSIO41007 | Callyspongia sp. (Sponge, Callyspongiidae) | Xuwen, Guangdong, China | [61] |
| C. cladosporioides MCCC 3A00182 | Marine Sediment | Southwest Pacific Ocean | [75] | |||
| 3β,5α-Dihydroxy-(22E,24R)-ergosta-7,22-diene-6-one (266) | 428 | C28H44O3 | C. cladosporioides MCCC 3A00182 | Marine Sediment | Southwest Pacific Ocean | [75] |
| Stigma-5-en-3-O-β-glucopyranoside (267) | 576 | C35H60O6 | Cladosporium sp. WZ-2008-0042 | Dichotella gemmacea (Gorgonian, Ellisellidae) | Weizhou Island coral reef, South China Sea | [100] |
| 3α-Hydroxy-pregna-7-ene-6,20-dione = Cladosporisteroid B (268) | 330 | C21H30O3 | Cladosporium sp. WZ-2008-0042 | Dichotella gemmacea (Gorgonian, Ellisellidae) | Weizhou Island coral reef, South China Sea | [100] |
| Cladosporium sp. SCSIO41007 | Callyspongia sp. (Sponge, Callyspongiidae) | Xuwen, Guangdong, China | [61] | |||
| C. cladosporioides MCCC 3A00182 | Marine Sediment | Southwest Pacific Ocean | [75] | |||
| C. sphaerospermum SW67 | Hydractinia echinata (Hydroid, Hydractiniidae) | South Korea | [101] | |||
| Cladosporisteroid C (269) | 374 | C23H34O4 | Cladosporium sp. SCSIO41007 | Callyspongia sp. (Sponge, Callyspongiidae) | Xuwen, Guangdong, China | [61] |
| Pregn-7-dien-3,6,20-trione (270) | 328 | C21H28O3 | Cladosporium sp. SCSIO41007 | Callyspongia sp. (Sponge, Callyspongiidae) | Xuwen, Guangdong, China | [61] |
| 18. Alcohols and aldehydes | 70.43 μg/mL (EC50) | |||||
| Compound (271) | 434 | C30H58O | Cladosporium sp. | Marine sediment | San Antonio Oeste, Río Negro, Argentina | [102] |
| Compound (272) | 458 | C32H58O | Cladosporium sp. | Marine sediment | San Antonio Oeste, Río Negro, Argentina | [102] |
| Compound (273) | 458 | C32H58O | Cladosporium sp. | Marine sediment | San Antonio Oeste, Río Negro, Argentina | [102] |
| Compound (274) | 458 | C32H58O | Cladosporium sp. | Marine sediment | San Antonio Oeste, Río Negro, Argentina | [102] |
| Compound (275) | 460 | C32H60O | Cladosporium sp. | Marine sediment | San Antonio Oeste, Río Negro, Argentina | [102] |
| Compound (276) | 460 | C32H60O | Cladosporium sp. | Marine sediment | San Antonio Oeste, Río Negro, Argentina | [102] |
| Compound (277) | 462 | C32H62O | Cladosporium sp. | Marine sediment | San Antonio Oeste, Río Negro, Argentina | [102] |
| Compound (278) | 462 | C32H62O | Cladosporium sp. | Marine sediment | San Antonio Oeste, Río Negro, Argentina | [102] |
| Compound (279) | 482 | C34H58O | Cladosporium sp. | Marine sediment | San Antonio Oeste, Río Negro, Argentina | [102] |
| Compound (280) | 484 | C34H60O | Cladosporium sp. | Marine sediment | San Antonio Oeste, Río Negro, Argentina | [102] |
| Compound (281) | 484 | C34H60O | Cladosporium sp. | Marine sediment | San Antonio Oeste, Río Negro, Argentina | [102] |
| Compound (282) | 484 | C34H60O | Cladosporium sp. | Marine sediment | San Antonio Oeste, Río Negro, Argentina | [102] |
| Compound (283) | 484 | C34H60O | Cladosporium sp. | Marine sediment | San Antonio Oeste, Río Negro, Argentina | [102] |
| Compound (284) | 486 | C34H62O | Cladosporium sp. | Marine sediment | San Antonio Oeste, Río Negro, Argentina | [102] |
| (2S,3S,4E)-Hepta-4,6-diene-2,3-diol (285) | 128 | C7H12O2 | Cladosporium sp. OUCMDZ-302 | Excoecaria agallocha (Mangrove plant, Euphorbiaceae) | Wenchang, Hainan, China | [95] |
| (3E,8E,6S)-Undeca-3,8,10-trien-1,6-diol (286) | 182 | C11H18O2 | Cladosporium sp. OUCMDZ-302 | Excoecaria agallocha (Mangrove plant, Euphorbiaceae) | Wenchang, Hainan, China | [95] |
| Compound Name | Biological Activity | Assay, Organism, or Cell Line | Biological Results | Positive Control | Ref. |
|---|---|---|---|---|---|
| Cladosin C (3) | Antiviral | Neuraminidase inhibition assay/Influenza A H1N1 virus | 276.0 µM (IC50) | Ribavirin 131.0 µM (IC50) | [42] |
| Cladosin I (8) | Cytotoxicity | MTT/K562 | 4.1 µM (IC50) | Doxorubicin 0.3 µM (IC50) | [55] |
| Cytotoxicity | MTT/HL-60 | 2.8 µM (IC50) | Doxorubicin 0.2 µM (IC50) | [55] | |
| Cytotoxicity | SEB/HCT-116 | 11.0 µM (IC50) | Doxorubicin 0.2 µM (IC50) | [55] | |
| Cytotoxicity | SRB/PC-3 | 13.0 µM (IC50) | Doxorubicin 1.0 µM (IC50) | [55] | |
| Cytotoxicity | SRB/SH-SY5Y | 12.0 µM (IC50) | Doxorubicin 0.1 µM (IC50) | [55] | |
| Cytotoxicity | SRB/MGC-803 | 19.0 µM (IC50) | Doxorubicin 0.2 µM (IC50) | [55] | |
| Cladosin K (10) | Cytotoxicity | MTT/K562 | 5.9 µM (IC50) | Doxorubicin 0.3 µM (IC50) | [55] |
| Cytotoxicity | MTT/HL-60 | 7.5 µM (IC50) | Doxorubicin 0.2 µM (IC50) | [55] | |
| Cytotoxicity | SEB/HCT-116 | 14.0 µM (IC50) | Doxorubicin 0.2 µM (IC50) | [55] | |
| Cytotoxicity | SRB/PC-3 | 18.0 µM (IC50) | Doxorubicin 1.0 µM (IC50) | [55] | |
| Cladosporicin A (12) | Cytotoxicity | SRB/Bt549 | 70.88 µM (IC50) | Etoposide 1.82 µM (IC50) | [38] |
| Cytotoxicity | SRB/HCC70 | 74.48 µM (IC50) | Etoposide 1.76 µM (IC50) | [38] | |
| Cytotoxicity | SRB/MDA-MB-231 | 75.54 µM (IC50) | Etoposide 2.27 µM (IC50) | [38] | |
| Cytotoxicity | SRB/MDA-MB-468 | 79.36 µM (IC50) | Etoposide 2.08 µM (IC50) | [38] | |
| Cladodionen (13) | Cytotoxicity | MTT/K562 | 4.5 µM (IC50) | Doxorubicin 0.3 µM (IC50) | [55] |
| Cytotoxicity | MTT/HL-60 | 6.6 µM (IC50) | Doxorubicin 0.2 µM (IC50) | [55] | |
| Cytotoxicity | SRB/HCT-116 | 12.0 µM (IC50) | Doxorubicin 0.2 µM (IC50) | [55] | |
| Cytotoxicity | SRB/PC-3 | 11.0 µM (IC50) | Doxorubicin 1.0 µM (IC50) | [55] | |
| Cytotoxicity | SRB/SH-SY5Y | 15.0 µM (IC50) | Doxorubicin 0.1 µM (IC50) | [55] | |
| Cytotoxicity | SRB/MGC-803 | 22.0 µM (IC50) | Doxorubicin 0.2 µM (IC50) | [55] | |
| Cytotoxicity | MTT/MCF-7 | 18.7 µM (IC50) | Adriamycin 0.67 µM (IC50) | [56] | |
| Cytotoxicity | MTT/HeLa | 19.1 µM (IC50) | Adriamycin 0.32 µM (IC50) | [56] | |
| Cytotoxicity | CCK-8/HCT-116 | 17.9 µM (IC50) | Adriamycin 0.21 µM (IC50) | [56] | |
| Cytotoxicity | MTT/HL-60 | 9.0 µM (IC50) | Adriamycin 0.02 µM (IC50) | [56] | |
| Cladosporiumin I (29) | Cytotoxicity | SRB/Bt549 | 76.18 µM (IC50) | Etoposide 1.82 µM (IC50) | [38] |
| Cytotoxicity | SRB/HCC70 | 85.29 µM (IC50) | Etoposide 1.76 µM (IC50) | [38] | |
| Cytotoxicity | SRB/MDA-MB-231 | 82.37 µM (IC50) | Etoposide 2.27 µM (IC50) | [38] | |
| Cytotoxicity | SRB/MDA-MB-468 | 81.44 µM (IC50) | Etoposide 2.08 µM (IC50) | [38] | |
| Cladosporiumin J (30) | Cytotoxicity | SRB/Bt549 | 78.96 µM (IC50) | Etoposide 1.82 µM (IC50) | [38] |
| Cytotoxicity | SRB/HCC70 | 76.41 µM (IC50) | Etoposide 1.76 µM (IC50) | [38] | |
| Cytotoxicity | SRB/MDA-MB-231 | 79.27 µM (IC50) | Etoposide 2.27 µM (IC50) | [38] | |
| Cytotoxicity | SRB/MDA-MB-468 | 74.64 µM (IC50) | Etoposide 2.08 µM (IC50) | [38] | |
| Cyclo-(Val-Pro) (32) | Insecticidal | Inhibitinon 50%/B. amphitrite | 37.82 μg/mL (EC50) | FSW with DMSO | [60] |
| Lethality 50%/B. amphitrite | >200 μg/mL (LC50) | FSW with DMSO | [60] | ||
| Inhibitinon 50%/B. neritina | >200 μg/mL (EC50) | FSW with DMSO | [60] | ||
| Lethality 50%/B. neritina | >200 μg/mL (LC50) | FSW with DMSO | [60] | ||
| Cyclo-(Val-Pro) (32) | Antimicrobial | Serial dilution/L. hongkongensis | 80 μg/mL (MIC) | Streptomycin 250 μg/mL (MIC) Penicillin 0.25 μg/mL (MIC) | [60] |
| Cyclo-(Phe-Pro) (33) | Insecticidal | Inhibitinon 50%/B. amphitrite | 68.57 μg/mL (EC50) | FSW with DMSO | [60] |
| Lethality 50%/B. amphitrite | 115.04 μg/mL (LC50) | FSW with DMSO | [60] | ||
| Inhibitinon 50%/B. neritina | 70.43 μg/mL (EC50) | FSW with DMSO | [60] | ||
| Lethality 50%/B. neritina | >200 μg/mL (LC50) | FSW with DMSO | [60] | ||
| Cyclo-(Phe-Pro) (33) | Antimicrobial | Serial dilution/L. hongkongensis | 200 μg/mL (MIC) | Streptomycin 1.0 250 μg/mL (MIC) | [60] |
| Serial dilution/M. luteus | 200 μg/mL (MIC) | Streptomycin 250 μg/mL (MIC) Penicillin 0.5 μg/mL (MIC) | [60] | ||
| Serial dilution/Ruegeria sp. | 100 μg/mL (MIC) | Streptomycin 500 μg/mL (MIC) Penicillin 0.25 μg/mL (MIC) | [60] | ||
| Glyantrypine (42) | Antiviral | CPE inhibition assay/Influenza A H1N1 virus | 150 µM (IC50) | Ribavirin 87.0 µM (IC50) | [64] |
| 3-Hydroxyglyantrypine (43) | Antiviral | CPE inhibition assay/Influenza A H1N1 virus | 110 µM (IC50) | Ribavirin 87.0 µM (IC50) | [64] |
| 14R-Oxoglyantrypine (44) | Antiviral | CPE inhibition assay/Influenza A H1N1 virus | 130 µM (IC50) | Ribavirin 87.0 µM (IC50) | [64] |
| 14S-Oxoglyantrypine (45) | Antiviral | CPE inhibition assay/Influenza A H1N1 virus | 85 µM (IC50) | Ribavirin 87.0 µM (IC50) | [64] |
| Cladoquinazoline (47) | Antiviral | CPE inhibition assay/Influenza A H1N1 virus | 150 µM (IC50) | Ribavirin 87.0 µM (IC50) | [64] |
| Epi-Cladoquinazoline (48) | Antiviral | CPE inhibition assay/Influenza A H1N1 virus | 140 µM (IC50) | Ribavirin 87.0 µM (IC50) | [64] |
| Norquinadoline A (49) | Antiviral | CPE inhibition assay/Influenza A H1N1 virus | 82 µM (IC50) | Ribavirin 87.0 µM (IC50) | [64] |
| Quinadoline A (50) | Antiviral | CPE inhibition assay/Influenza A H1N1 virus | 130 µM (IC50) | Ribavirin 87.0 µM (IC50) | [64] |
| Deoxynortryptoquivaline (51) | Antiviral | CPE inhibition assay/Influenza A H1N1 virus | 87 µM (IC50) | Ribavirin 87.0 µM (IC50) | [64] |
| Deoxytryptoquivaline (52) | Antiviral | CPE inhibition assay/Influenza A H1N1 virus | 85 µM (IC50) | Ribavirin 87.0 µM (IC50) | [64] |
| Tryptoquivaline (53) | Antiviral | CPE inhibition assay/Influenza A H1N1 virus | 89 µM (IC50) | Ribavirin 87.0 µM (IC50) | [64] |
| CS-C (54) | Antiviral | CPE inhibition assay/Influenza A H1N1 virus | 140 µM (IC50) | Ribavirin 87.0 µM (IC50) | [64] |
| Quinadoline B (55) | Antiviral | CPE inhibition assay/Influenza A H1N1 virus | 82 µM (IC50) | Ribavirin 87.0 µM (IC50) | [64] |
| Quinolactacin A2 (58) | Cytotoxicity | MTT/HepG-2 | 96.54 µM (IC50) | Curcumin 61.38 µM (IC50) | [66] |
| MTT/HL-60 | 54.47 µM (IC50) | Curcumin 13.78 µM (IC50) | [66] | ||
| MTT/MCF-7 | 94.49 µM (IC50) | Curcumin 20.68 µM (IC50) | [66] | ||
| MTT/LNCap | 45.71 µM (IC50) | Curcumin 6.15 µM (IC50) | [66] | ||
| Anti-malarial | Flow cytometry/SYBR Green I fluorescence/P. falciparum chloroquine sensitive (3D7) | 24.8 µM (EC50) | Artesunate 0.074 µM (EC50) | [66] | |
| Citrinadin A (68) | Cytotoxicity | MTT/HepG-2 | 82.15 µM (IC50) | Curcumin 61.38 µM (IC50) | [66] |
| MTT/HL-60 | 57.23 µM (IC50) | Curcumin 13.78 µM (IC50) | [66] | ||
| MTT/MCF-7 | 66.07 µM (IC50) | Curcumin 20.68 µM (IC50) | [66] | ||
| MTT/LNCap | 41.42 µM (IC50) | Curcumin 6.15 µM (IC50) | [66] | ||
| Anti-malarial | Flow cytometry/SYBR Green I fluorescence/P. falciparum chloroquine sensitive (3D7) | >25.0 µM (EC50) | Artesunate 0.074 µM (EC50) | [66] | |
| Butrecitrinadin (70) | Cytotoxicity | MTT/HepG-2 | 78.57 µM (IC50) | Curcumin 61.38 µM (IC50) | [66] |
| MTT/HL-60 | 60.31 µM (IC50) | Curcumin 13.78 µM (IC50) | [66] | ||
| MTT/MCF-7 | 51.32 µM (IC50) | Curcumin 20.68 µM (IC50) | [66] | ||
| MTT/LNCap | 32.94 µM (IC50) | Curcumin 6.15 µM (IC50) | [66] | ||
| Anti-malarial | Flow cytometry/SYBR Green I fluorescence/P. falciparum chloroquine sensitive (3D7) | >25.0 µM (EC50) | Artesunate 0.074 µM (EC50) | [66] | |
| Cladosporitin B (74) | Cytotoxicity | MTT/BEL-7042 | 29.4 µM (IC50) | Adriamycin 11.9 µM (IC50) | [67] |
| Cytotoxicity | MTT/K562 | 25.6 µM (IC50) | Adriamycin 14.2 µM (IC50) | [67] | |
| Cytotoxicity | MTT/SGC-7901 | 41.7 µM (IC50) | Adriamycin 6.66 µM (IC50) | [67] | |
| Talaroconvolutin A (76) | Cytotoxicity | MTT/HeLa | 14.9 µM (IC50) | Adriamycin 11.5 µM (IC50) | [67] |
| Cytotoxicity | MTT/BEL-7042 | 26.7 µM (IC50) | Adriamycin 11.9 µM (IC50) | [67] | |
| Talaroconvolutin A (76) | α-Glucosidase inhibitory | Glucose oxidase method | 78.2 µM (IC50) | Acarbose 275.7 µM (IC50) | [67] |
| Cladosporamide A (77) | Protein tyrosine phosphatase 1B inhibitory | PTP1B/Spectrophotometry | 48.0 µM (IC50) | Oleanolic acid 0.9 µM (IC50) | [68] |
| TCPTP/Spectrophotometry | 54.0 µM (IC50) | Oleanolic acid 0.8 µM (IC50) | [68] | ||
| Cladosporilactam A (78) | Cytotoxicity | MTT/HeLa | 0.76 µM (IC50) | Adriamycin | [69] |
| MTT/HT-29 | 2.48 µM (IC50) | Adriamycin | [69] | ||
| SRB/P388 | 1.35 µM (IC50) | Adriamycin | [69] | ||
| SRB/A549 | 3.11 µM (IC50) | Adriamycin | [69] | ||
| 2-Methylacetate-3,5,6-trimethylpyrazine (84) | Insecticidal | CM/Helicoverpa armigera Hubner larvae | 100.0 μg/mL (IC50) | Azadirachtin 25.0 μg/mL (IC50) | [71] |
| Antimicrobial | Microplate assay/S. aureus | 12.5 μg/mL (MIC) | Ciprofloxacin 0.39 μg/mL (MIC) | [71] | |
| Cytochalasin D (85) | Antimicrobial | Microplate assay/S. aureus | 25.0 μg/mL (MIC) | Ciprofloxacin 0.39 μg/mL (MIC) | [71] |
| Pandangolide 3 (96) | Antimicrobial | Microplate assay/C. glecosporioides | 2.0 µg/mL (MIC) | Amphotericin B 0.5 µg/mL (MIC) | [39] |
| Microplate assay/B. sorokiniana | 8.0 µg/mL (MIC) | Amphotericin B 0.5 µg/mL (MIC) | [39] | ||
| Thiocladospolide A (101) | Antimicrobial | Microplate assay/E. tarda | 1.0 µg/mL (MIC) | Chloramphenicol 0.5 µg/mL (MIC) | [39] |
| Microplate assay/E. ictarda | 8.0 µg/mL (MIC) | Chloramphenicol 0.5 µg/mL (MIC) | [39] | ||
| Microplate assay/C. glecosporioides | 2.0 µg/mL (MIC) | Amphotericin B 0.5 µg/mL (MIC) | [39] | ||
| Thiocladospolide B (102) | Antimicrobial | Microplate assay/C. glecosporioides | 2.0 µg/mL (MIC) | Amphotericin B 0.5 µg/mL (MIC) | [39] |
| Microplate assay/P. piricola Nose | 32.0 µg/mL (MIC) | Amphotericin B 2.0 µg/mL (MIC) | [39] | ||
| Microplate assay/F. oxysporum f. sp. cucumerinum | 1.0 µg/mL (MIC) | Amphotericin B 0.5 µg/mL (MIC) | [39] | ||
| Thiocladospolide C (103) | Antimicrobial | Microplate assay/C. glecosporioides | 1.0 µg/mL (MIC) | Amphotericin B 0.5 µg/mL (MIC) | [39] |
| Microplate assay/P. piricola Nose | 32.0 µg/mL (MIC) | Amphotericin B 2.0 µg/mL (MIC) | [39] | ||
| Microplate assay/F. oxysporum f. sp. cucumerinum | 32.0 µg/mL (MIC) | Amphotericin B 0.5 µg/mL (MIC) | [39] | ||
| Thiocladospolide D (104) | Antimicrobial | Microplate assay/E. ictarda | 1.0 µg/mL (MIC) | Chloramphenicol 0.5 µg/mL (MIC) | [39] |
| Microplate assay/C. glecosporioides | 1.0 µg/mL (MIC) | Amphotericin B 0.5 µg/mL (MIC) | [39] | ||
| Microplate assay/P. piricola Nose | 32.0 µg/mL (MIC) | Amphotericin B 2.0 µg/mL (MIC) | [39] | ||
| Microplate assay/F. oxysporum f. sp. cucumerinum | 1.0 µg/mL (MIC) | Amphotericin B 0.5 µg/mL (MIC) | [39] | ||
| Thiocladospolide F (106) | Antimicrobial | Microplate assay/E. tarda | 2.0 µg/mL (MIC) | Chloramphenicol 0.5 µg/mL (MIC) | [79] |
| Antimicrobial | Microplate assay/H. maydis | 4.0 µg/mL (MIC) | Amphotericin B 0.5 µg/mL (MIC) | [79] | |
| Thiocladospolide G (108) | Antimicrobial | Microplate assay/E. tarda | 2.0 µg/mL (MIC) | Chloramphenicol 0.5 µg/mL (MIC) | [79] |
| Thiocladospolide G (109) | Antimicrobial | Microplate assay/E. tarda | 4.0 μg/mL (MIC) | Chloramphenicol 1.0 μg/mL (MIC) | [78] |
| Thiocladospolide H (110) | Antimicrobial | Microplate assay/E. ictarda | 8.0 μg/mL (MIC) | Chloramphenicol 1.0 μg/mL (MIC) | [78] |
| Sporiolide A (113) | Cytotoxicity | MTT/L1210 | 0.13 µM (IC50) | - | [80] |
| Sporiolide B (114) | Cytotoxicity | MTT/L1210 | 0.81 µM (IC50) | - | [80] |
| Dendrodolide A (117) | Antimicrobial | Broth dilution assay/B. cereus | 12.5 μM (MIC) | Ciprofloxacin 1.56 μM (MIC) | [69] |
| Broth dilution assay/T. halophilus | 3.13 μM (MIC) | Ciprofloxacin 1.56 μM (MIC) | [69] | ||
| Broth dilution assay/S. epidermidis | 6.25 μM (MIC) | Ciprofloxacin 0.78 μM (MIC) | [69] | ||
| Broth dilution assay/S. aureus | 6.25 μM (MIC) | Ciprofloxacin 0.39 μM (MIC) | [69] | ||
| Broth dilution assay/E. coli | 12.5 μM (MIC) | Ciprofloxacin 1.56 μM (MIC) | [69] | ||
| Broth dilution assay/P. putida | 12.5 μM (MIC) | Ciprofloxacin 0.39 μM (MIC) | [69] | ||
| Broth dilution assay/N. brasiliensis | 6.25 μM (MIC) | Ciprofloxacin 0.78 μM (MIC) | [69] | ||
| Broth dilution assay/V. parahaemolyticus | 12.5 μM (MIC) | Ciprofloxacin 1.56 μM (MIC) | [69] | ||
| Dendrodolide C (118) | Antimicrobial | Broth dilution assay/B. cereus | 25.0 μM (MIC) | Ciprofloxacin 1.56 μM (MIC) | [69] |
| Broth dilution assay/T. halophilus | 3.13 μM (MIC) | Ciprofloxacin 1.56 μM (MIC) | [69] | ||
| Broth dilution assay/S. epidermidis | 25.0 μM (MIC) | Ciprofloxacin 0.78 μM (MIC) | [69] | ||
| Broth dilution assay/S. aureus | 25.0 μM (MIC) | Ciprofloxacin 0.39 μM (MIC) | [69] | ||
| Broth dilution assay/E. coli | 12.5 μM (MIC) | Ciprofloxacin 1.56 μM (MIC) | [69] | ||
| Broth dilution assay/P. putida | 25.0 μM (MIC) | Ciprofloxacin 0.39 μM (MIC) | [69] | ||
| Broth dilution assay/N. brasiliensis | 12.5 μM (MIC) | Ciprofloxacin 0.78 μM (MIC) | [69] | ||
| Broth dilution assay/V. parahaemolyticus | 25.0 μM (MIC) | Ciprofloxacin 1.56 μM (MIC) | [69] | ||
| Dendrodolide M (120) | Antimicrobial | Broth dilution assay/B. cereus | 6.25 μM (MIC) | Ciprofloxacin 1.56 μM (MIC) | [69] |
| Broth dilution assay/T. halophilus | 25.0 μM (MIC) | Ciprofloxacin 1.56 μM (MIC) | [69] | ||
| Broth dilution assay/S. epidermidis | 25.0 μM (MIC) | Ciprofloxacin 0.78 μM (MIC) | [69] | ||
| Broth dilution assay/S. aureus | 12.5 μM (MIC) | Ciprofloxacin 0.39 μM (MIC) | [69] | ||
| Dendrodolide C (118) | Antimicrobial | Broth dilution assay/B. cereus | 25.0 μM (MIC) | Ciprofloxacin 1.56 μM (MIC) | [69] |
| Broth dilution assay/E. coli | 25.0 μM (MIC) | Ciprofloxacin 1.56 μM (MIC) | [69] | ||
| Broth dilution assay/P. putida | 6.25 μM (MIC) | Ciprofloxacin 0.39 μM (MIC) | [69] | ||
| Broth dilution assay/N. brasiliensis | 25.0 μM (MIC) | Ciprofloxacin 0.78 μM (MIC) | [69] | ||
| Broth dilution assay/V. parahaemolyticus | 25.0 μM (MIC) | Ciprofloxacin 1.56 μM (MIC) | [69] | ||
| Cladocladosin A (121) | Antimicrobial | Microplate assay/E. tarda | 1.0 µg/mL (MIC) | Chloramphenicol 0.5 µg/mL (MIC) | [79] |
| Antimicrobial | Microplate assay/P. aeruginosa | 4.0 µg/mL (MIC) | Chloramphenicol 2.0 µg/mL (MIC) | [79] | |
| Ent-cladospolide F (123) | AchE inhibitory | Modified Ellman’s enzyme/Immunosorbent assay | 40.26 µM (IC50) | Tacrine 0.5 µM (IC50) | [40] |
| Iso-cladospolide B (126) | Antimicrobial | Broth dilution assay/B. cereus | 6.25 μM (MIC) | Ciprofloxacin 1.56 μM (MIC) | [69] |
| Broth dilution assay/T. halophilus | 6.25 μM (MIC) | Ciprofloxacin 1.56 μM (MIC) | [69] | ||
| Broth dilution assay/S. epidermidis | 25.0 μM (MIC) | Ciprofloxacin 0.78 μM (MIC) | [69] | ||
| Broth dilution assay/S. aureus | 25.0 μM (MIC) | Ciprofloxacin 0.39 μM (MIC) | [69] | ||
| Broth dilution assay/E. coli | 25.0 μM (MIC) | Ciprofloxacin 1.56 μM (MIC) | [69] | ||
| Broth dilution assay/P. putida | 6.25 μM (MIC) | Ciprofloxacin 0.39 μM (MIC) | [69] | ||
| Broth dilution assay/N. brasiliensis | 12.5 μM (MIC) | Ciprofloxacin 0.78 μM (MIC) | [69] | ||
| Broth dilution assay/V. parahaemolyticus | 25.0 μM (MIC) | Ciprofloxacin 1.56 μM (MIC) | [69] | ||
| Microplate assay/C. mandshurica Miura | 8.0 μg/mL (MIC) | Nystatin 1.0 μg/mL (MIC) | [78] | ||
| Cladosporol C (143) | Cytotoxicity | Trypan blue-cell viability assay/K562 | ˃30.0 µM (IC50) | Trichostatin A 0.24 µM (IC50) | [82] |
| Trypan blue-cell viability assay/A549 | 33.9 µM (IC50) | Trichostatin A 0.05 µM (IC50) | [82] | ||
| Trypan blue-cell viability assay/Huh-7 | ˃30.0 µM (IC50) | Trichostatin A 0.08 µM (IC50) | [82] | ||
| Trypan blue-cell viability assay/H1975 | 45.6 µM (IC50) | Trichostatin A 0.09 µM (IC50) | [82] | ||
| Trypan blue-cell viability assay/MCF-7 | ˃30.0 µM (IC50) | Trichostatin A 0.78 µM (IC50) | [82] | ||
| Trypan blue-cell viability assay/U937 | ˃30.0 µM (IC50) | Trichostatin A 0.06 µM (IC50) | [82] | ||
| Trypan blue-cell viability assay/BGC823 | ˃30.0 µM (IC50) | Trichostatin A 0.09 µM (IC50) | [82] | ||
| Trypan blue-cell viability assay/HL-60 | 72.5 µM (IC50) | Trichostatin A 0.09 µM (IC50) | [82] | ||
| Trypan blue-cell viability assay/A549 | ˃30.0 µM (IC50) | Trichostatin A 0.11 µM (IC50) | [82] | ||
| Trypan blue-cell viability assay/MOLT-4 | 14.4 µM (IC50) | Trichostatin A 0.03 µM (IC50) | [82] | ||
| MTT/A549 | 14.0 µM (IC50) | Cisplatin 1.3 µM (IC50) | [84] | ||
| MTT/HeLa | 4.0 µM (IC50) | Paclitaxel 4.9 µM (IC50) | [84] | ||
| Antimicrobial | Microplate assay/E. coli | 8.0 μg/mL (MIC) | Chloramphenicol 0.025 μg/mL (MIC) | [84] | |
| Microplate assay/M. luteus | 32.0 μg/mL (MIC) | Chloramphenicol 0.5 μg/mL (MIC) | [84] | ||
| Microplate assay/V. harveyi | 16.0 μg/mL (MIC) | Chloramphenicol 2.0 μg/mL (MIC) | [84] | ||
| Microplate assay/S. aureus | 6.25 μg/mL (MIC) | Ciprofloxacin 0.39 μg/mL (MIC) | [71] | ||
| Microplate assay/M. luteus | 12.5 μg/mL (MIC) | Ciprofloxacin 0.39 μg/mL (MIC) | [71] | ||
| Cladosporol D (144) | Anti-inflammatory | Spectrophotometry/Anti-COX-2/PGF2α inhibition | 60.2 µM (IC50) | Indomethacin 18.3 µM (IC50) NS-398 1.0 µM (IC50) | [82] |
| Cladosporol E (145) | Insecticidal | Measuring the corrected mortality (CM) | 150.0 μg/mL (IC50) | Azadirachtin 25.0 μg/mL (IC50) | [71] |
| Antimicrobial | Microplate assay/S. aureus | 1.56 μg/mL (MIC) | Ciprofloxacin 0.39 μg/mL (MIC) | [71] | |
| Microplate assay/M. luteus | 12.5 μg/mL (MIC) | Ciprofloxacin 0.39 μg/mL (MIC) | [71] | ||
| Cladosporol F (146) | Cytotoxicity | MTT/K562 | 23.0 µM (IC50) | Doxorubicin 0.6 µM (IC50) | [83] |
| SRB/HeLa | 13.8 µM (IC50) | Doxorubicin 0.5 µM (IC50) | [83] | ||
| SRB/HCT-116 | 23.0 µM (IC50) | Doxorubicin 0.2 µM (IC50) | [83] | ||
| MTT/A549 | 15.0 µM (IC50) | Cisplatin 1.3 µM (IC50) | [84] | ||
| MTT/HeLa | 10.0 µM (IC50) | Paclitaxel 4.9 µM (IC50) | [84] | ||
| Antimicrobial | Microplate assay/E. coli | 32.0 μg/mL (MIC) | Chloramphenicol 0.025 μg/mL (MIC) | [84] | |
| Microplate assay/M. luteus | 64.0 μg/mL (MIC) | chloramphenicol 0.5 μg/mL (MIC) | [84] | ||
| Microplate assay/V. harveyi | 32.0 μg/mL (MIC) | Chloramphenicol 2.0 μg/mL (MIC) | [84] | ||
| Cladosporol G (147) | Cytotoxicity | MTT/K562 | 8.8 µM (IC50) | Doxorubicin 0.6 µM (IC50) | [83] |
| SRB/HeLa | 3.9 µM (IC50) | Doxorubicin 0.5 µM (IC50) | [83] | ||
| SRB/HCT-116 | 19.4 µM (IC50) | Doxorubicin 0.2 µM (IC50) | [83] | ||
| Cladosporol G (148) | Cytotoxicity | MTT/A549 | 13.0 µM (IC50) | Cisplatin 1.3 µM (IC50) | [84] |
| MTT/H446 | 11.0 µM (IC50) | Adriamycin 4.0 µM (IC50) | [84] | ||
| MTT/Huh7 | 10.0 µM (IC50) | Fluorouracil 6.2 µM (IC50) | [84] | ||
| MTT/L02 | 11.0 µM (IC50) | Cisplatin 13.0 µM (IC50) | [84] | ||
| MTT/LM3 | 14.0 µM (IC50) | Cisplatin 9.1 µM (IC50) | [84] | ||
| MTT/SW1990 | 15.0 µM (IC50) | Gemcitabine 2.2 µM (IC50) | [84] | ||
| Antimicrobial | Microplate assay/E. coli | 64.0 μg/mL (MIC) | Chloramphenicol 0.025 μg/mL (MIC) | [84] | |
| Microplate assay/M. luteus | 128.0 μg/mL (MIC) | Chloramphenicol 0.5 μg/mL (MIC) | [84] | ||
| Microplate assay/V. harveyi | 64.0 μg/mL (MIC) | Chloramphenicol 2.0 μg/mL (MIC) | [84] | ||
| Cladosporol H (149) | Cytotoxicity | MTT/A549 | 5.0 µM (IC50) | Cisplatin 1.3 µM (IC50) | [84] |
| MTT/H446 | 10.0 µM (IC50) | Adriamycin 4.0 µM (IC50) | [84] | ||
| MTT/Huh7 | 1.0 µM (IC50) | Fluorouracil 6.2 µM (IC50) | [84] | ||
| MTT/LM3 | 4.1 µM (IC50) | Cisplatin 9.1 µM (IC50) | [84] | ||
| MTT/MCF-7 | 10.0 µM (IC50) | Paclitaxel 1.8 µM (IC50) | [84] | ||
| MTT/SW1990 | 15.0 µM (IC50) | Gemcitabine 2.2 µM (IC50) | [84] | ||
| Antimicrobial | Microplate assay/E. coli | 32.0 μg/mL (MIC) | Chloramphenicol 0.025 μg/mL (MIC) | [84] | |
| Microplate assay/M. luteus | 64.0 μg/mL (MIC) | Chloramphenicol 0.5 μg/mL (MIC) | [84] | ||
| Microplate assay/V. harveyi | 4.0 μg/mL (MIC) | Chloramphenicol 2.0 μg/mL (MIC) | [84] | ||
| Cladosporol I (150) | Cytotoxicity | MTT/HeLa | 10.8 µM (IC50) | Paclitaxel 4.9 µM (IC50) | [84] |
| Antimicrobial | Microplate assay/E. coli | 64.0 μg/mL (MIC) | Chloramphenicol 0.025 μg/mL (MIC) | [84] | |
| Microplate assay/M. luteus | 64.0 μg/mL (MIC) | Chloramphenicol 0.5 μg/mL (MIC) | [84] | ||
| Microplate assay/V. harveyi | 16.0 μg/mL (MIC) | Chloramphenicol 2.0 μg/mL (MIC) | [84] | ||
| Cladosporol J (151) | MTT/A549 | 15.0 µM (IC50) | Cisplatin 1.3 µM (IC50) | [84] | |
| MTT/H446 | 11.0 µM (IC50) | Adriamycin 4.0 µM (IC50) | [84] | ||
| MTT/HeLa | 15.0 µM (IC50) | Paclitaxel 4.9 µM (IC50) | [84] | ||
| MTT/Huh7 | 20.0 µM (IC50) | Fluorouracil 6.2 µM (IC50) | [84] | ||
| MTT/MCF-7 | 12.0 µM (IC50) | Paclitaxel 1.8 µM (IC50) | [84] | ||
| Antimicrobial | Microplate assay/E. coli | 16.0 μg/mL (MIC) | Chloramphenicol 0.025 μg/mL (MIC) | [84] | |
| Microplate assay/M. luteus | 64.0 μg/mL (MIC) | Chloramphenicol 0.5 μg/mL (MIC) | [84] | ||
| Microplate assay/V. harveyi | 32.0 μg/mL (MIC) | Chloramphenicol 2.0 μg/mL (MIC) | [84] | ||
| Cladosporone A (152) | Cytotoxicity | Trypan blue-cell viability assay/K562 | 14.3 µM (IC50) | Trichostatin A 0.24 µM (IC50) | [82] |
| Trypan blue-cell viability assay/A549 | 15.7 µM (IC50) | Trichostatin A 0.05 µM (IC50) | [82] | ||
| Trypan blue-cell viability assay/Huh-7 | 29.9 µM (IC50) | Trichostatin A 0.08 µM (IC50) | [82] | ||
| Trypan blue-cell viability assay/H1975 | 40.6 µM (IC50) | Trichostatin A 0.09 µM (IC50) | [82] | ||
| Trypan blue-cell viability assay/MCF-7 | 21.3 µM (IC50) | Trichostatin A 0.78 µM (IC50) | [82] | ||
| Trypan blue-cell viability assay/U937 | 10.5 µM (IC50) | Trichostatin A 0.06 µM (IC50) | [82] | ||
| Trypan blue-cell viability assay/BGC823 | 17.0 µM (IC50) | Trichostatin A 0.09 µM (IC50) | [82] | ||
| Trypan blue-cell viability assay/HL-60 | 10.1 µM (IC50) | Trichostatin A 0.09 µM (IC50) | [82] | ||
| Trypan blue-cell viability assay/A549 | 53.7 µM (IC50) | Trichostatin A 0.11 µM (IC50) | [82] | ||
| Trypan blue-cell viability assay/MOLT-4 | 14.6 µM (IC50) | Trichostatin A 0.03 µM (IC50) | [82] | ||
| Anti-inflammatory | Spectrophotometry/Anti-COX-2/PGF2α inhibition | 49.1 µM (IC50) | Indomethacin 18.3 µM (IC50) NS-398 1.0 µM (IC50) | [82] | |
| Aladothalen (160) | Antimicrobial | Agar dilution method/B. cereus | 50.0 µM (MIC) | Ciprofloxacin ˂ 0.4 µM (MIC) | [89] |
| Agar dilution method/M. phlei | 25.0 µM (MIC) | Ciprofloxacin ˂ 0.4 µM (MIC) | [89] | ||
| Agar dilution method/MRCNS | 25.0 µM (MIC) | Ciprofloxacin 25.0 µM (MIC) | [89] | ||
| Cladonaphchrom A (169) | Antimicrobial | Microplate assay/S. albus | 1.25 µg/mL (MIC) | Ciprofloxacin 0.6 µg/mL (MIC) | [90] |
| Microplate assay/E. coli | 2.5 µg/mL (MIC) | Ciprofloxacin 0.3 µg/mL (MIC) | [90] | ||
| Microplate assay/B. subtilis | 10.0 µg/mL (MIC) | Ciprofloxacin 0.6 µg/mL (MIC) | [90] | ||
| Microplate assay/M. tetragenus | 5.0 µg/mL (MIC) | Ciprofloxacin 0.3µg/mL (MIC) | [90] | ||
| Microplate assay/M. luteus | 10.0 µg/mL (MIC) | Ciprofloxacin 0.3 µg/mL (MIC) | [90] | ||
| Microplate assay/A. brassicicola | 50.0 µg/mL (MIC) | Prochloraz 12.5 µg/mL (MIC) | [90] | ||
| Microplate assay/P. parasitica var. nicotianae | 50.0 µg/mL (MIC) | Prochloraz 50.0 µg/mL (MIC) | [90] | ||
| Microplate assay/C. capsici | 25.0 µg/mL (MIC) | Prochloraz 12.5 µg/mL (MIC) | [90] | ||
| Microplate assay/B. oryzae | 100.0 µg/mL (MIC) | Prochloraz 50.0 µg/mL (MIC) | [90] | ||
| Microplate assay/D. medusaea | 50.0 µg/mL (MIC) | Prochloraz 50.0 µg/mL (MIC) | [90] | ||
| Microplate assay/C. paradoxa | 50.0 µg/mL (MIC) | Prochloraz 25.0 µg/mL (MIC) | [90] | ||
| Cladonaphchrom B (170) | Antimicrobial | Microplate assay/S. albus | 2.5 µg/mL (MIC) | Ciprofloxacin 0.6 µg/mL (MIC) | [90] |
| Microplate assay/E. coli | 2.5 µg/mL (MIC) | Ciprofloxacin 0.3 µg/mL (MIC) | [90] | ||
| Microplate assay/B. subtilis | 5.0 µg/mL (MIC) | Ciprofloxacin 0.6 µg/mL (MIC) | [90] | ||
| Microplate assay/M. tetragenus | 5.0 µg/mL (MIC) | Ciprofloxacin 0.3µg/mL (MIC) | [90] | ||
| Microplate assay/M. luteus | 10.0 µg/mL (MIC) | Ciprofloxacin 0.3 µg/mL (MIC) | [90] | ||
| Microplate assay/A. brassicicola | 25.0 µg/mL (MIC) | Prochloraz 12.5 µg/mL (MIC) | [90] | ||
| Microplate assay/P. parasitica var. nicotianae | 50.0 µg/mL (MIC) | Prochloraz 50.0 µg/mL (MIC) | [90] | ||
| Microplate assay/C. capsici | 25.0 µg/mL (MIC) | Prochloraz 12.5 µg/mL (MIC) | [90] | ||
| Microplate assay/D. medusaea | 100.0 µg/mL (MIC) | Prochloraz 50.0 µg/mL (MIC) | [90] | ||
| Microplate assay/C. paradoxa | 50.0 µg/mL (MIC) | Prochloraz 25.0 µg/mL (MIC) | [90] | ||
| Malettinin A (178) | Antimicrobial | Microplate assay/T. rubrum | 33.1 µM (IC50) | Clotrimazole 0.2 µM (IC50) | [93] |
| Malettinin B (179) | Microplate assay/X. campestris | 28.3 µM ((IC50) | Chloramphenicol 2.1 µM (IC50) | [93] | |
| Microplate assay/T. rubrum | 60.6 µM (IC50) | Clotrimazole 0.2 µM (IC50) | [93] | ||
| Malettinin C (180) | Antimicrobial | Microplate assay/T. rubrum | 37.9 µM (IC50) | Clotrimazole 0.2 µM (IC50) | [93] |
| Microplate assay/X. campestris | 83.2 µM (IC50) | Chloramphenicol 2.1 µM (IC50) | [93] | ||
| Malettinin E (181) | Antimicrobial | Microplate assay/T. rubrum | 28.7 µM (IC50) | Clotrimazole 0.2 µM (IC50) | [93] |
| Microplate assay/X. campestris | 30.7 µM (IC50) | Chloramphenicol 2.1 µM ((IC50) | [93] | ||
| Cladosporinone (182) | Broth dilution assay/S. aureus | 64.0 μg/mL(MIC) | - | [94] | |
| Viriditoxin (183) | Broth dilution assay/S. aureus | 0.015 μg/mL (MIC) 0.023 μM (MIC) | - | [94] | |
| Viriditoxin derivative 1 (184) | Broth dilution assay/S. aureus | 2.0 μg/mL (MIC) | - | [94] | |
| Viriditoxin derivative 2 (185) | Broth dilution assay/S. aureus | 16.0 μg/mL (MIC) | - | [94] | |
| (2S,4S)-4-Methoxy-2-methylchroman-5-ol (201) | Antioxidant | DPPH assay | 5.66 µM (IC50) | Ascorbic acid 3.29 µM (IC50) | [95] |
| (2S,4S)-2-Methylchroman-4,5-diol (203) | Antioxidant | DPPH assay | 6.67 µM (IC50) | Ascorbic acid 3.29 µM (IC50) | [95] |
| Citrinin H1 (206) | Insecticidal | Measuring the corrected mortality (CM) | 100.0 μg/mL (IC50) | Azadirachtin 25.0 μg/mL (IC50) | [71] |
| Antimicrobial | Microplate assay/S. aureus | 6.25 μg/mL (MIC) | Ciprofloxacin 0.39 μg/mL (MIC) | [71] | |
| Microplate assay/E. coli | 12.5 μg/mL (MIC) | Ciprofloxacin 0.19 μg/mL (MIC) | [71] | ||
| Microplate assay/B. cereus | 12.5 μg/mL (MIC) | Ciprofloxacin 0.19 μg/mL (MIC) | [71] | ||
| Vermistatin (214) | Insecticidal | CM/Helicoverpa armigera Hubner larvae | 150.0 μg/mL (IC50) | Azadirachtin 25.0 μg/mL (IC50) | [71] |
| Antimicrobial | Microplate assay/S. aureus | 25.0 μg/mL (MIC) | Ciprofloxacin 0.39 μg/mL (MIC) | [71] | |
| Microplate assay/B. cereus | 25.0 μg/mL (MIC) | Ciprofloxacin 0.39 μg/mL (MIC) | [71] | ||
| Cladoscyclitol B (219) | α-Glucosidase inhibitory | Colorimetric assay | 2.95 µM (IC50) | Acarbose 2.35 µM (IC50) | [97] |
| 3-Phenyl-2-propenoic acid (232) | Insecticidal | Inhibitinon 50%/B. amphitrite | 84.28 μg/mL (EC50) | FSW with DMSO | [60] |
| Cladosporinone (182) | Broth dilution assay/S. aureus | 64.0 μg/mL(MIC) | - | [94] | |
| Lethality 50%/B. amphitrite | >200 μg/mL (LC50) | FSW with DMSO | [60] | ||
| Inhibitinon 50%/B. neritina | 11.15 μg/mL (EC50) | FSW with DMSO | [60] | ||
| Lethality 50%/B. neritina | >200 μg/mL (LC50) | FSW with DMSO | [60] | ||
| Antimicrobial | Serial dilution/L. hongkongensis | 80 μg/mL (MIC) | Streptomycin 250 μg/mL (MIC) Penicillin 0.25 μg/mL (MIC) | [60] | |
| 3-(2,3-Dihydroxy phenoxy) butanoic acid (233) | Antioxidant | DPPH assay | 0.24 µM (IC50) | Ascorbic acid 3.29 µM (IC50) | [95] |
| Methyl (3S)-3-(2,3-dihydroxy phenyloxy)butanoate (235) | Antioxidant | DPPH assay | 2.65 µM (IC50) | Ascorbic acid 3.29 µM (IC50) | [95] |
| 2-Phenylethanol (238) | Insecticidal | Inhibitinon 50%/B. amphitrite | 53.65 μg/mL (EC50) | FSW with DMSO | [60] |
| Lethality 50%/B. amphitrite | >200 μg/mL (LC50) | FSW with DMSO | [60] | ||
| Inhibitinon 50%/B. neritina | 102.23 μg/mL (EC50) | FSW with DMSO | [60] | ||
| Lethality 50%/B. neritina | >200 μg/mL (LC50) | FSW with DMSO | [60] | ||
| 4-O-α-D-Ribofuranose-2-pentyl-3-phemethylol (240) | α-Glucosidase inhibitory | Colorimetric assay | 2.05 µM (IC50) | Acarbose 2.35 µM (IC50) | [97] |
| Cladosporin D (247) | Antioxidant | DPPH assay | 16.4 µM (IC50) | Ascorbic acid 4.9 µM (IC50) | [36] |
| (2S)-7,4′-dihydroxy-5-methoxy-8-(γ,γ-dimethylallyl)-flavanone (248) | Protein tyrosine phosphatase 1B inhibitory | PTP1B/Spectrophotometry | 11.0 µM (IC50) | Oleanolic acid 0.9 µM (IC50) | [68] |
| TCPTP/Spectrophotometry | 27.0 µM (IC50) | Oleanolic acid 0.8 µM (IC50) | [68] | ||
| Bis(2-ethylhexyl)phthalate (249) | Insecticidal | Inhibitinon 50%/B. amphitrite | 9.18 μg/mL (EC50) | FSW with DMSO | [60] |
| Lethality 50%/B. amphitrite | >200 μg/mL (LC50) | FSW with DMSO | [60] | ||
| Inhibitinon 50%/B. neritina | 77.85 μg/mL (EC50) | FSW with DMSO | [60] | ||
| Lethality 50%/B. neritina | >200 μg/mL (LC50) | FSW with DMSO | [60] | ||
| 1,1′-Dioxine-2,2′-dipropionic acid (252) | Insecticidal | Measuring the corrected mortality (CM)/Helicoverpa armigera Hubner larvae | 150.0 μg/mL (IC50) | Azadirachtin 25.0 μg/mL (IC50) | [70,71] |
| Antimicrobial | Microplate assay/S. aureus | 25.0 μg/mL (MIC) | Ciprofloxacin 0.39 μg/mL (MIC) | [71] | |
| Microplate assay/E. coli | 25.0 μg/mL (MIC) | Ciprofloxacin 0.19 μg/mL (MIC) | [71] | ||
| Microplate assay/B. cereus | 12.5 μg/mL (MIC) | Ciprofloxacin 0.39 μg/mL (MIC) | [71] | ||
| 5α,8α-Epidioxy-ergosta-6,22E-dien-3β-ol (256) | Antiviral | Neuraminidase inhibition assay/RSV | 0.11 µM (IC50) | Ribavirin 0.08 µM (IC50) | [100] |
| 3β,5α-Dihydroxy-6β-methoxyergosta-7,22-diene (262) | Antiviral | Neuraminidase inhibition assay/RSV | 0.11 µM (IC50) | Ribavirin 0.08 µM (IC50) | [100] |
| 5α,8α-Epidioxy-ergosta-6,9,22E-triene-3β-ol (258) | Antiviral | Neuraminidase inhibition assay/RSV | 0.17 µM (IC50) | Ribavirin 0.08 µM (IC50) | [100] |
| 3α-Hydroxy-pregna-7-ene-6,20-dione = Cladosporisteroid B (268) | Antiviral | Neuraminidase inhibition assay/RSV | 0.12 µM (IC50) | Ribavirin 0.08 µM (IC50) | [100] |
| CPE inhibition assay/ Influenza A H3N3 virus | 16.2 µM (IC50) | Oseltamivir 34.0 nM (IC50) | [61] | ||
| (3E,8E,6S)-Undeca-3,8,10-trien-1,6-diol (286) | Cytotoxicity | SRB/H1975 | 10.0 µM (IC50) | Adriamycin 0.38 µM (IC50) | [95] |
3. Secondary Metabolites and Bioactivities of Marine-Associated Cladosporium Species
3.1. Tetramic Acid Derivatives
3.2. Diketopiperazines
3.3. Alkaloids
3.4. Macrolides
3.5. Butanolides and Butenolides
3.6. Seco-Acids
3.7. Tetralones (Napthalenones)
3.8. Perylenequinones
3.9. Naphthalene Derivatives
3.10. Xanthones
3.11. Tropolones
3.12. Binaphthopyrones
3.13. Benzopyranes, Benzopyrones, and Pyrones
3.14. Lactones, Cyclohexene, and Azaphilone Derivatives
3.15. Phenolics and Other Aromatic Compounds
3.16. Sterols and Terpenes
3.17. Alcohols and Aldehydes
3.18. Bioactivities of Cladosporium Species Extracts
4. Conclusions
5. Strategies for Activating Silencing Gene Clusters
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Townsed, M.; Davies, K.; Hanley, N.; Hewitt, J.E.; Lundquist, C.J.; Lohrer, A.M. The challenge of implementing the marine ecosystem service concept. Front. Mar. Sci. 2018, 5, 359. [Google Scholar] [CrossRef]
- Boeuf, G. Marine biodiversity characteristics. C. R. Biol. 2011, 334, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Donato, P.D.; Buono, A.; Poli, A.; Finore, I.; Abbamondi, G.R.; Nicolaus, B.; Lama, L. Exploring marine environments for the identification of extremophiles and their enzymes for sustainable and green bioprocesses. Sustainability 2019, 11, 149. [Google Scholar] [CrossRef]
- Bonugli-Santos, R.C.; Dos Santos Vasconcelos, M.R.; Passarini, M.R.; Vieira, G.A.; Lopes, V.C.; Mainardi, P.H.; Dos Santos, J.A.; de Azevedo Duarte, L.; Otero, I.V.; da Silva Yoshida, A.M.; et al. Marine-derived fungi: Diversity of enzymes and biotechnological applications. Front. Microbiol. 2015, 6, 269. [Google Scholar] [CrossRef] [PubMed]
- Varrella, S.; Barone, G.; Tangherlini, M.; Rastelli, E.; Dell’Anno, A.; Corinaldesi, C. Diversity, Ecological Role and Biotechnological Potential of Antarctic Marine Fungi. J. Fungi 2021, 7, 391. [Google Scholar] [CrossRef] [PubMed]
- Lindequist, U. Marine-derived pharmaceuticals-challenges and opportunities. Biomol. Ther. 2016, 24, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2014, 31, 160–258. [Google Scholar]
- Gerwick, W.H.; Moore, B.S. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar] [CrossRef]
- Retter, A.; Nilsson, R.H.; Bourlat, S.J. Exploring the taxonomic composition of two fungal communities on the Swedish west coast through metabarcoding. Biodivers. Data J. 2019, 7, e35332. [Google Scholar] [CrossRef]
- Fan, B.; Parrot, D.; Blümel, M.; Labes, A.; Tasdemir, D. Influence of OSMAC-based cultivation in metabolome and anticancer activity of fungi associated with the brown alga Fucus Vesiculosus. Mar. Drugs 2019, 17, 67. [Google Scholar] [CrossRef]
- Furbino, L.E.; Pellizzari, F.M.; Neto, P.C.; Rosa, C.A.; Rosa, L.H. Isolation of fungi associated with macroalgae from maritime Antarctica and their production of agarolytic and carrageenolytic activities. Polar Biol. 2018, 41, 527–535. [Google Scholar] [CrossRef]
- Panno, L.; Bruno, M.; Voyron, S.; Anastasi, A.; Gnavi, G.; Miserere, L.; Varese, G.C. Diversity, ecological role and potential biotechnological applications of marine fungi associated to the seagrass Posidonia Oceanica. New Biotechnol. 2013, 30, 685–694. [Google Scholar] [CrossRef]
- Jones, E.B.G.; Sakayaroj, J.; Suetrong, S.; Somrithipol, S.; Pang, K.L. Classification of marine Ascomycota, anamorphic taxa and Basidiomycota. Fungal Divers. 2009, 35, 187. [Google Scholar]
- Jones, E.B.G. Are there more marine fungi to be described? Bot. Mar. 2011, 54, 343–354. [Google Scholar] [CrossRef]
- Marinefungi. Available online: https://www.marinefungi.org/ (accessed on 9 November 2021).
- Hasan, S.; Ansari, M.I.; Ahmad, A.; Mishra, M. Major bioactive metabolites from marine fungi: A Review. Bioinformation 2015, 11, 176–181. [Google Scholar] [CrossRef]
- Jayawardena, R.S.; Hyde, K.D.; Chen, Y.J.; Papp, V.; Palla, B.; Papp, D.; Bhunjun, C.S.; Hurdeal, V.G.; Senwanna, C.; Manawasinghe, I.S.; et al. One stop shop IV: Taxonomic update with molecular phylogeny for important phytopathogenic genera: 76–100 (2020). Fungal Divers. 2020, 103, 87–218. [Google Scholar] [CrossRef]
- Bensch, K.; Braun, U.; Groenewald, J.Z.; Crous, P.W. The genus Cladosporium. Stud. Mycol. 2012, 72, 1–401. [Google Scholar] [CrossRef]
- Levetin, E.; Dorsey, K. Contribution of leaf surface fungi to the air spora. Aerobiologia 2006, 22, 3–12. [Google Scholar] [CrossRef]
- Heuchert, B.; Braun, U.; ScHuBert, K. Morphotaxonomic revision of fungi-colous Cladosporium species (hyphomycetes). Schlechtendalia 2005, 13, 1–78. [Google Scholar]
- Bensch, K.; Groenewald, J.Z.; Meijer, M.; Dijksterhuis, J.; Jurjević, Ž.; Andersen, B.; Houbraken, J.; Crous, P.W.; Samson, R.A. Cladosporium species in indoor environments. Stud. Mycol. 2018, 89, 177–301. [Google Scholar]
- El-Dawy, E.G.A.E.M.; Gherbawy, Y.A.; Hussein, M.A. Morphological, molecular characterization, plant pathogenicity and biocontrol of Cladosporium complex groups associated with faba beans. Sci. Rep. 2021, 11, 14183. [Google Scholar] [CrossRef] [PubMed]
- Denning, D.W.; O’Driscoll, B.R.; Hogaboam, C.M.; Bowyer, P.; Niven, R.M. The link between fungi and severe asthma: A summary of the evidence. Eur. Resp. J. 2006, 27, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Van Kan, J.A.; Van den Ackerveken, G.F.J.M.; De Wit, P.J.G.M. Cloning and characterization of cDNA of avirulence gene avr9 of the fungal pathogen Cladosporium fulvum, causal agent of tomato leaf mold. Mol. Plant Microbe Interact. 1991, 4, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Batra, N.; Kaur, H.; Mohindra, S.; Singh, S.; Shamanth, A.S.; Rudramurthy, S.M. Cladosporium sphaerospermum causing brain abscess, a saprophyte turning pathogen: Case and review of published reports. J. Mycol. Med. 2019, 29, 180–184. [Google Scholar] [CrossRef]
- Sandoval-Denis, M.; Gené, J.; Sutton, D.A.; Wiederhold, N.P.; Cano-Lira, J.F.; Guarro, J. New species of Cladosporium associated with human and animal infections. Persoonia 2016, 36, 281–298. [Google Scholar] [CrossRef]
- Simon-Nobbe, B.; Denk, U.; Pöll, V.; Rid, R.; Breitenbach, M. The spectrum of fungal allergy. Int. Arch. Allergy Immunol. 2008, 145, 58–86. [Google Scholar] [CrossRef]
- De Hoog, G.S.; Guarro, J.; Gené, J.; Figueras, M.J. Atlas of Clinical Fungi, 2nd ed.; Centraalbureau voor Schimmelcultures (CBS): Utrecht, The Netherlands, 2000. [Google Scholar]
- Hamayun, M.; Khan, S.A.; Ahmad, N.; Tang, D.S.; Kang, S.M.; Na, C.I.; Sohn, E.Y.; Hwang, Y.H.; Shin, D.H.; Lee, B.H.; et al. Cladosporium sphaerospermum as a new plant growth-promoting endophyte from the roots of Glycine max (L.) Merr. World J. Microbiol. Biotechnol. 2009, 25, 627–632. [Google Scholar] [CrossRef]
- Sellart-Altisent, M.; Torres-Rodríguez, J.M.; Gómez de Ana, S.; Alvarado-Ramírez, E. Microbiota fúngica nasal en sujetos alérgicos y sanos [Nasal fungal microbiota in allergic and healthy subjects]. Rev. Iberoam. Micol. 2007, 24, 125–130. [Google Scholar] [CrossRef]
- Sandoval-Denis, M.; Sutton, D.A.; Martin-Vicente, A.; Cano-Lira, J.F.; Wiederhold, N.; Guarro, J.; Gené, J. Cladosporium species recovered from clinical samples in the United States. J. Clin. Microbiol. 2015, 53, 2990–3000. [Google Scholar] [CrossRef]
- Sun, J.Z.; Liu, X.Z.; McKenzie, E.H.C.; Jeewon, R.; Liu, J.K.; Zhang, X.L.; Zhao, Q.; Hyde, K.D. Fungicolous fungi: Terminology, diversity, distribution, evolution and species checklist. Fungal Divers. 2019, 95, 337–430. [Google Scholar] [CrossRef]
- Torres, D.E.; Rojas-Martínez, R.I.; Zavaleta-Mejia, E.; Guevara-Fefer, P.; Márquez-Guzmán, G.J.; Perez-Martinez, C. Cladosporium cladosporioides and Cladosporium pseudocladosporioides as potential new fungal antagonists of Puccinia horiana Henn., the causal agent of Chrysanthemum white rust. PLoS ONE 2017, 12, e0170782. [Google Scholar] [CrossRef]
- Jashni, M.K.; van der Burgt, A.; Battaglia, E.; Mehrabi, R.; Collemare, J.; de Wit, P.J. Transcriptome and proteome analyses of proteases in biotroph fungal pathogen Cladosporium fulvum. J. Plant Pathol. 2020, 102, 377–386. [Google Scholar] [CrossRef]
- Ge, J.; Yin, Y.; Jiang, X.; Liu, W.; Yao, B.; Luo, H. Gene cloning and characterization of a novel glucose oxidase from Cladosporium tianshanense SL19. J. Agric. Sci. Technol. 2019, 12, 49–57. [Google Scholar]
- Amin, M.; Zhang, X.Y.; Xu, X.Y.; Qi, S.H. New citrinin derivatives from the deep-sea-derived fungus Cladosporium sp. SCSIO z015. Nat. Prod. Res. 2020, 34, 1219–1226. [Google Scholar] [CrossRef]
- Pan, F.; El-Kashef, D.H.; Kalscheuer, R.; Müller, W.E.G.; Lee, J.; Feldbrügge, M.; Mándi, A.; Kurtán, T.; Liu, Z.; Wu, W.; et al. New hybrid polyketides from the endophytic fungus Cladosporium sphaerospermum WBS017. Eur. J. Med. Chem. 2020, 191, 112159. [Google Scholar] [CrossRef]
- Rischer, M.; Lee, S.R.; Eom, H.J.; Park, H.B.; Vollmers, J.; Kaster, A.-K.; Shin, Y.-H.; Oh, D.-C.; Kim, K.H.; Beemelmanns, C. Spirocyclic cladosporicin A and cladosporiumins I and J from a Hydractinia-associated Cladosporium sphaerospermum SW67. Org. Chem. Front. 2019, 6, 1084–1093. [Google Scholar] [CrossRef]
- Zhang, F.Z.; Li, X.M.; Yang, S.Q.; Meng, L.H.; Wang, B.G. Thiocladospolides A-D, 12-membered macrolides from the mangrove-derived endophytic fungus Cladosporium cladosporioides MA-299 and structure revision of pandangolide 3. J. Nat. Prod. 2019, 82, 1535–1541. [Google Scholar] [CrossRef]
- Zhang, F.Z.; Li, X.M.; Li, X.; Yang, S.Q.; Meng, L.H.; Wang, B.G. Polyketides from the mangrove-derived endophytic fungus Cladosporium cladosporioides. Mar. Drugs 2019, 17, 296. [Google Scholar] [CrossRef]
- Zhu, M.; Gao, H.; Wu, C.; Zhu, T.; Che, Q.; Gu, Q.; Guo, P.; Li, D. Lipid-lowering polyketides from a soft coral-derived fungus Cladosporium sp. TZP29. Bioorg. Med. Chem. Lett. 2015, 25, 3606–3609. [Google Scholar] [CrossRef]
- Wu, G.; Sun, X.; Yu, G.; Wang, W.; Zhu, T.; Gu, Q.; Li, D. Cladosins A-E, hybrid polyketides from a deep-sea-derived fungus, Cladosporium sphaerospermum. J. Nat. Prod. 2014, 77, 270–275. [Google Scholar] [CrossRef]
- Birolli, W.G.; Zanin, L.L.; Jimenez, D.E.Q.; Porto, A.L.M. Synthesis of Knoevenagel adducts under microwave irradiation and biocatalytic ene-reduction by the marine-derived fungus Cladosporium sp. CBMAI 1237 for the production of 2-cyano-3-phenylpropanamide derivatives. Mar. Biotechnol. 2020, 22, 317–330. [Google Scholar] [CrossRef]
- Birolli, W.G.; de Santos, A.D.; Alvarenga, N.; Garcia, A.C.F.S.; Romão, L.P.C.; Porto, A.L.M. Biodegradation of anthracene and several PAHs by the marine-derived fungus Cladosporium sp. CBMAI 1237. Mar. Pollut. Bull. 2018, 129, 525–533. [Google Scholar] [CrossRef]
- Gil-Durán, C.; Ravanal, M.C.; Ubilla, P.; Vaca, I.; Chávez, R. Heterologous expression, purification and characterization of a highly thermolabile endoxylanase from the Antarctic fungus Cladosporium sp. Fungal Biol. 2018, 122, 875–882. [Google Scholar] [CrossRef]
- Taskin, M.; Ortucu, S.; Unver, Y.; Tasar, O.C.; Ozdemir, M.; Kaymak, H.C. Invertase production and molasses decolourization by cold-adapted filamentous fungus Cladosporium herbarum ER-25 in non-sterile molasses medium. Process Saf. Environ. Prot. 2016, 103, 136–143. [Google Scholar] [CrossRef]
- Trivedi, N.; Reddy, C.; Radulovich, R.; Jha, B. Solid state fermentation (SSF)-derived cellulase for saccharification of the green seaweed Ulva for bioethanol production. Algal Res. 2015, 9, 48–54. [Google Scholar] [CrossRef]
- Del-Cid, A.; Ubilla, P.; Ravanal, M.C.; Medina, E.; Vaca, I.; Levicán, G.; Eyzaguirre, J.; Chávez, R. Cold-active xylanase produced by fungi associated with Antarctic marine sponges. Appl. Biochem. Biotechnol. 2014, 172, 524–532. [Google Scholar] [CrossRef]
- Bastos, S.C.; Pimenta, C.J.; Dias, D.R.; Chalfoun, S.M.; Ange’lico, C.L.; Tavares, L.S. Pectinases from a new strain of Cladosporium cladosporioides (Fres.) De Vries isolated from coffee bean. World J. Agric. Sci. 2013, 9, 167–172. [Google Scholar]
- Bonugli-Santos, R.C.; Durrant, L.R.; da Silva, M.; Sette, L.D. Production of laccase, manganese peroxidase and lignin peroxidase by Brazilian marine-derived fungi. Enzyme Microb. Technol. 2010, 46, 32–37. [Google Scholar] [CrossRef]
- Duarte, A.W.F.; Dayo-Owoyemi, I.; Nobre, F.S.; Pagnocca, F.C.; Chaud, L.C.S.; Pessoa, A.; Felipe, M.G.; Sette, L.D. Taxonomic assessment and enzymes production by yeasts isolated from marine and terrestrial Antarctic samples. Extremophiles 2013, 17, 1023–1035. [Google Scholar] [CrossRef]
- Ball, A.; Truskewycz, A. Polyaromatic hydrocarbon exposure: An ecological impact ambiguity. Environ. Sci. Pollut. Res. 2013, 20, 4311–4326. [Google Scholar] [CrossRef]
- Lee, S.R.; Lee, D.; Eom, H.J.; Rischer, M.; Ko, Y.-J.; Kang, K.S.; Kim, C.S.; Beemelmanns, C.; Kim, K.H. Hybrid polyketides from a Hydractinia-associated Cladosporium sphaerospermum SW67 and their putative biosynthetic origin. Mar. Drugs 2019, 17, 606. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.H.; Wu, G.W.; Zhu, T.J.; Gu, Q.Q.; Li, D.H. Cladosins F and G, two new hybrid polyketides from the deep-sea-derived Cladosporium sphaerospermum 2005-01-E3. J. Asian Nat. Prod. Res. 2015, 17, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; He, X.; Wu, G.; Liu, C.; Lu, C.; Gu, Q.; Che, Q.; Zhu, T.; Zhang, G.; Li, D. Aniline-tetramic acids from the deep-sea-derived fungus Cladosporium sphaerospermum L3P3 cultured with the HDAC inhibitor SAHA. J. Nat. Prod. 2018, 81, 1651–1657. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Kong, F.; Wang, Y.; Fu, P.; Zhum, W. Cladodionen, a cytotoxic hybrid polyketide from the marine-derived Cladosporium sp. OUCMDZ-1635. Mar. Drugs 2018, 16, 71. [Google Scholar] [CrossRef]
- Liang, X.; Huang, Z.H.; Ma, X.; Qi, S.H. Unstable tetramic acid derivatives from the deep-sea-derived fungus Cladosporium sphaerospermum EIODSF 008. Mar. Drugs 2018, 16, 448. [Google Scholar] [CrossRef]
- Huang, Z.-H.; Nong, X.-H.; Liang, X.; Qi, S.-H. New tetramic acid derivatives from the deep-sea-derived fungus Cladosporium sp. SCSIO z0025. Tetrahedron 2018, 74, 2620–2626. [Google Scholar] [CrossRef]
- Ding, L.; Qin, S.; Li, F.; Chi, X.; Laatsch, H. Isolation, antimicrobial activity, and metabolites of fungus Cladosporium sp. associated with red alga Porphyra yezoensis. Curr. Microbiol. 2008, 56, 229–235. [Google Scholar] [CrossRef]
- Qi, S.-H.; Xu, Y.; Xiong, H.-R.; Qian, P.-Y.; Zhang, S. Antifouling and antibacterial compounds from a marine fungus Cladosporium sp. F14. World J. Microbiol. Biotechnol. 2009, 25, 399–406. [Google Scholar] [CrossRef]
- Pang, X.; Lin, X.; Wang, J.; Liang, R.; Tian, Y.; Salendra, L.; Luo, X.; Zhou, X.; Yang, B.; Tu, Z.; et al. Three new highly oxygenated sterols and one new dihydroisocoumarin from the marine sponge-derived fungus Cladosporium sp. SCSIO41007. Steroids 2018, 129, 41–46. [Google Scholar] [CrossRef]
- Gu, B.; Zhang, Y.; Ding, L.; He, S.; Wu, B.; Dong, J.; Zhu, P.; Chen, J.; Zhang, J.; Yan, X. Preparative separation of sulfur-containing diketopiperazines from marine fungus Cladosporium sp. using high-speed counter-current chromatography in stepwise elution mode. Mar. Drugs 2015, 13, 354–365. [Google Scholar] [CrossRef]
- Manríquez, V.; Galdámeza, A.; Veliz, B.; Rovirosa, J.; Díaz-Marrero, A.R.; Cueto, M.; Darias, J.; Martínez, C.; San-Martín, A. N-methyl-1H-indole-2-carboxamide from the marine fungus Cladosporium cladosporioides. J. Chil. Chem. Soc. 2009, 54, 314–316. [Google Scholar] [CrossRef][Green Version]
- Peng, J.; Lin, T.; Wang, W.; Xin, Z.; Zhu, T.; Gu, Q.; Li, D. Antiviral alkaloids produced by the mangrove-derived fungus Cladosporium sp. PJX-41. J. Nat. Prod. 2013, 76, 1133–1140. [Google Scholar] [CrossRef]
- Yang, G.; Nenkep, V.N.; Siwe, X.N.; Leutou, A.S.; Feng, Z.H.; Zhang, D.; Choi, H.D.; Kang, J.S.; Son, B.W. An acetophenone derivative, clavatol, and a benzodiazepine alkaloid, circumdatin A, from the marine-derived fungus Cladosporium. Nat. Prod. Sci. 2009, 15, 130–133. [Google Scholar]
- Kyeremeh, K.; Owusu, K.B.-A.; Ofosuhene, M.; Ohashi, M.; Agyapong, J.; Camas, A.S.; Camas, M. Anti-proliferative and antiplasmodia activity of quinolactacin A2, citrinadin A and butrecitrinadin co-isolated from a Ghanaian mangrove endophytic fungus Cladosporium oxysporum strain BRS2A-AR2F. J. Chem. Appl. 2017, 3, 12. [Google Scholar]
- Wang, P.; Cui, Y.; Cai, C.; Chen, H.; Dai, Y.; Chen, P.; Kong, F.; Yuan, J.; Song, X.; Mei, W.; et al. Two new succinimide derivatives cladosporitins A and B from the mangrove-derived fungus Cladosporium sp. HNWSW-1. Mar. Drugs 2019, 17, 4. [Google Scholar] [CrossRef]
- Rotinsulu, H.; Yamazaki, H.; Sugai, S.; Iwakura, N.; Wewengkang, D.S.; Sumilat, D.A.; Namikoshi, M. Cladosporamide A, a new protein tyrosine phosphatase 1B inhibitor, produced by an Indonesian marine sponge-derived Cladosporium sp. J. Nat. Med. 2018, 72, 779–783. [Google Scholar] [CrossRef]
- Cao, F.; Yang, Q.; Shao, C.L.; Kong, C.J.; Zheng, J.J.; Liu, Y.F.; Wang, C.Y. Bioactive 7-oxabicyclic[6.3.0]lactam and 12-membered macrolides from a gorgonian-derived Cladosporium sp. fungus. Mar. Drugs 2015, 13, 4171–4178. [Google Scholar] [CrossRef]
- Huang, C.; Chen, T.; Yan, Z.; Guo, H.; Hou, X.; Jiang, L.; Long, Y. Thiocladospolide E and cladospamide A, novel 12-membered macrolide and macrolide lactam from mangrove endophytic fungus Cladosporium sp. SCNU-F0001. Fitoterapia 2019, 137, 104246. [Google Scholar] [CrossRef]
- Bai, M.; Zheng, C.J.; Tang, D.Q.; Zhang, F.; Wang, H.Y.; Chen, G.Y. Two new secondary metabolites from a mangrove-derived fungus Cladosporium sp. JS1-2. J. Antibiot. 2019, 72, 779–782. [Google Scholar] [CrossRef]
- Zhang, H.; Tomoda, H.; Tabata, N.; Miura, H.; Namikoshi, M.; Yamaguchi, Y.; Masuma, R.; Omura, S. Cladospolide D, a new 12-membered macrolide antibiotic produced by Cladosporium sp. FT-0012. J. Antibiot. 2001, 54, 635–641. [Google Scholar] [CrossRef]
- Wuringege Guo, Z.K.; Wei, W.; Jiao, R.H.; Yan, T.; Zang, L.Y.; Jiang, R.; Tan, R.X.; Ge, H.M. Polyketides from the plant endophytic fungus Cladosporium sp. IFB3lp-2. J. Asian Nat. Prod. Res. 2013, 15, 928–933. [Google Scholar] [CrossRef]
- Jadulco, R.; Proksch, P.; Wray, V.; Sudarsono Berg, A.; Gräfe, U. New macrolides and furan carboxylic acid derivative from the sponge-derived fungus Cladosporium herbarum. J. Nat. Prod. 2001, 64, 527–530. [Google Scholar] [CrossRef]
- He, Z.H.; Zhang, G.; Yan, Q.X.; Zou, Z.B.; Xiao, H.X.; Xie, C.L.; Tang, X.X.; Luo, L.Z.; Yang, X.W. Cladosporactone A, a unique polyketide with 7-methylisochromen-3-one skeleton from the deep-sea-derived fungus Cladosporium cladosporioides. Chem. Biodivers. 2020, 17, e2000158. [Google Scholar] [CrossRef]
- Gao, C.H.; Nong, X.H.; Qi, S.H.; Luo, X.M.; Zhang, S.; Xiong, H.R. A new nine-membdered lactone from a marine fungus Cladosporium sp. F14. Chin. Chem. Lett. 2010, 21, 1355–1357. [Google Scholar] [CrossRef]
- Gesner, S.; Cohen, N.; Ilan, M.; Yarden, O.; Carmeli, S. Pandangolide 1a, a metabolite of the sponge-associated fungus Cladosporium sp., and the absolute stereochemistry of pandangolide 1 and iso-cladospolide B. J. Nat. Prod. 2005, 68, 1350–1353. [Google Scholar] [CrossRef]
- Wang, W.; Feng, H.; Sun, C.; Che, Q.; Zhang, G.; Zhu, T.; Li, D. Thiocladospolides F-J, antibacterial sulfur containing 12-membered macrolides from the mangrove endophytic fungus Cladosporium oxysporum HDN13-314. Phytochemistry 2020, 178, 112462. [Google Scholar] [CrossRef]
- Zhang, F.Z.; Li, X.M.; Meng, L.H.; Wang, B.G. Cladocladosin A, an unusual macrolide with bicyclo 5/9 ring system, and two thiomacrolides from the marine mangrove-derived endophytic fungus, Cladosporium cladosporioides MA-299. Bioorg. Chem. 2020, 101, 103950. [Google Scholar] [CrossRef]
- Shigemori, H.; Kasai, Y.; Komatsu, K.; Tsuda, M.; Mikami, Y.; Kobayashi, J. Sporiolides A and B, new cytotoxic twelve-membered macrolides from a marine-derived fungus Cladosporium pecies. Mar. Drugs 2004, 2, 164–169. [Google Scholar] [CrossRef]
- Peng, X.; Wang, Y.; Zhu, G.; Zhu, W. Fatty acid derivatives from the halotolerant fungus Cladosporium cladosporioides. Magn. Reson. Chem. 2018, 56, 18–24. [Google Scholar] [CrossRef]
- Ai, W.; Lin, X.; Wang, Z.; Lu, X.; Mangaladoss, F.; Yang, X.; Zhou, X.; Tu, Z.; Liu, Y. Cladosporone A, a new dimeric tetralone from fungus Cladosporium sp. KcFL6’ derived of mangrove plant Kandelia candel. J. Antibiot. 2015, 68, 213–215. [Google Scholar] [CrossRef]
- Zhang, Z.; He, X.; Liu, C.; Che, Q.; Zhu, T.; Gu, Q.; Li, D. Clindanones A and B and cladosporols F and G, polyketides from the deep-sea derived fungus Cladosporium cladosporioides HDN14-342. RSC Adv. 2016, 6, 76498–76504. [Google Scholar] [CrossRef]
- Li, H.L.; Li, X.M.; Mándi, A.; Antus, S.; Li, X.; Zhang, P.; Liu, Y.; Kurtán, T.; Wang, B.G. Characterization of cladosporols from the marine algal-derived endophytic fungus Cladosporium cladosporioides en-399 and configurational revision of the previously reported cladosporol derivatives. J. Org. Chem. 2017, 82, 9946–9954. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhou, L.; Kong, F.; Ma, Q.; Xie, Q.; Li, J.; Dai, H.; Guo, L.; Zhao, Y. Altertoxins with quorum sensing inhibitory activities from the marine-derived fungus Cladosporium sp. KFD33. Mar. Drugs 2020, 18, 67. [Google Scholar] [CrossRef] [PubMed]
- Zhen, F.; Zhang-hua, S.; Yu-chan, C.; Hao-hua, L.; Wei-min, Z. Cladosperanol A, a new dimeric tetralone from marine-derived fungus cladosporium perangustum. Nat. Prod. Res. Dev. 2016, 28, 486–489. [Google Scholar]
- Fan, Z.; Sun, Z.H.; Liu, H.X.; Chen, Y.C.; Li, H.H.; Zhang, W.M. Perangustols A and B, a pair of new azaphilone epimers from a marine sediment-derived fungus Cladosporium perangustm FS62. J. Asian Nat. Prod. Res. 2016, 18, 1024–1029. [Google Scholar] [CrossRef]
- Wu, J.T.; Zheng, C.J.; Zhang, B.; Zhou, X.M.; Zhou, Q.; Chen, G.Y.; Zeng, Z.E.; Xie, J.L.; Han, C.R.; Lyu, J.X. Two new secondary metabolites from a mangrove-derived fungus Cladosporium sp. JJM22. Nat. Prod. Res. 2019, 33, 34–40. [Google Scholar] [CrossRef]
- Fan, C.; Zhou, G.; Wang, W.; Zhang, G.; Zhu, T.; Che, Q.; Li, D. Tetralone derivatives from a deep-sea derived fungus Cladosporium sp. HDN17-58. Nat. Prod. Commun. 2021, 16, 1934578X211008322. [Google Scholar] [CrossRef]
- Ma, R.-Z.; Zheng, C.-J.; Zhang, B.; Yang, J.-Y.; Zhou, X.-M.; Song, X.-M. Two New naphthalene-chroman coupled derivatives from the mangrove-derived fungus Cladosporium sp. JJM22. Phytochem. Lett. 2021, 43, 114–116. [Google Scholar] [CrossRef]
- Li, Z.; Yang, J.Y.; Caj, J.; Ouyang, Z.J.; Zhou, C.H.; Chen, G.Y.; Zhou, X.M. Study on secondary metabolites of endophytic fungus Cladosporium sp. JJM22 hosted in Ceriops tagal. Zhongguo Zhong Yao Za Zhi 2021, 46, 2079–2083. [Google Scholar]
- Wang, C.N.; Lu, H.M.; Gao, C.H.; Guo, L.; Zhan, Z.Y.; Wang, J.J.; Liu, Y.H.; Xiang, S.T.; Wang, J.; Luo, X.W. Cytotoxic benzopyranone and xanthone derivatives from a coral symbiotic fungus Cladosporium halotolerans GXIMD 02502. Nat. Prod. Res. 2020, 1–8. [Google Scholar] [CrossRef]
- Silber, J.; Ohlendorf, B.; Labes, A.; Wenzel-Storjohann, A.; Näther, C.; Imhoff, J.F. Malettinin E, an antibacterial and antifungal tropolone produced by a marine Cladosporium strain. Front. Mar. Sci. 2014, 1, 35. [Google Scholar] [CrossRef]
- Liu, Y.; Kurtán, T.; Yun Wang, C.; Han Lin, W.; Orfali, R.; Müller, W.E.; Daletos, G.; Proksch, P. Cladosporinone, a new viriditoxin derivative from the hypersaline lake derived fungus Cladosporium cladosporioides. J. Antibiot. 2016, 69, 702–706. [Google Scholar] [CrossRef]
- Wang, L.; Han, X.; Zhu, G.; Wang, Y.; Chairoungdua, A.; Piyachaturawat, P.; Zhum, W. Polyketides from the endophytic fungus Cladosporium sp. isolated from the mangrove plant Excoecaria agallocha. Front. Chem. 2018, 6, 344. [Google Scholar] [CrossRef]
- Jadulco, R.; Brauers, G.; Edrada, R.A.; Ebel, R.; Wray, V.; Sudarsono, S.; Proksch, P. New metabolites from sponge-derived fungi Curvularia lunata and Cladosporium herbarum. J. Nat. Prod. 2002, 65, 730–733. [Google Scholar] [CrossRef]
- Zhang, B.; Wu, J.T.; Zheng, C.J.; Zhou, X.M.; Yu, Z.X.; Li, W.S.; Chen, G.Y.; Zhu, G.Y. Bioactive cyclohexene derivatives from a mangrove-derived fungus Cladosporium sp. JJM22. Fitoterapia 2021, 149, 104823. [Google Scholar] [CrossRef]
- San-Martin, A.; Painemal, K.; Diaz, Y.; Martinez, C.; Rovirosa, J. Metabolites from the marine fungus Cladosporium cladosporioides. J. Argent. Chem. Soc. 2005, 93, 247–251. [Google Scholar]
- Kuznetsova, T.A.; Afiyatullov, S.A.; Denisenko, V.A.; Pivkin, M.V.; Elyakov, G.B. Sterols from a marine isolate of the fungus Cladosporium sphaerospermum Penz. Biochem. Syst. Ecol. 1998, 26, 365–366. [Google Scholar] [CrossRef]
- Yu, M.L.; Guan, F.F.; Cao, F.; Jia, Y.L.; Wang, C.Y. A new antiviral pregnane from a gorgonian-derived Cladosporium sp. fungus. Nat. Prod. Res. 2018, 32, 1260–1266. [Google Scholar] [CrossRef]
- Lee, S.R.; Kang, H.; Yoo, M.J.; Yu, J.S.; Lee, S.; Yi, S.A.; Beemelmanns, C.; Lee, J.; Kim, K.H. Anti-adipogenic pregnane steroid from a Hydractinia-associated fungus, Cladosporium sphaerospermum SW67. Nat. Prod. Sci. 2020, 26, 230–235. [Google Scholar]
- Gallo, M.L.; Seldes, A.M.; Cabrera, G.M. Antibiotic long-chain and α,β-unsaturated aldehydes from the culture of the marine fungus Cladosporium sp. Biochem. Syst. Ecol. 2004, 32, 545–551. [Google Scholar] [CrossRef]
- AlMatar, M.; Makky, E.A. Cladosporium cladosporioides from the perspectives of medical and biotechnological approaches. 3 Biotech 2016, 6, 4. [Google Scholar] [CrossRef]
- Armisén, R.; Galatas, F.; Hispanagar, A.S. Agar. In Handbook of Hydrocolloids; Phillips, G.O., Williams, P.A., Eds.; Woodhead Publishing: Cambridge, UK, 2000; pp. 21–40. [Google Scholar]
- Wang, J.; Jiang, X.; Mou, H.; Guan, H. Anti-oxidation of agar oligosaccharides produced by agarase from a marine bacterium. J. Appl. Phycol. 2004, 16, 333–340. [Google Scholar] [CrossRef]
- Necas, J.; Bartosikova, L. Carrageenan: A review. Vet. Med. 2013, 58, 187–205. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Mohamed, S.G.A.; Altyar, A.E.; Mohamed, G.A. Natural products of the fungal genus Humicola: Diversity, biological activity, and industrial importance. Curr. Microbiol. 2021, 78, 2488–2509. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Mohamed, S.G.A.; Sindi, I.A.; Mohamed, G.A. Biologically active secondary metabolites and biotechnological applications of species of the family Chaetomiaceae (Sordariales): An updated review from 2016 to 2021. Mycol. Prog. 2021, 20, 595–639. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Mohamed, S.G.A.; Altyar, A.E.; Mohamed, G.A. Genus Thielavia: Phytochemicals, industrial importance and biological relevance. Nat. Prod. Res. 2021, 1–16. [Google Scholar] [CrossRef]
- Collins, T.; Gerday, C.; Feller, G. Xylanases, xylanase families and extremophilic xylanases. FEMS Microb. Rev. 2005, 29, 3–23. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, D.T.; Tiwari, R.; Sah, A.K.; Raghukumara, C. Enhanced production of laccase by a marine fungus during treatment of colored effluents and synthetic dyes. Enzym. Microb. Technol. 2006, 38, 504–511. [Google Scholar] [CrossRef]
- Kuwahara, M.; Glenn, J.K.; Morgan, M.A.; Gold, M.H. Separation and characterization of two extracellular H2O2 dependent oxidases from ligninolytic cultures of Phanerochaete chrysosporium. FEBS Lett. 1984, 169, 247–250. [Google Scholar] [CrossRef]
- Raghukumar, C.; Muraleedharan, U.; Gaud, V.R.; Mishra, R. Simultaneous detoxification and decolorization of molasses spent wash by the immobilized white-rot fungus Flavodon flavusisolated from a marine habitat. Enzym. Microb. Technol. 2004, 35, 197–202. [Google Scholar] [CrossRef]
- Da Silva, M.; Passarini, M.R.Z.; Bonugli, R.C.; Sette, L.D. Cnidarian-derived filamentous fungi from Brazil: Isolation, characterisation and RBBR decolourisation screening. Environ. Technol. 2008, 29, 1331–1339. [Google Scholar] [CrossRef]
- Panno, L.; Voyron, S.; Anastasi, A.; Mussat Sarto, R.; Varese, G.C. Biodiversity of marine fungi associated with the seagrass Posidonia oceanica: An ecological and biotechnological perspective. Biol. Mar. Mediterr. 2011, 18, 85–88. [Google Scholar]
- Taskin, M.; Esim, N.; Genisel, M.; Ortucu, S.; Hasenekoglu, I.; Canli, O.; Erdal, S. Enhancement of invertase production by Aspergillus niger OZ-3 using low-intensity static magnetic fields. Prep. Biochem. Biotechnol. 2013, 43, 177–188. [Google Scholar] [CrossRef]
- Veana, F.; Martínez-Hernández, J.L.; Aguilar, N.; Rodríguez-Herrera, R.; Michelena, G. Utilization of molasses and sugar cane bagasse for production of fungal invertase in solid state fermentation using Aspergillus niger GH1. Braz. J. Microbiol. 2014, 5, 373–377. [Google Scholar] [CrossRef]
- Yadav, S.; Chandra, R. Biodegradation of organic compounds of molasses melanoidin (MM) from biomethanated distillery spent wash (BMDS) during the decolourisation by a potential bacterial consortium. Biodegradation 2012, 23, 609–620. [Google Scholar] [CrossRef]
- Gavrilescua, M.; Chisti, Y. Biotechnology-a sustainable alternative for chemical industry. Biotechnol. Adv. 2005, 23, 471–499. [Google Scholar] [CrossRef]
- Winkler, C.K.; Faber, K.; Hall, M. Biocatalytic reduction of activated C=C-bonds and beyond: Emerging trends. Curr. Opin. Chem. Biol. 2018, 43, 97–105. [Google Scholar] [CrossRef]
- Garzon-Posse, F.; Becerra-Figueroa, L.; Hernandez-Arias, J.; Gamba-Sanchez, D. Whole cells as biocatalysts in organic transformations. Molecules 2018, 23, 1265. [Google Scholar] [CrossRef]
- Khare, R.; Pandey, J.; Smriti, R.R. The importance and applications of Knoevenagel reaction (brief review). Orient. J. Chem. 2019, 35, 423–429. [Google Scholar] [CrossRef]
- Rocha, L.C.; Ferreira, H.V.; Luiz, R.F.; Sette, L.D.; Porto, A.L. Stereoselective bioreduction of 1-(4-methoxyphenyl) ethanone by whole cells of marine-derived fungi. Mar. Biotechnol. 2012, 14, 358–362. [Google Scholar] [CrossRef]
- Rocha, L.C.; Seleghim, M.H.R.; Comasseto, J.V.; Sette, L.D.; Porto, A.L.M. Stereoselective bioreduction of alpha-azido ketones by whole cells of marine-derived fungi. Mar. Biotechnol. 2015, 17, 736–742. [Google Scholar] [CrossRef] [PubMed]
- Athanasellis, G.; Igglessi-Markopoulou, O.; Markopoulos, J. Tetramic and tetronic acids as scaffolds in bioinorganic and bioorganic chemistry. Bioinorg. Chem. Appl. 2010, 2010, 315056. [Google Scholar] [CrossRef] [PubMed]
- Royles, B.J.L. Naturally occurring tetramic acids: Structure, isolation, and synthesis. Chem. Rev. 1995, 95, 1981–2001. [Google Scholar] [CrossRef]
- Mo, X.; Li, Q.; Ju, J. Naturally occurring tetramic acid products: Isolation, structure elucidation and biological activity. RSC Adv. 2014, 4, 50566–50593. [Google Scholar] [CrossRef]
- Jiang, M.; Chen, S.; Li, J.; Liu, L. The Biological and Chemical Diversity of Tetramic Acid Compounds from Marine-Derived Microorganisms. Mar. Drugs 2020, 18, 114. [Google Scholar] [CrossRef]
- Gomes, N.G.M.; Pereira, R.B.; Andrade, P.B.; Valentao, P. Double the chemistry, double the fun: Structural diversity and biological activity of marine-derived diketopiperazine dimers. Mar. Drugs 2019, 17, 551. [Google Scholar] [CrossRef]
- Huang, R.M.; Yi, X.X.; Zhou, Y.Y.; Su, X.D.; Peng, Y.; Gao, C.H. An update on 2,5-diketopiperazines from marine organisms. Mar. Drugs 2014, 12, 6213–6235. [Google Scholar] [CrossRef]
- Borgman, P.; Lopez, R.D.; Lane, A.L. The expanding spectrum of diketopiperazine natural product biosynthetic pathways containing cyclodipeptide synthases. Org. Biomol. Chem. 2019, 17, 2305–2314. [Google Scholar] [CrossRef]
- Song, Z.; Hou, Y.; Yang, Q.; Li, X.; Wu, S. Structures and Biological Activities of Diketopiperazines from Marine Organisms: A Review. Mar. Drugs 2021, 19, 403. [Google Scholar] [CrossRef]
- Urban, S.; Hickford, S.J.H.; Blunt, J.W.; Munro, M.H.G.; Yue, Y.F.; Chang, Q.H.; Zhu, H.J.; Cao, F. Bioactive marine alkaloids. Curr. Org. Chem. 2000, 4, 765–807. [Google Scholar] [CrossRef]
- Meng, Z.H.; Sun, T.T.; Zhao, G.Z.; Yue, Y.-F.; Chang, Q.-H.; Zhu, H.-J.; Cao, F. Marine-derived fungi as a source of bioactive indole alkaloids with diversified structures. Mar. Life Sci. Technol. 2021, 3, 44–61. [Google Scholar] [CrossRef]
- Islam, M.T.; Mubarak, M.S. Pyrrolidine alkaloids and their promises in pharmacotherapy. Adv. Tradit. Med. 2020, 20, 13–22. [Google Scholar] [CrossRef]
- Netz, N.; Opatz, T. Marine indole alkaloids. Mar. Drugs 2015, 13, 4814–4914. [Google Scholar] [CrossRef]
- Woodward, R.B. Struktur und biogenese der makrolide. Angew. Chem. 1957, 69, 50–58. [Google Scholar] [CrossRef]
- Janas, A.; Przybylski, P. 14- and 15-membered lactone macrolides and their analogues and hybrids: Structure, molecular mechanism of action and biological activity. Eur. J. Med. Chem. 2019, 182, 111662. [Google Scholar] [CrossRef]
- Zhang, H.; Zou, J.; Yan, X.; Chen, J.; Cao, X.; Wu, J.; Liu, Y.; Wang, T. Marine-Derived Macrolides 1990–2020: An Overview of Chemical and Biological Diversity. Mar. Drugs 2021, 19, 180. [Google Scholar] [CrossRef]
- Ray Choudhury, A.; Mukherjee, S. Deconjugated butenolide: A versatile building block for asymmetric catalysis. Chem. Soc. Rev. 2020, 49, 6755–6788. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, V.; Ghosh, S. Butenolide: A Novel Synthesis and Biological Activities, 1st ed.; LAP Lambert Academic Publishing: Saarbrücken, Germany, 2012. [Google Scholar]
- Gibson, M.Z.; Nguyen, M.A.; Zingales, S.K. Design, synthesis, and evaluation of (2-(pyridinyl)methylene)-1-tetralone chalcones for anticancer and antimicrobial activity. Med. Chem. 2018, 14, 333–343. [Google Scholar] [CrossRef]
- Gauni, B.; Mehariya, K.; Shah, A.; Duggirala, S.M. Tetralone scaffolds and their potential therapeutic applications. Lett. Drug Des. Disc. 2021, 18, 222–238. [Google Scholar] [CrossRef]
- Zurlo, D.; Leone, C.; Assante, G.; Salzano, S.; Renzone, G.; Scaloni, A.; Foresta, C.; Colantuoni, V.; Lupo, A. Cladosporol a stimulates G1-phase arrest of the cell cycle by up-regulation of p21waf1/cip1 expression in human colon carcinoma HT-29 cells. Mol. Carcinog. 2013, 52, 1–17. [Google Scholar] [CrossRef]
- Zurlo, D.; Assante, G.; Moricca, S.; Colantuoni, V.; Lupo, A. Cladosporol A, a new peroxisome proliferator-activated receptor γ (PPARγ) ligand, inhibits colorectal cancer cells proliferation through β-catenin/TCF pathway inactivation. Biochim. Biophys. Acta 2014, 1840, 2361–2372. [Google Scholar] [CrossRef]
- Mulrooey, C.A.; O’Brien, E.M.; Morgan, B.J.; Kozlowski, M.C. Perylenequinones: Isolation, synthesis, and biological activity. Eur. J. Org. Chem. 2012, 2012, 3887–3904. [Google Scholar] [CrossRef]
- Stack, M.E.; Prival, M.J. Mutagenicity of the Alternaria metabolites altertoxins I., II, and III. Appl. Environ. Microbiol. 1986, 52, 718–722. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Mohamed, G.A. Naturally occurring naphthalenes: Chemistry, biosynthesis, structural elucidation, and biological activities. Phytochem. Rev. 2016, 15, 279–295. [Google Scholar] [CrossRef]
- Vieira, L.; Kijjoa, A. Naturally-occurring xanthones: Recent developments. Curr. Med. Chem. 2005, 12, 2413–2446. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Abdallah, H.M.; El-Halawany, A.M.; Radwan, M.F.; Shehata, I.A.; Al-Harshany, E.M.; Zayed, M.F.; Mohamed, G.A. Garcixanthones B and C, new xanthones from the pericarps of Garcinia mangostana and their cytotoxic activity. Phytochem. Lett. 2018, 25, 12–16. [Google Scholar] [CrossRef]
- Mohamed, G.A.; Al-Abd, A.M.; El-Halawany, A.M.; Abdallah, H.M.; Ibrahim, S.R.M. New xanthones and cytotoxic constituents from Garcinia mangostana fruit hulls against human hepatocellular, breast, and colorectal cancer cell lines. J. Ethnopharmacol. 2017, 198, 302–312. [Google Scholar] [CrossRef]
- Guo, H.; Roman, D.; Beemelmanns, C. Tropolone natural products. Nat. Prod. Rep. 2019, 36, 1137–1155. [Google Scholar] [CrossRef]
- Choque, E.; El Rayess, Y.; Raynal, J.; Mathieu, F. Fungal naphtho-γ-pyrones—Secondary metabolites of industrial interest. Appl. Microbiol. Biotechnol. 2015, 99, 1081–1096. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Nozawa, K.; Nakajima, S.; Kawai, K. Structure revision of mycotoxin, viriditoxin, and its derivatives. Chem. Pharm. Bull. 1990, 38, 3180–3181. [Google Scholar] [CrossRef]
- De Rosa, S.; De Giulio, A.; Iodice, C.; Alcaraz, M.J.; Paya, M. Long-chain aldehydes from the red alga, Corallina Mediterr. Phytochemistry 1995, 40, 995–996. [Google Scholar] [CrossRef]
- Suzuki, M.; Kurosawa, E.; Kurata, K. (E)-2-Tridecyl-2-heptadecenal, an unusual metabolite from the red alga Laurencia sp. Bull. Chem. Soc. Jpn. 1987, 60, 3793–3794. [Google Scholar] [CrossRef]
- Wulandari, A.P.; Examinati, R.R.I.N.; Madihah; Huspa, D.H.P.; Andayaningsih, P.; Andayaningsih, P. Cytotoxicity of metabolites produced by endophytic fungus Cladosporium sp. isolated from marine macroalgae on in-vitro MCF-7, HeLa, and DU-145 cell lines. Int. J. Pharm. Pharm. Sci. 2018, 10, 72–76. [Google Scholar] [CrossRef]
- Hulikere, M.M.; Joshi, C.G.; Ananda, D.; Poyya, J.; Nivya, T. Antiangiogenic, wound healing and antioxidant activity of Cladosporium cladosporioides (Endophytic Fungus) isolated from seaweed (Sargassum wightii). Mycology 2016, 7, 203–211. [Google Scholar] [CrossRef]
- Xiong, H.; Qi, S.; Xu, Y.; Miao, L.; Qian, P.Y. Antibiotic and antifouling compound production by the marine-derived fungus Cladosporium sp. F14. J. Hydro-Environ. Res. 2009, 2, 264–270. [Google Scholar] [CrossRef]
- Hulikere, M.M.; Joshi, C.G.; Danagoudar, A.; Poyya, J.; Kudva, A.K.; Dhananjaya, B.L. Biogenic synthesis of gold nanoparticles by marine endophytic fungus Cladosporium cladosporioides isolated from seaweed and evaluation of their antioxidant and antimicrobial properties. Process Biochem. 2017, 63, 137–144. [Google Scholar]
- Ameen, F.; Al-Homaidan, A.A.; Al-Sabr, A.; Almansob, A.; AlNAdhari, S. Anti-oxidant, anti-fungal and cytotoxic effects of silver nanoparticles synthesized using marine fungus Cladosporium halotolerans. Appl. Nanosci. 2021. [Google Scholar] [CrossRef]
- Arnone, A.; Assante, G.; Modugno, V.D.; Merlini, L.; Nasini, G. Perylenequinones from cucumber seedlings infected with Cladosporium cucumerinum. Phytochemisry 1988, 27, 1675–1678. [Google Scholar] [CrossRef]
- Arnone, A.; Camarda, L.; Nasini, G.; Merlini, L. Secondary mould metabolites. Part 13. Fungal perylenequinones: Phleichrome, isophleichrome, and their endoperoxides. J. Chem. Soc. Perkin Trans. 1 1985, 1387–1392. [Google Scholar] [CrossRef]
- Arone, A.; Assante, G.; Merlini, L.; Nasini, G. Structure and stereochemistry of cladochrome D and E, novel perylenequinone pigment from Cladosporium cladosporioides. Gazetta Chem. Ital. 1989, 119, 557–559. [Google Scholar]
- Mino, Y.; Idonuma, T.; Sakai, R. Effect of Phleichrome Produced by the Timothy Leaf Spot Fungus, Cladosporium phlei on the Invertases from the Host Leaves. Ann. Phytopathol. Soc. Jpn. 1979, 45, 463–467. [Google Scholar] [CrossRef]
- Sassa, T.; Negoro, T.; Ueki, H. Production and Characterization of a new fungal metabolite, cotylenol. Agric. Biol. Chem. 1972, 36, 2281–2285. [Google Scholar] [CrossRef][Green Version]
- Sassa, T.; Ooi, T.; Nukina, M.; Ikeda, M.; Kato, N. Structural confirmation of cotylenin A, a novel fusicoccane-diterpene glycoside with potent plant growth-regulating activity from Cladosporium fungus sp. 501-7W. Biosci. Biotechnol. Biochem. 1998, 62, 1815–1818. [Google Scholar] [CrossRef]
- Sassa, T.; Togashi, M.; Kitaguchi, T. The structures of cotylenins A., B., C., D and E. Agric. Biol. Chem. 1975, 39, 1735–1744. [Google Scholar] [CrossRef]
- Sassa, T. Cotylenins, 13C NMR of cotylenins. Agric. Biol. Chem. 1979, 43, 385–387. [Google Scholar]
- Sassa, T. Cotylenins, leaf growth substances produced by a fungus. Part, I. Isolation and characterization of cotylenins A and B. Agric. Biol. Chem. 1971, 35, 1415–1418. [Google Scholar]
- Baral, B.; Akhgari, A.; Metsä-Ketelä, M. Activation of microbial secondary metabolic pathways: Avenues and challenges. Synth. Syst. Biotechnol. 2018, 3, 163–178. [Google Scholar] [CrossRef]
- Reich, M.; Labes, A. How to boost marine fungal research: A first step towards a multidisciplinary approach by combining molecular fungal ecology and natural products chemistry. Mar. Genom. 2017, 36, 57–75. [Google Scholar] [CrossRef]
- Scharf, D.H.; Brakhage, A.A. Engineering fungal secondary metabolism: A roadmap to novel compounds. J. Biotechnol. 2013, 163, 179–183. [Google Scholar] [CrossRef]
- Reen, F.J.; Romano, S.; Dobson, A.D.W.; O’Gara, F. The sound of silence: Activating silent biosynthetic gene clusters in marine microorganisms. Mar. Drugs 2015, 13, 4754–4783. [Google Scholar] [CrossRef]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Scherlach, K.; Hertweck, C. Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 2009, 7, 1753–1760. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.C.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J. Nat. Prod. 2007, 70, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Yeh, H.H.; Ahuja, M.; Chiang, Y.M.; Oakley, C.E.; Moore, S.; Yoon, O.; Hajovsky, H.; Bok, J.W.; Keller, N.P.; Wang, C.C.; et al. Resistance Gene-Guided Genome Mining: Serial Promoter Exchanges in Aspergillus nidulans Reveal the Biosynthetic Pathway for Fellutamide B, a Proteasome Inhibitor. ACS Chem. Biol. 2016, 11, 2275–2284. [Google Scholar] [CrossRef]
- Williams, R.B.; Henrikson, J.C.; Hoover, A.R.; Lee, A.E.; Cichewicz, R.H. Epigenetic remodeling of the fungal secondary metabolome. Org. Biomol. Chem. 2008, 6, 1895–1897. [Google Scholar] [CrossRef]
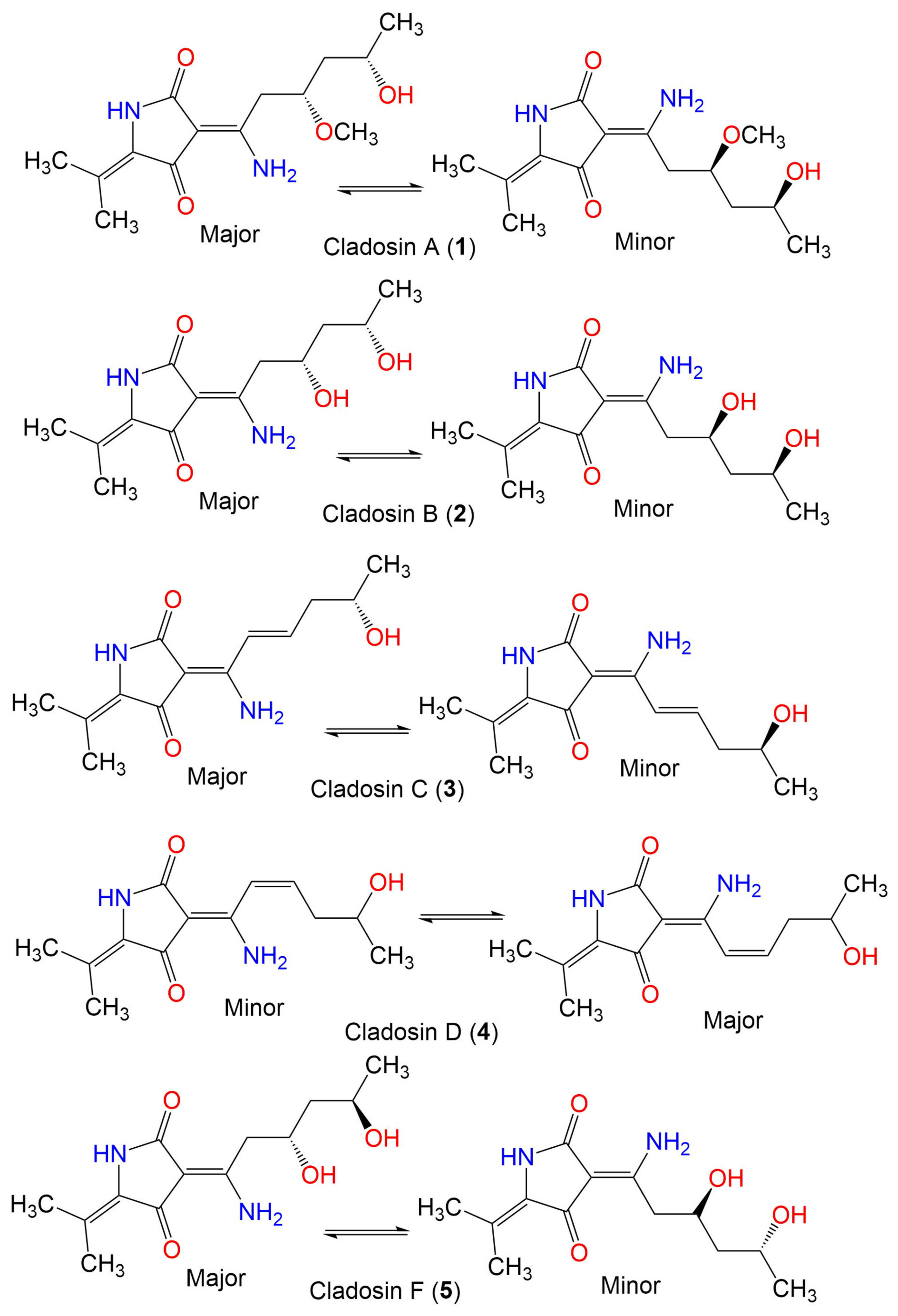
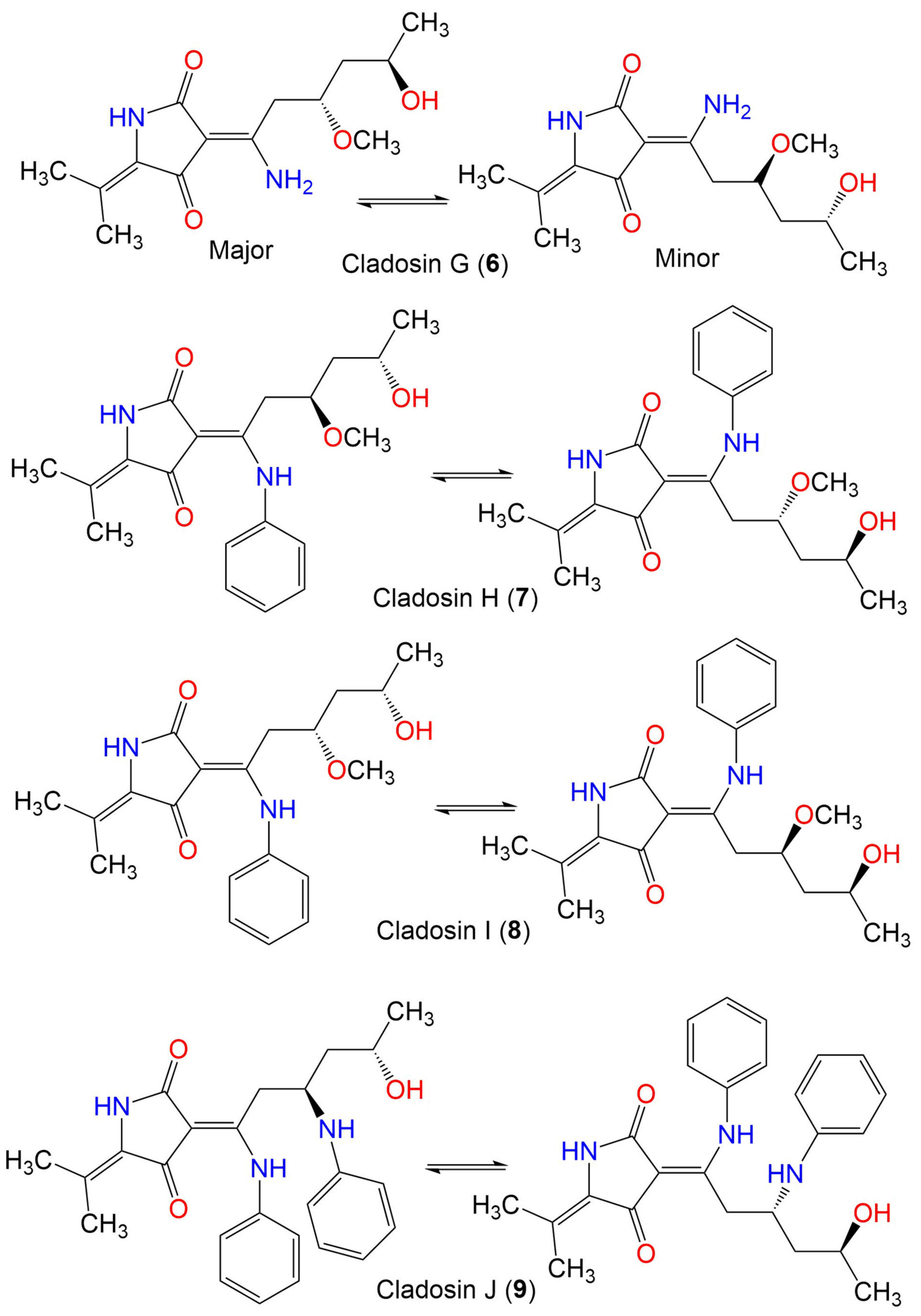

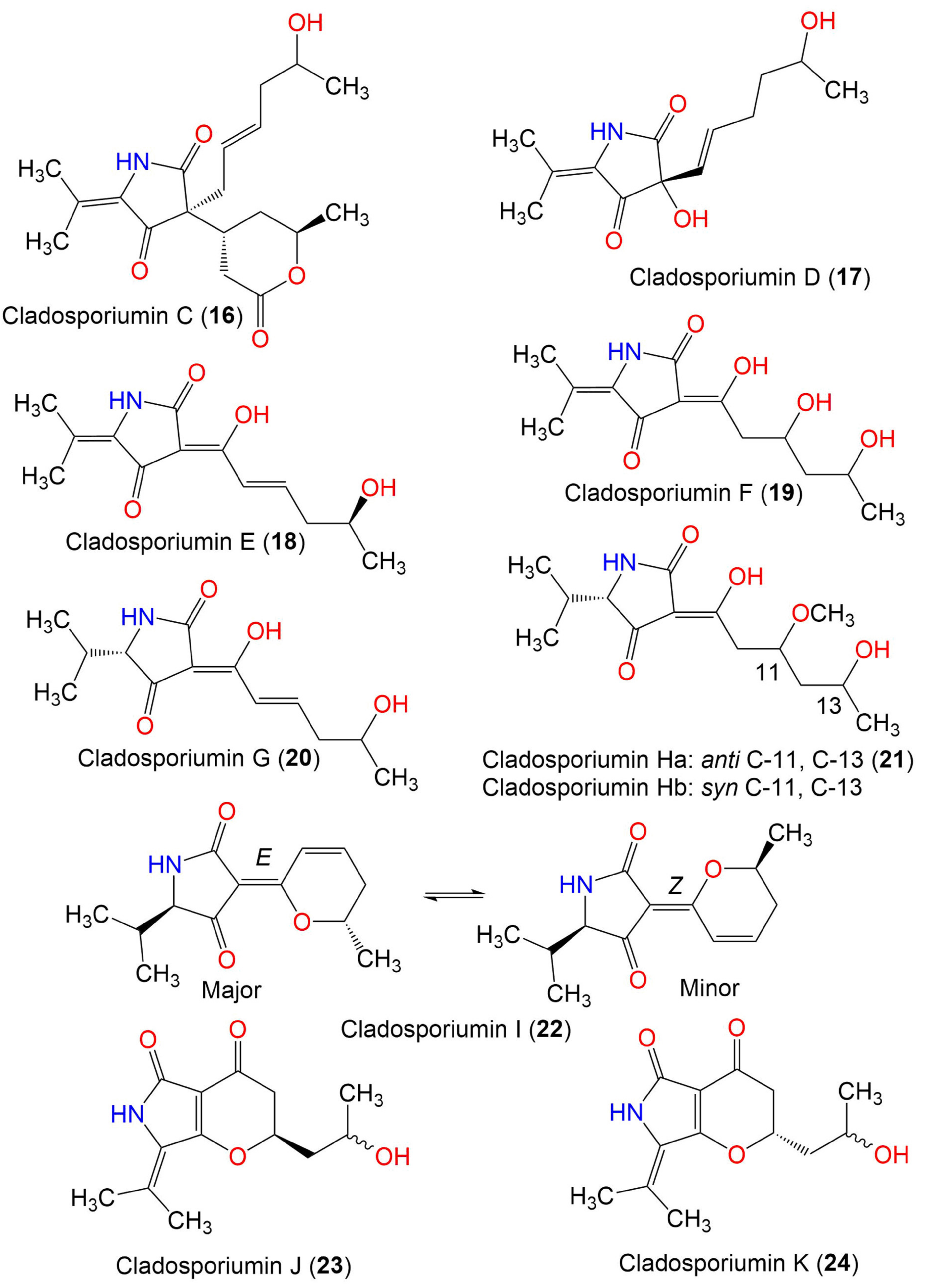
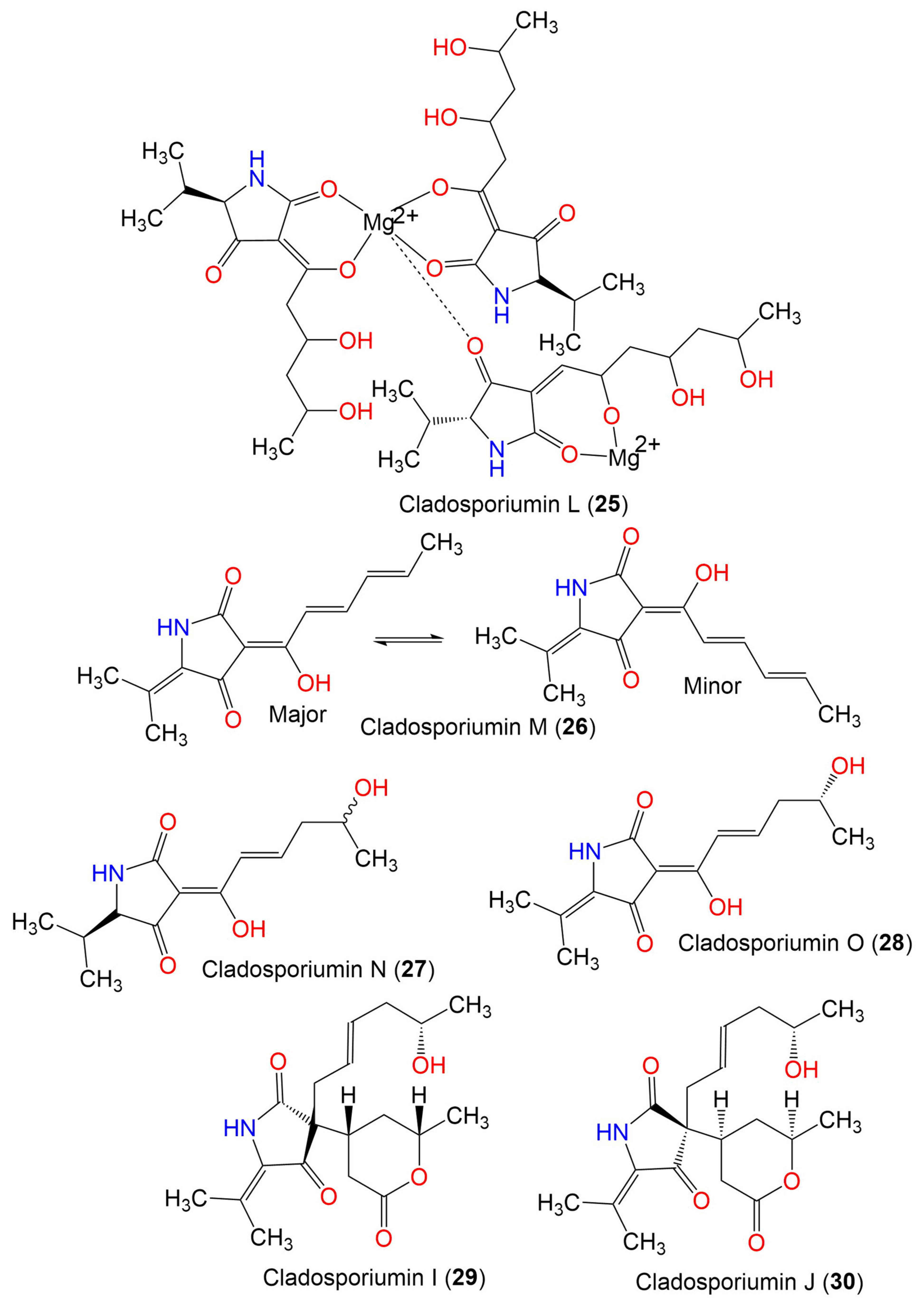
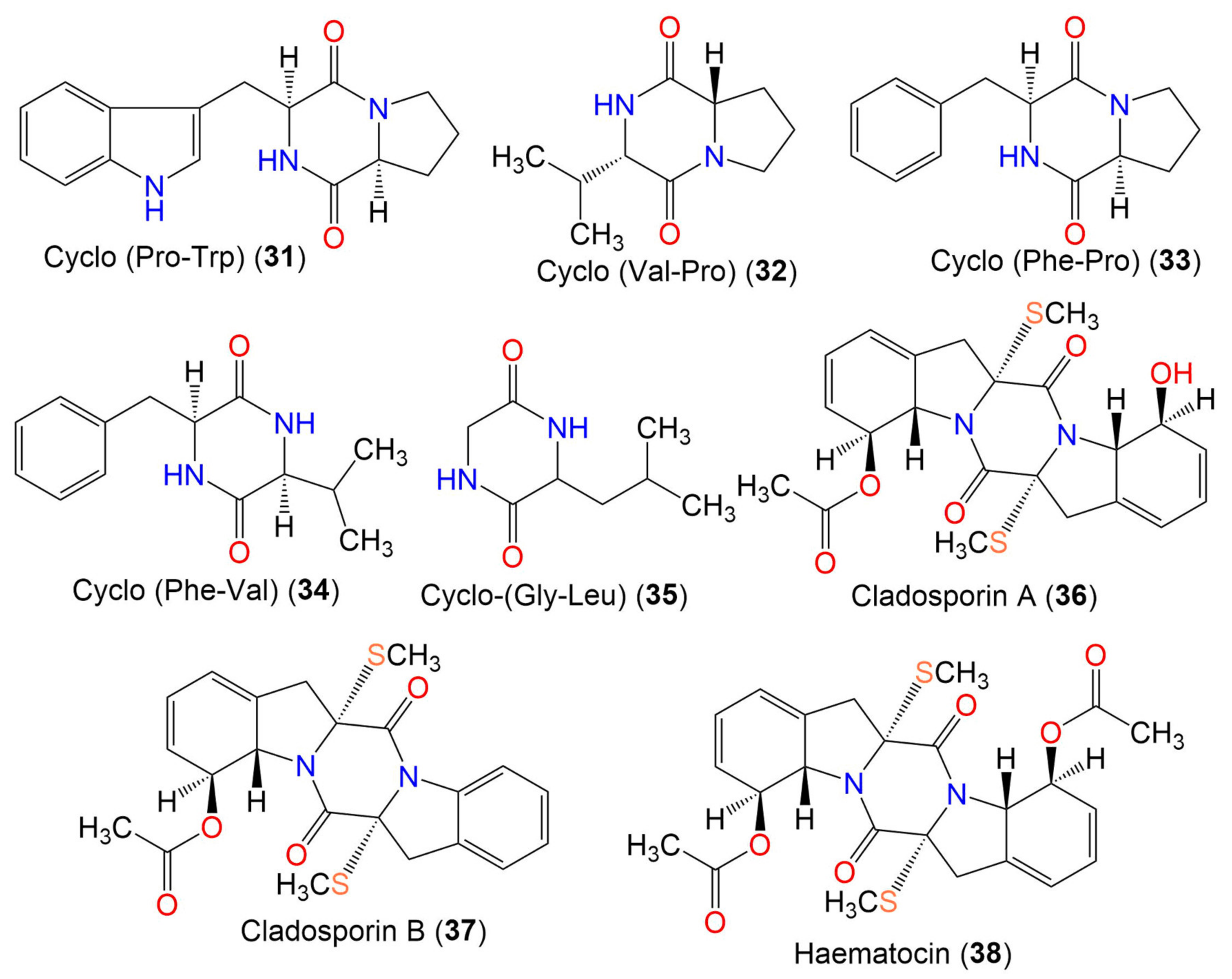

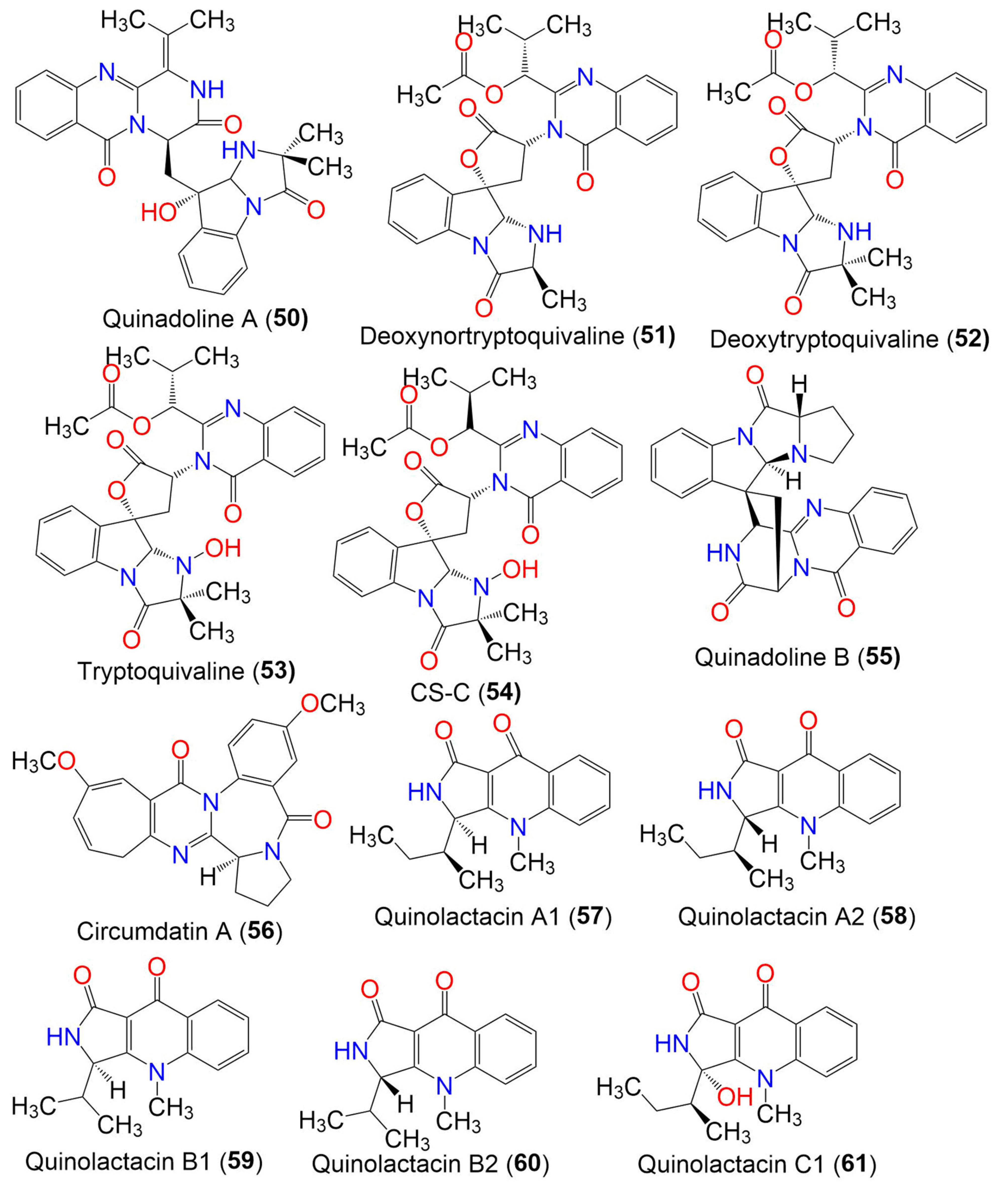

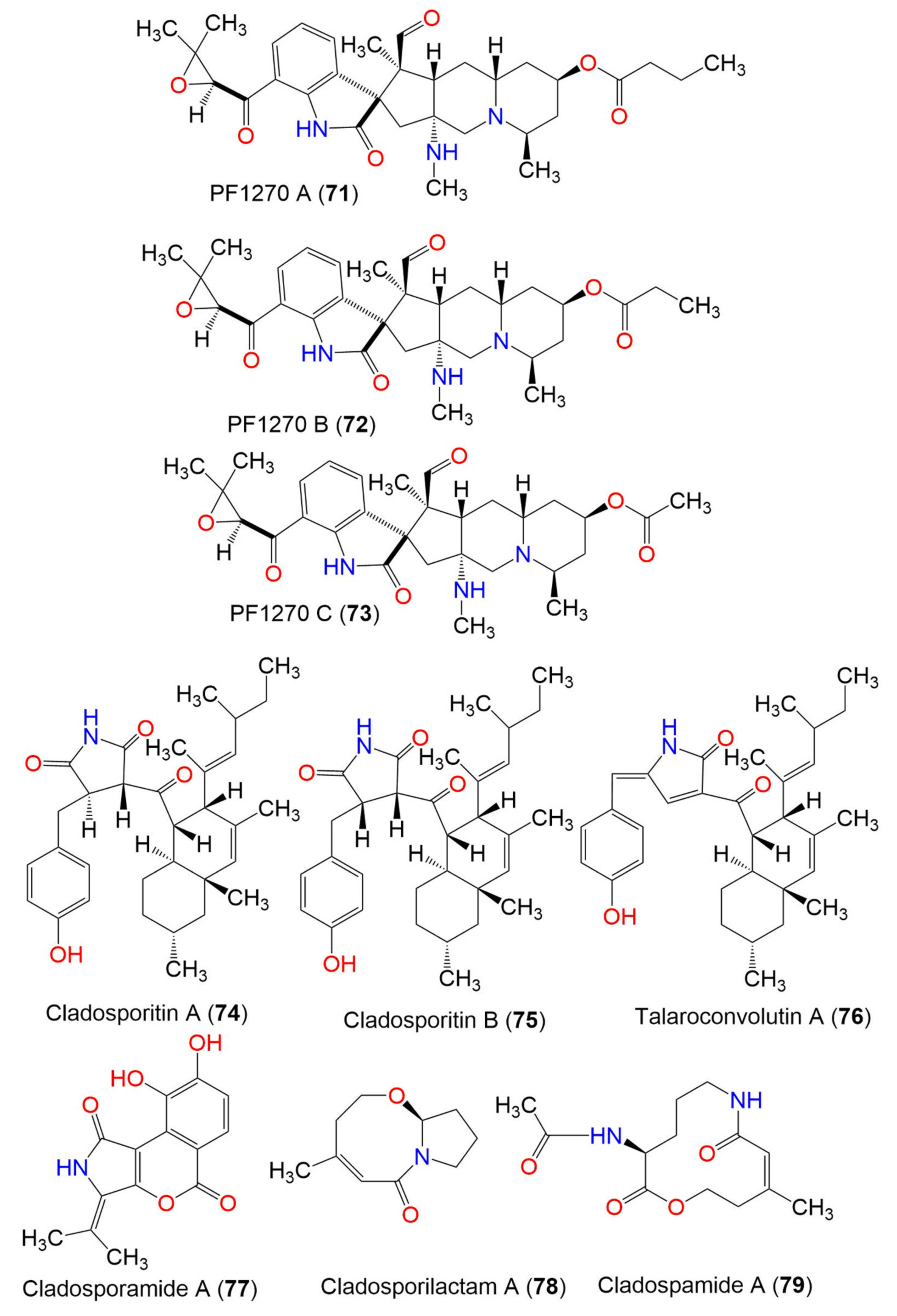
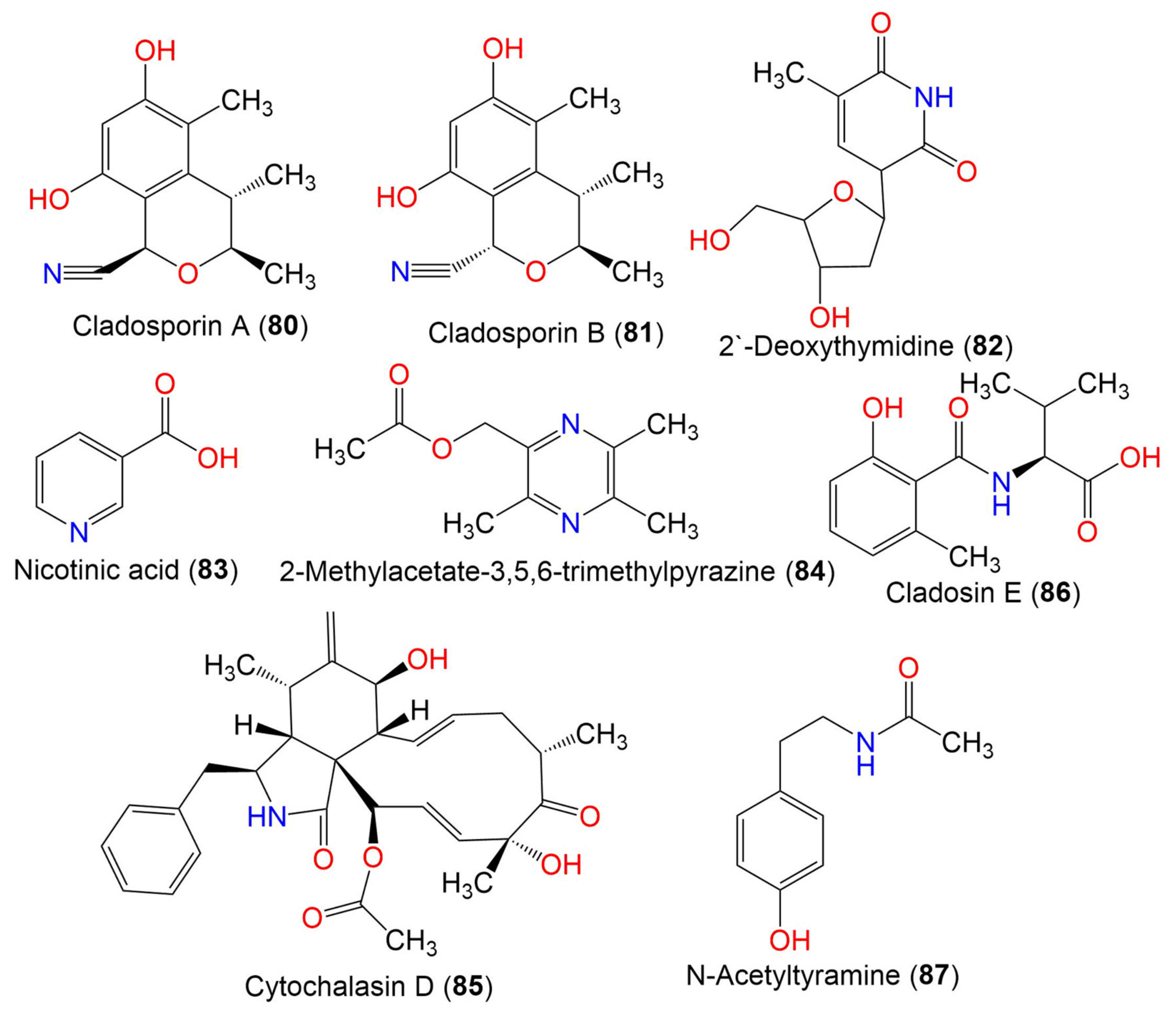

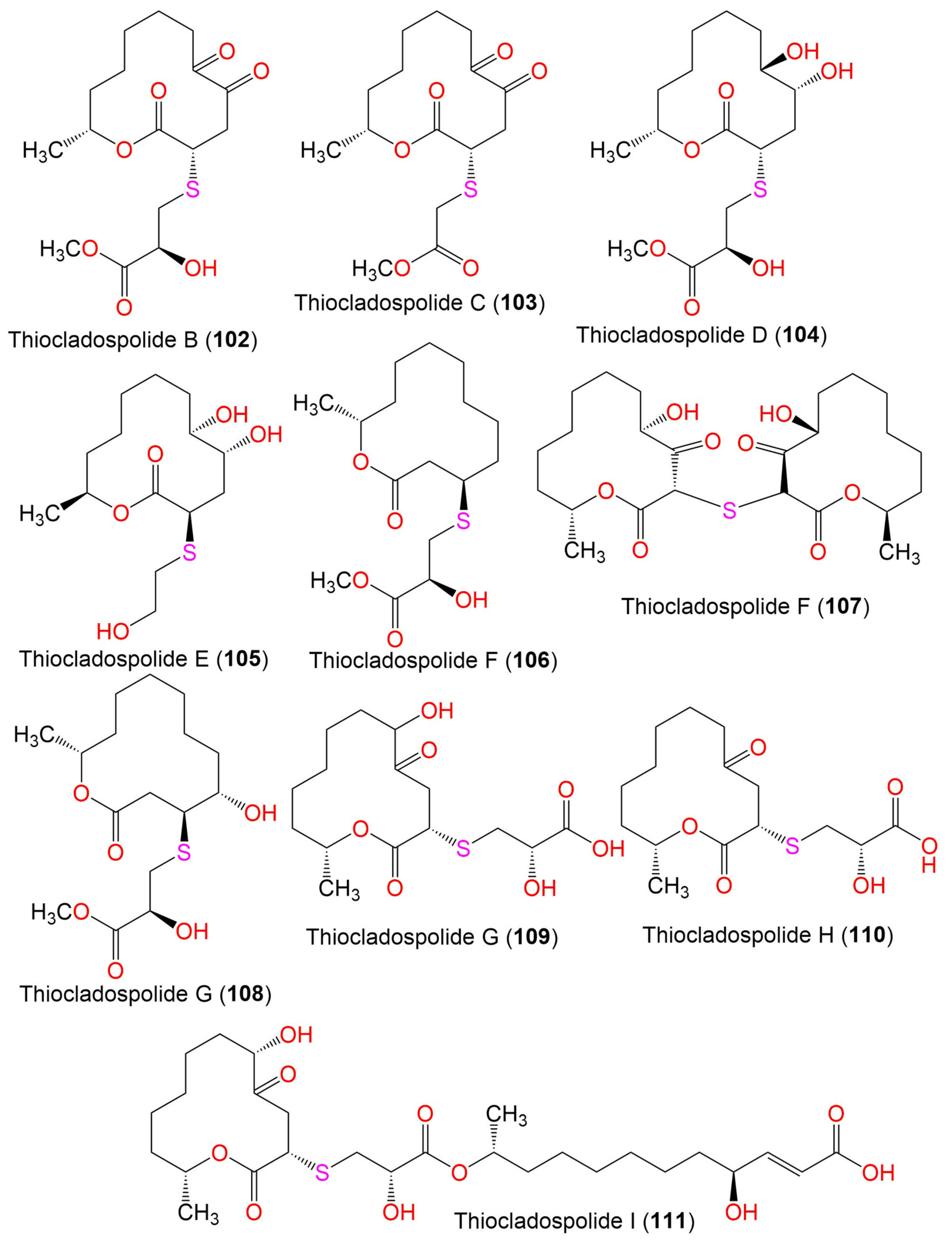
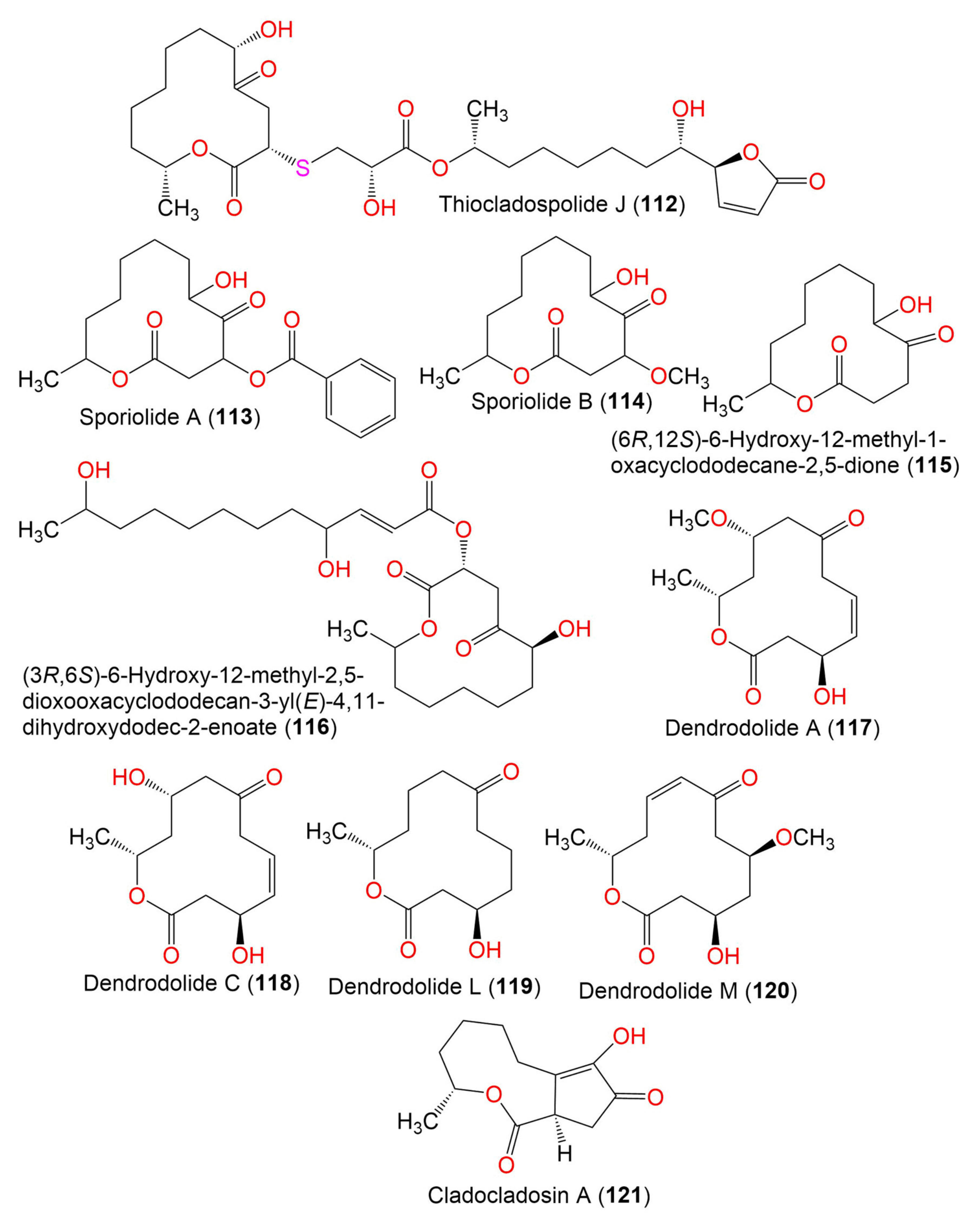
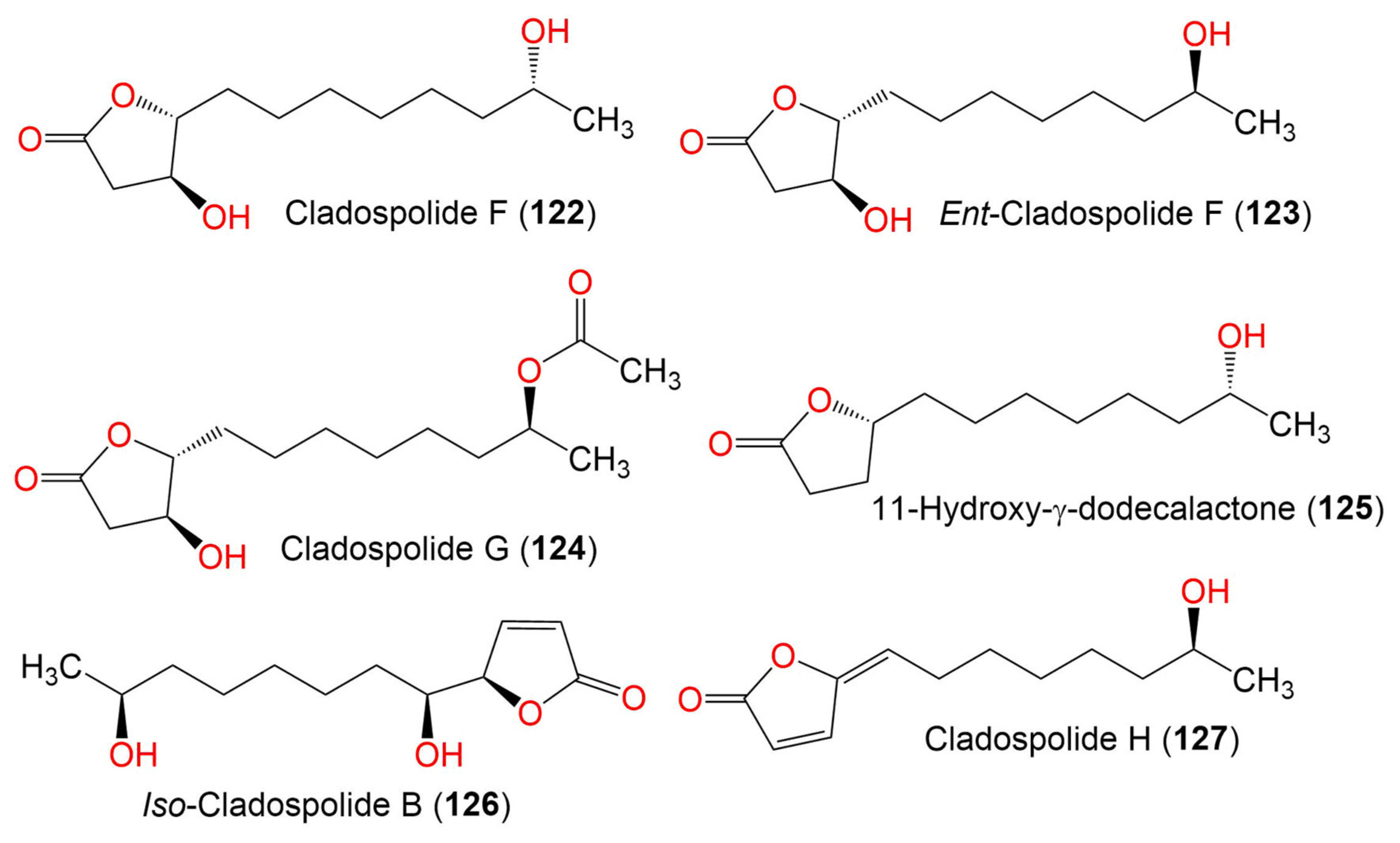
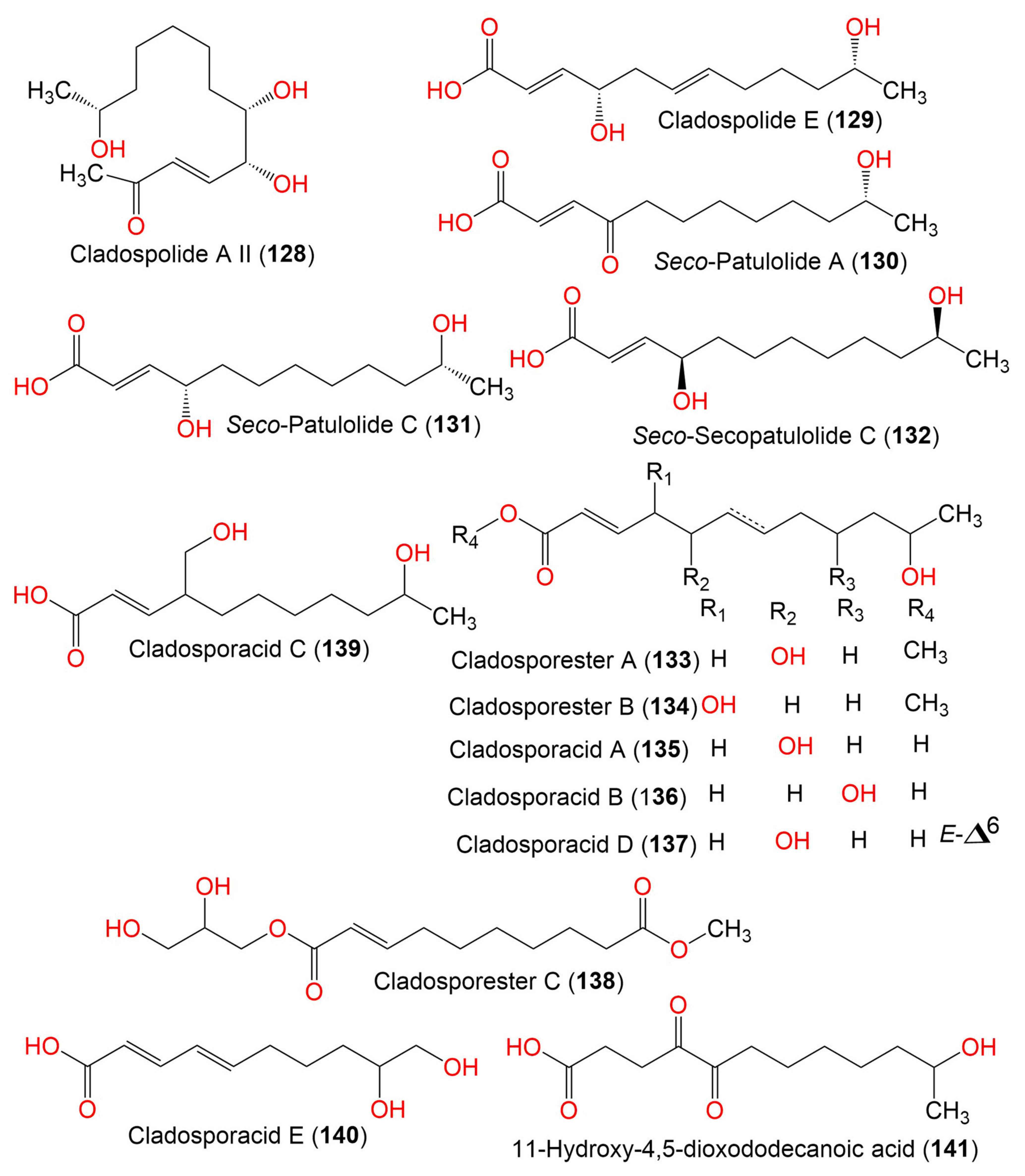
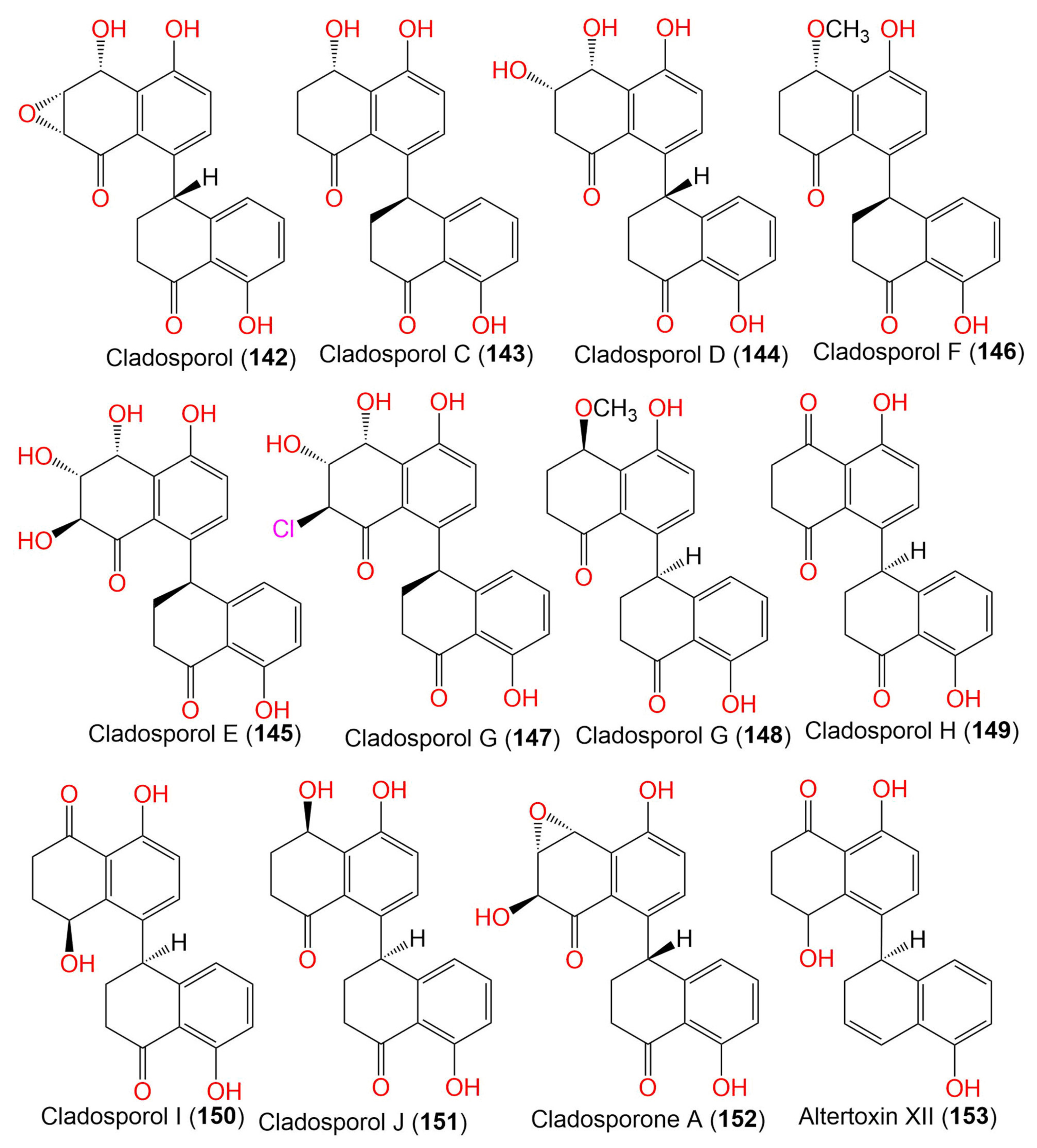

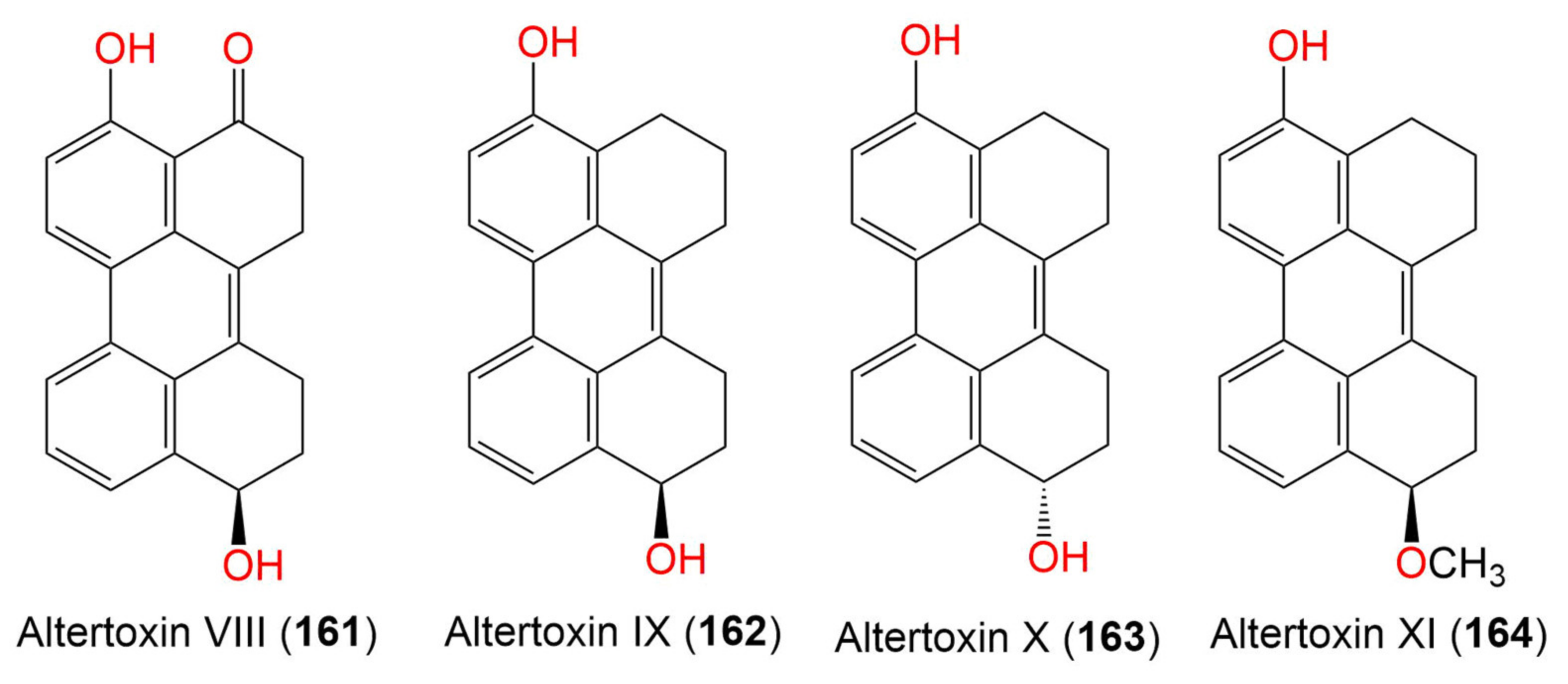
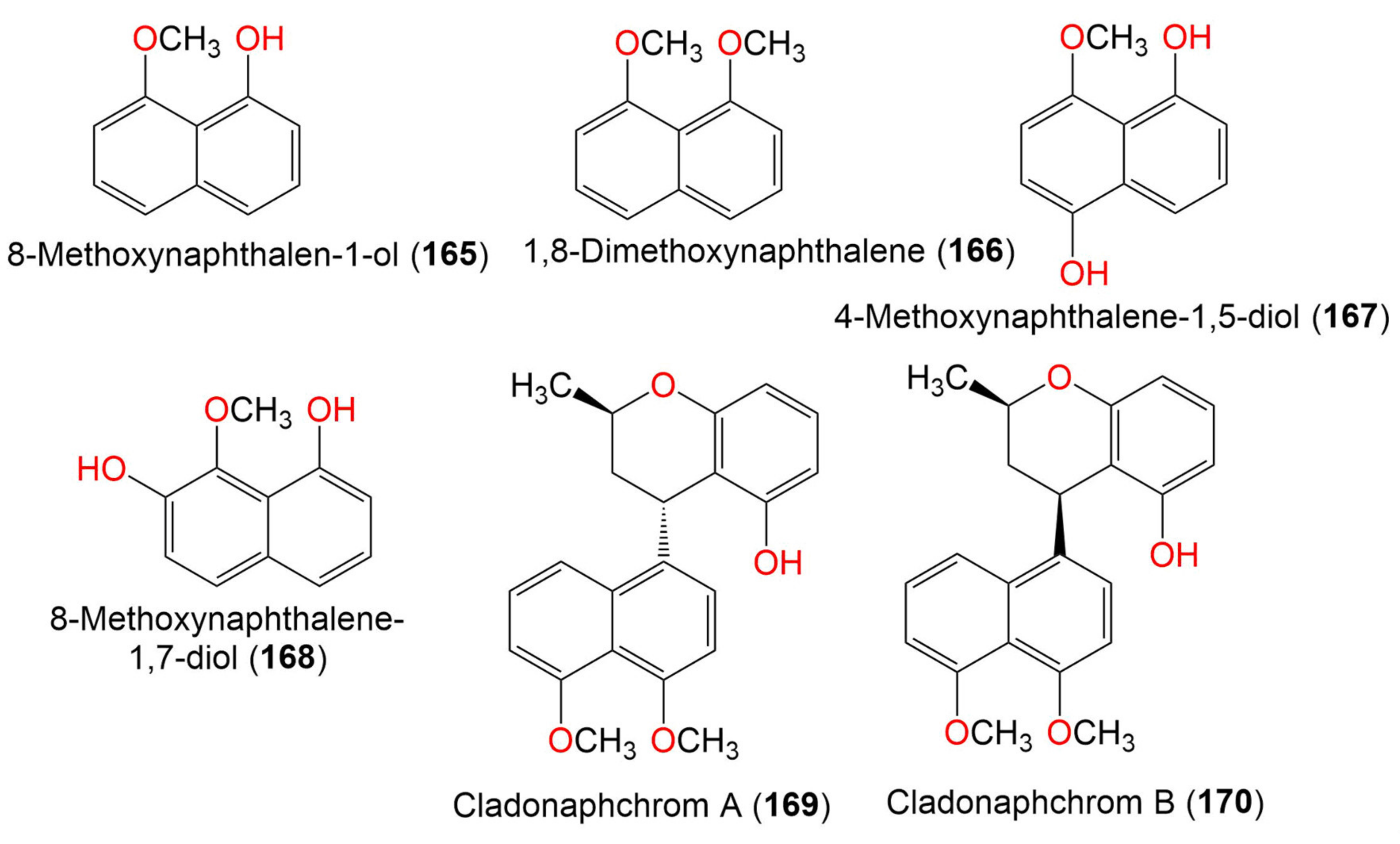
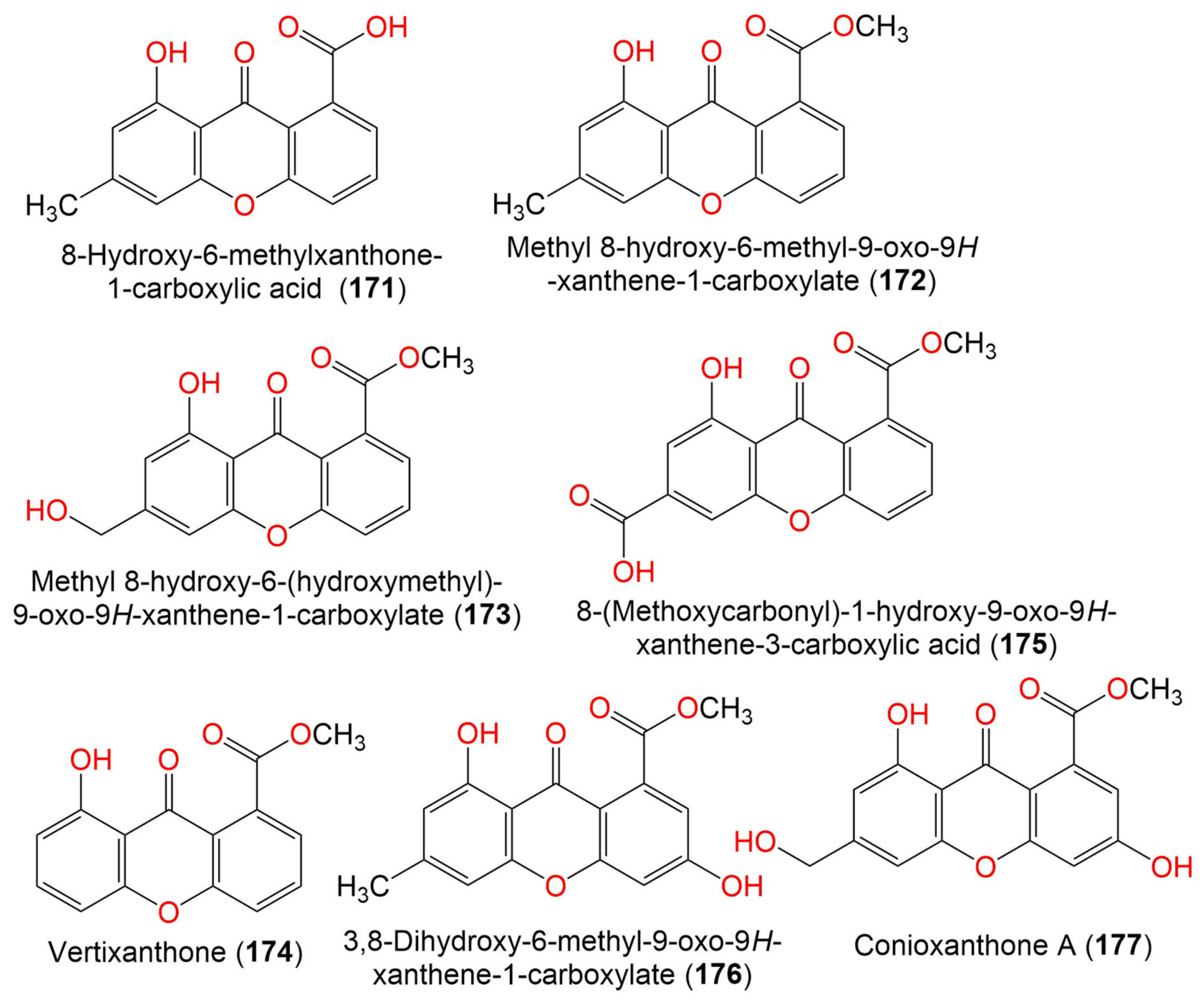
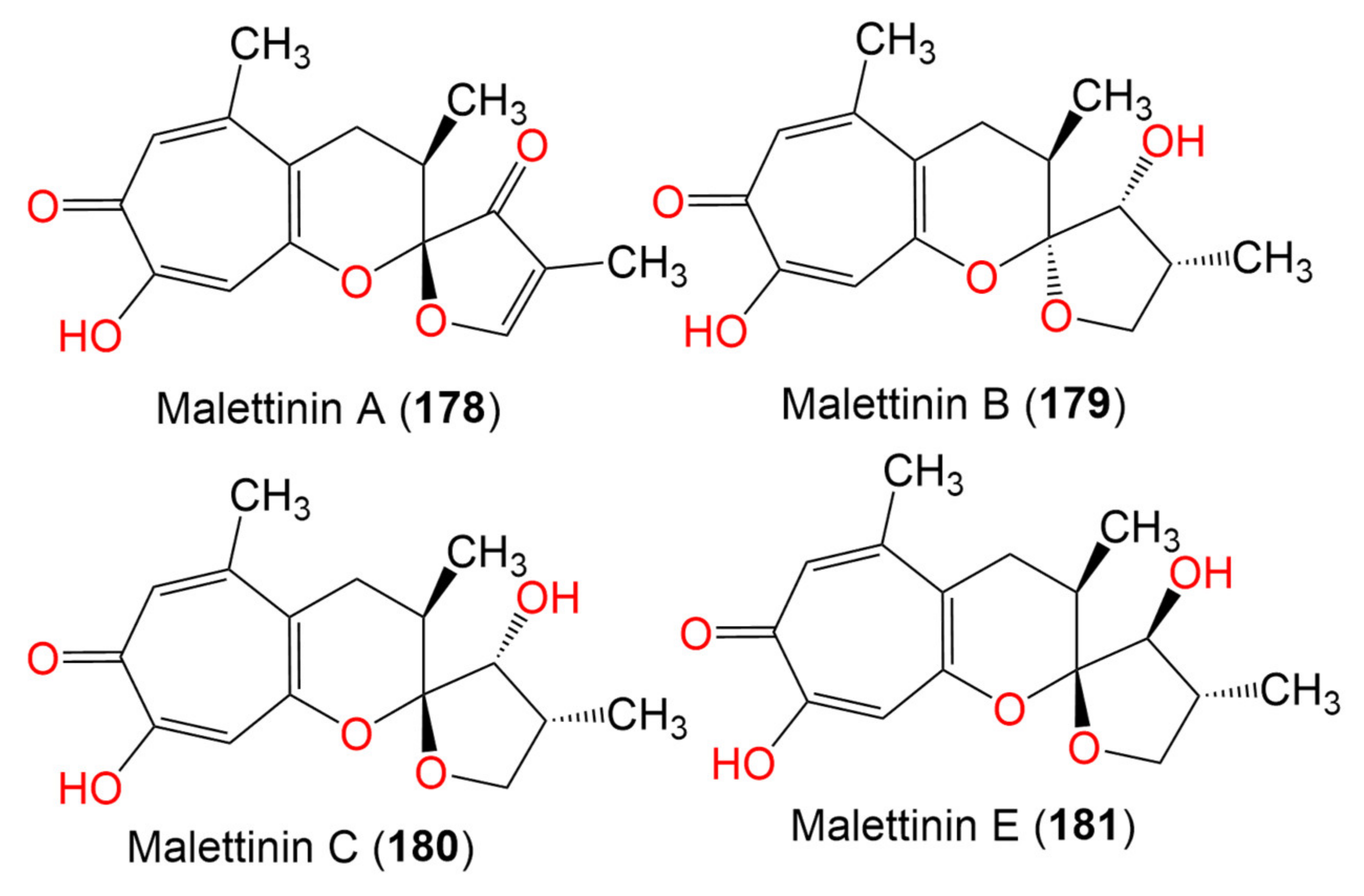


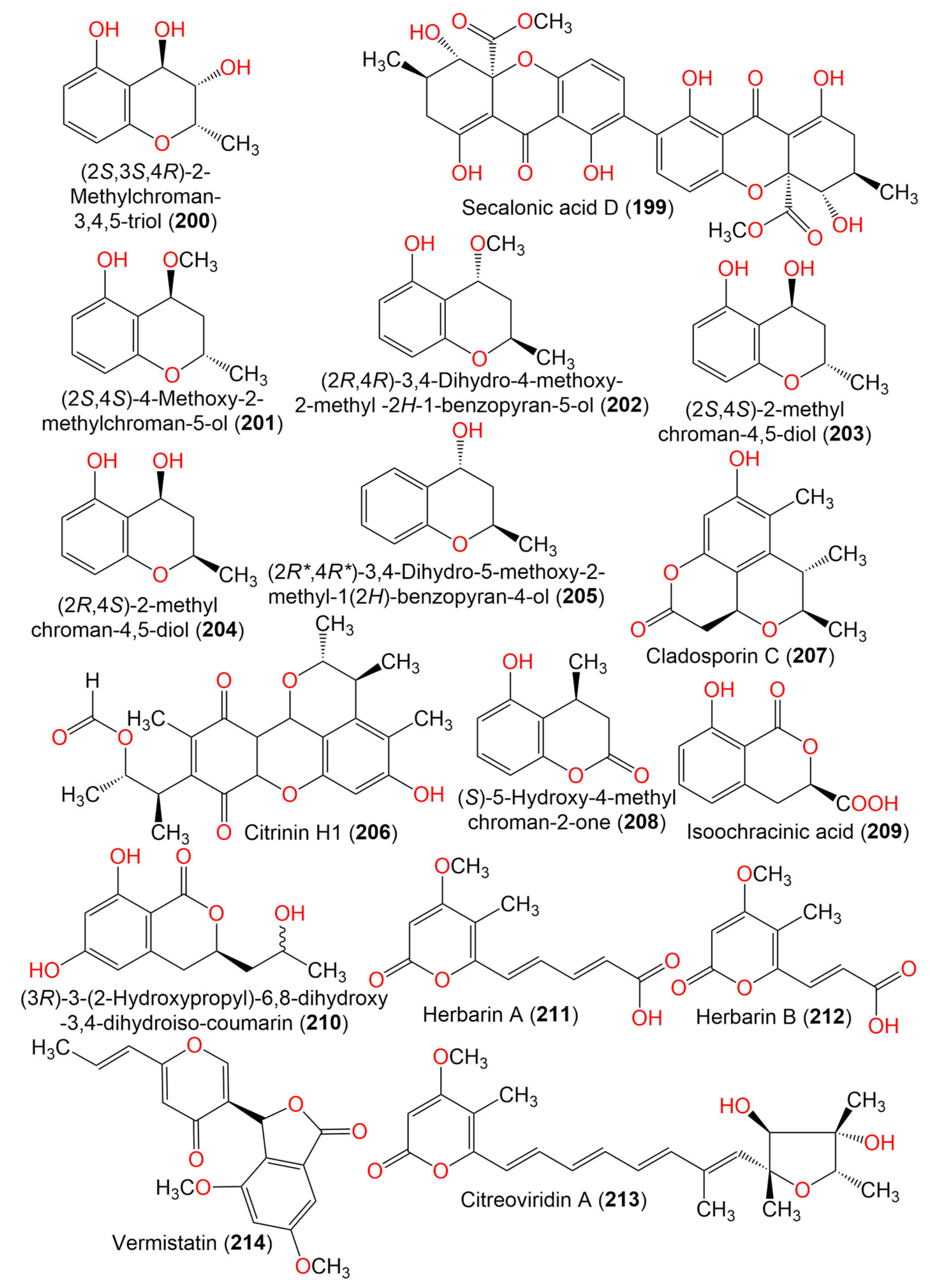
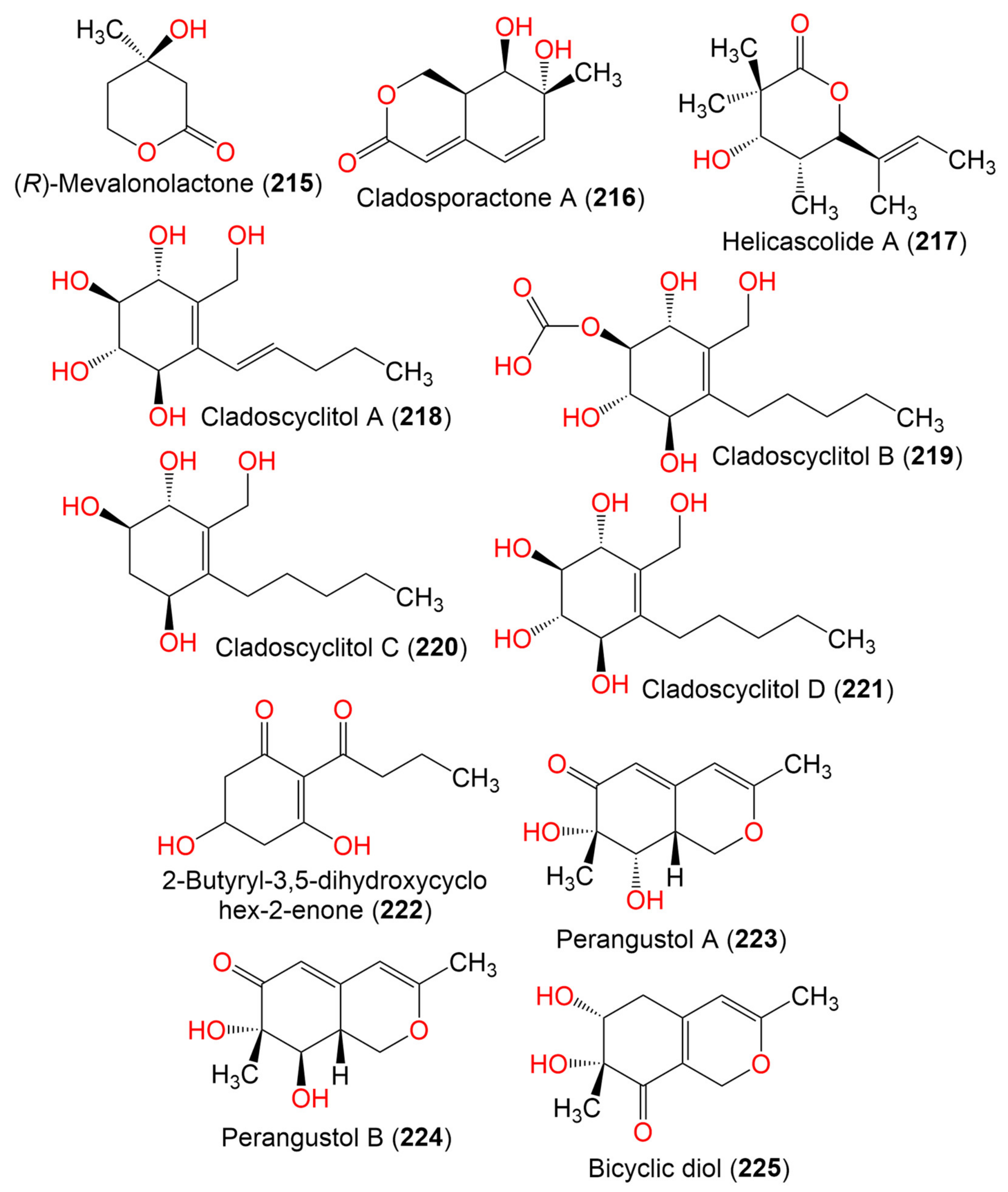
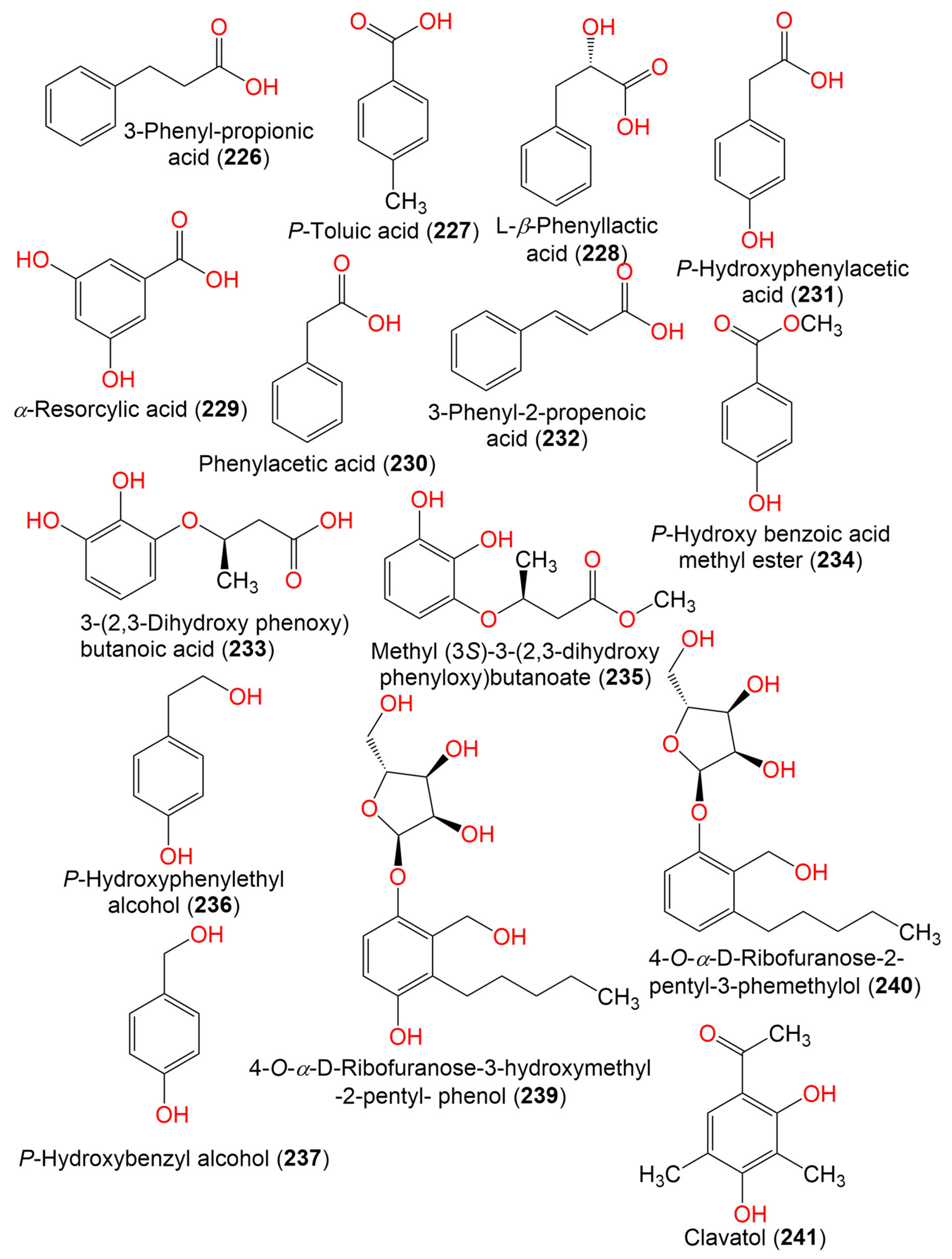


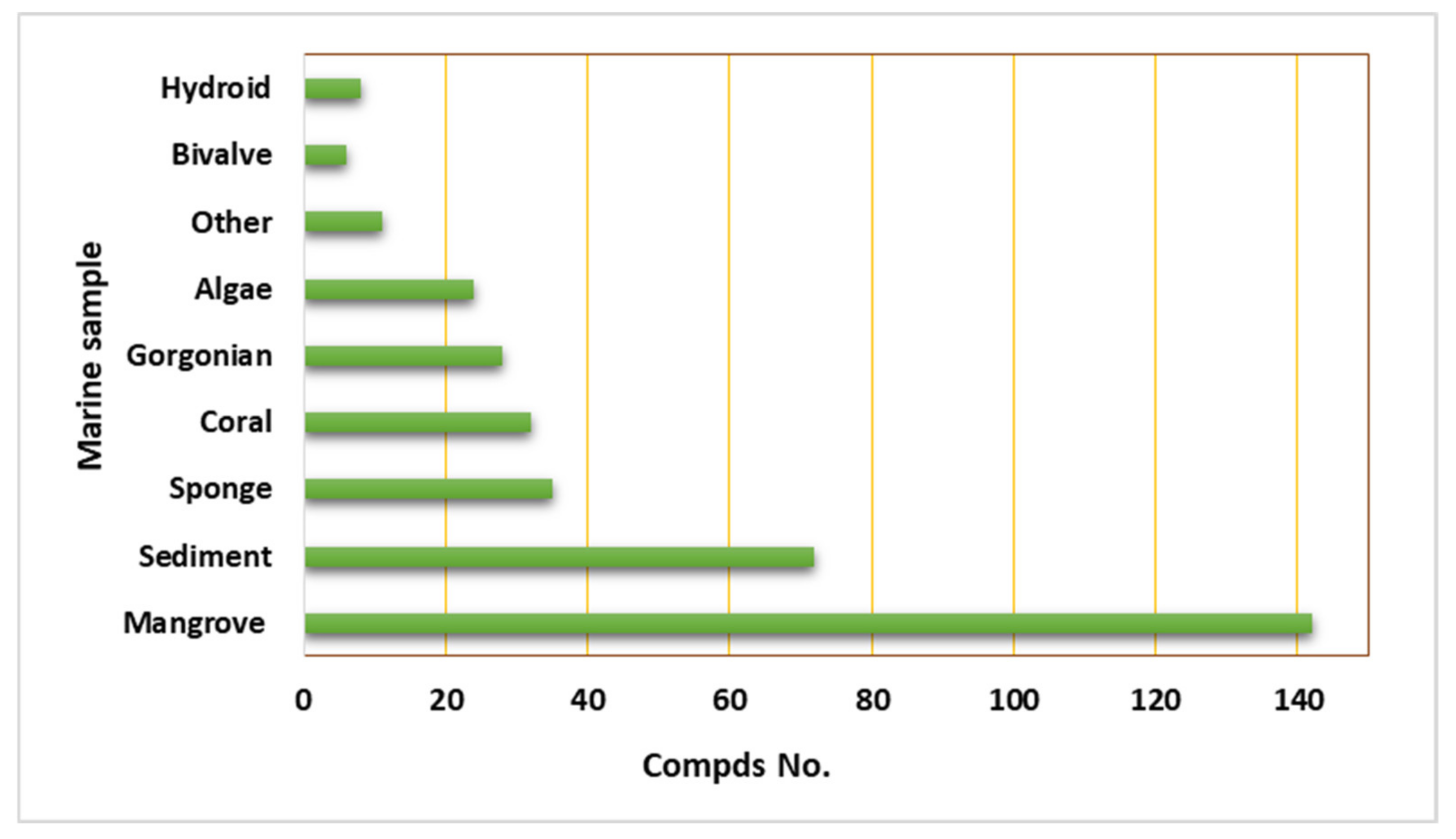
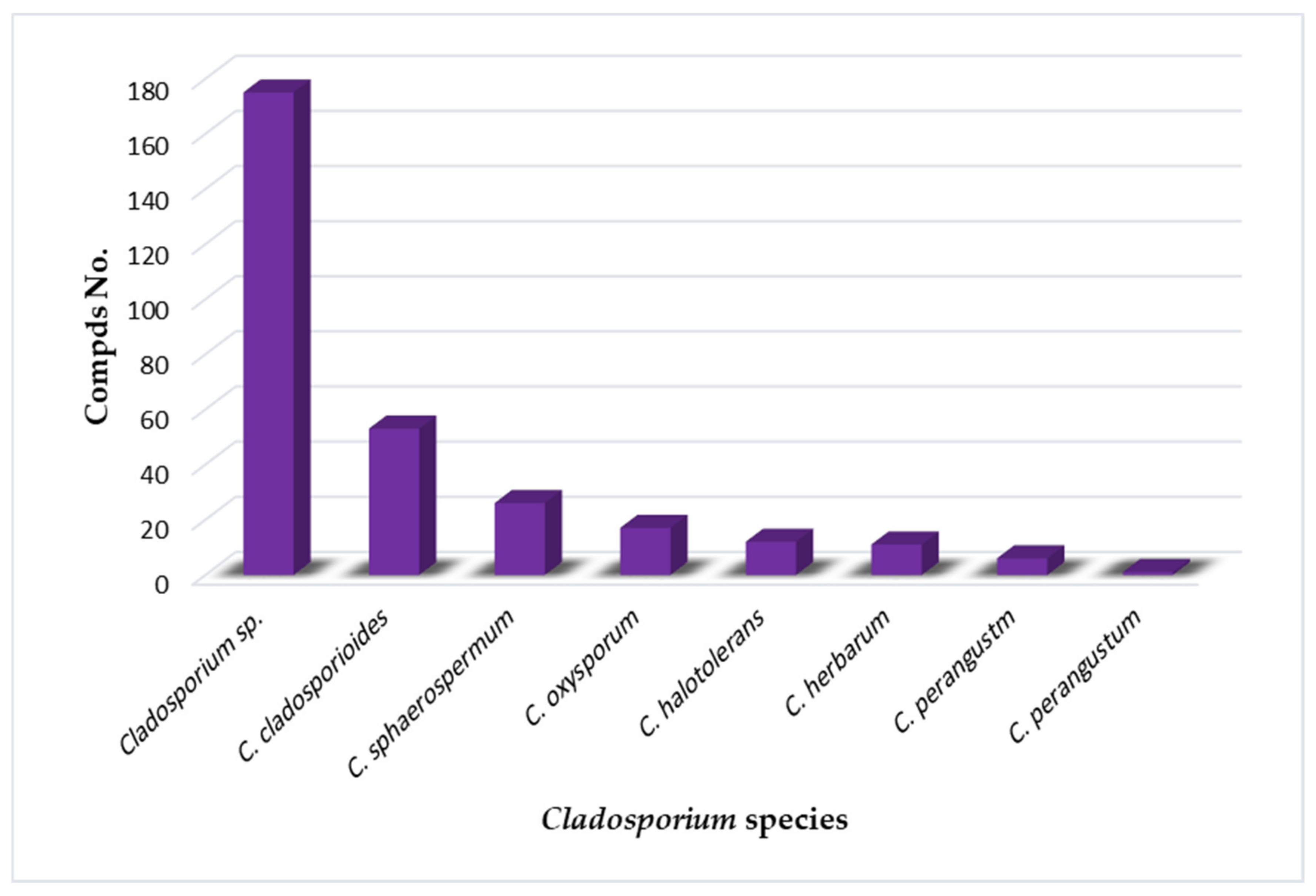
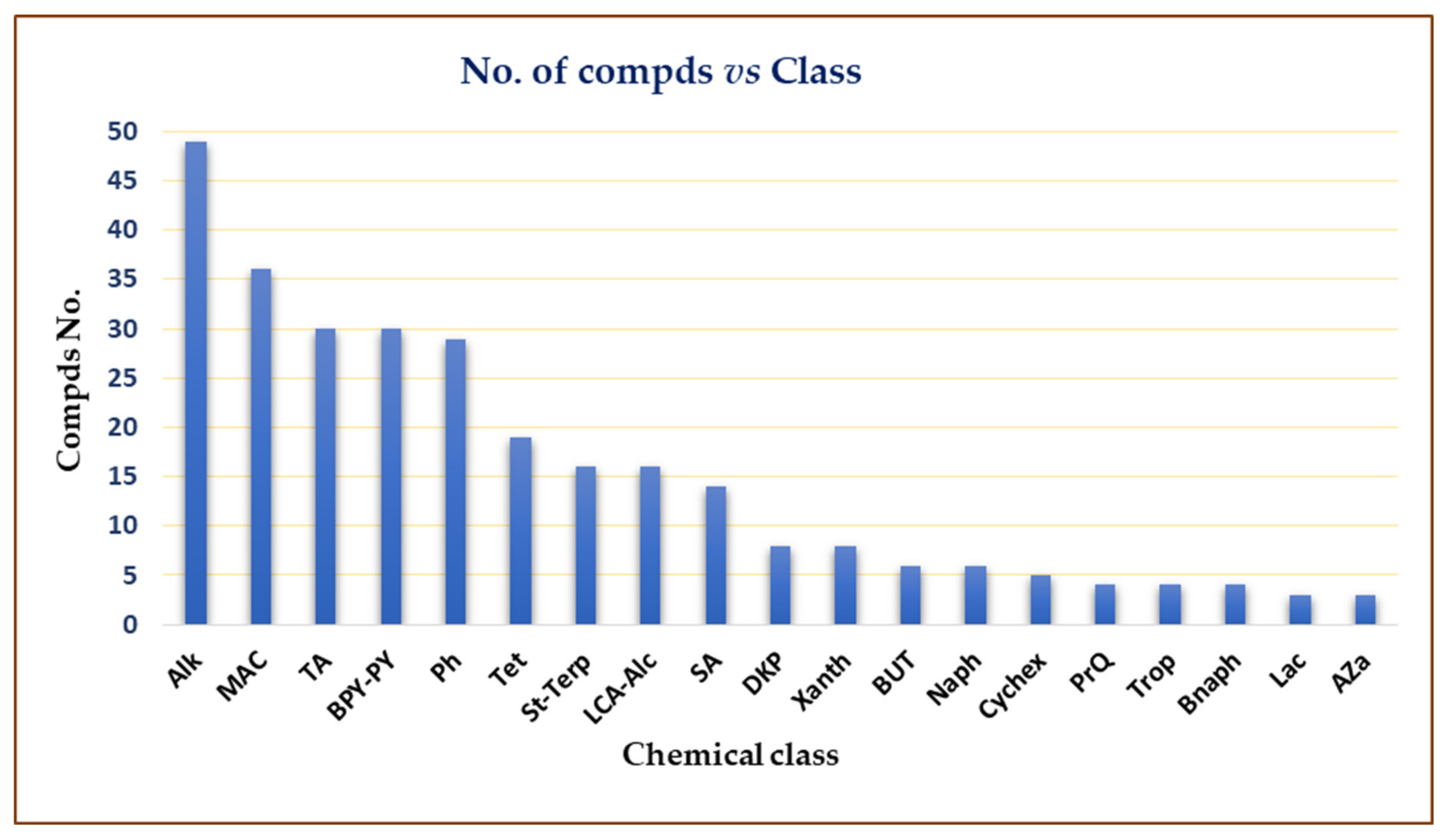
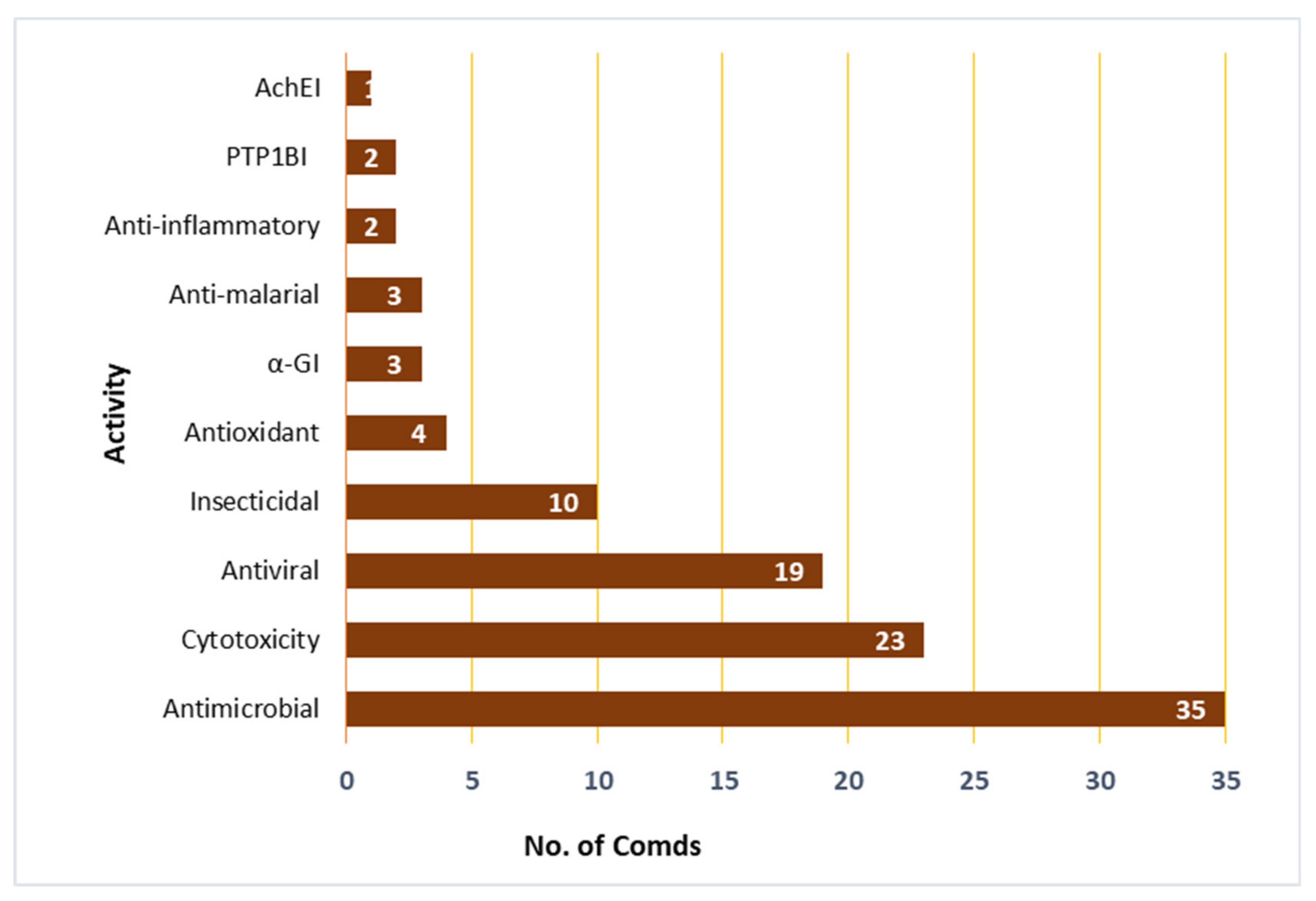
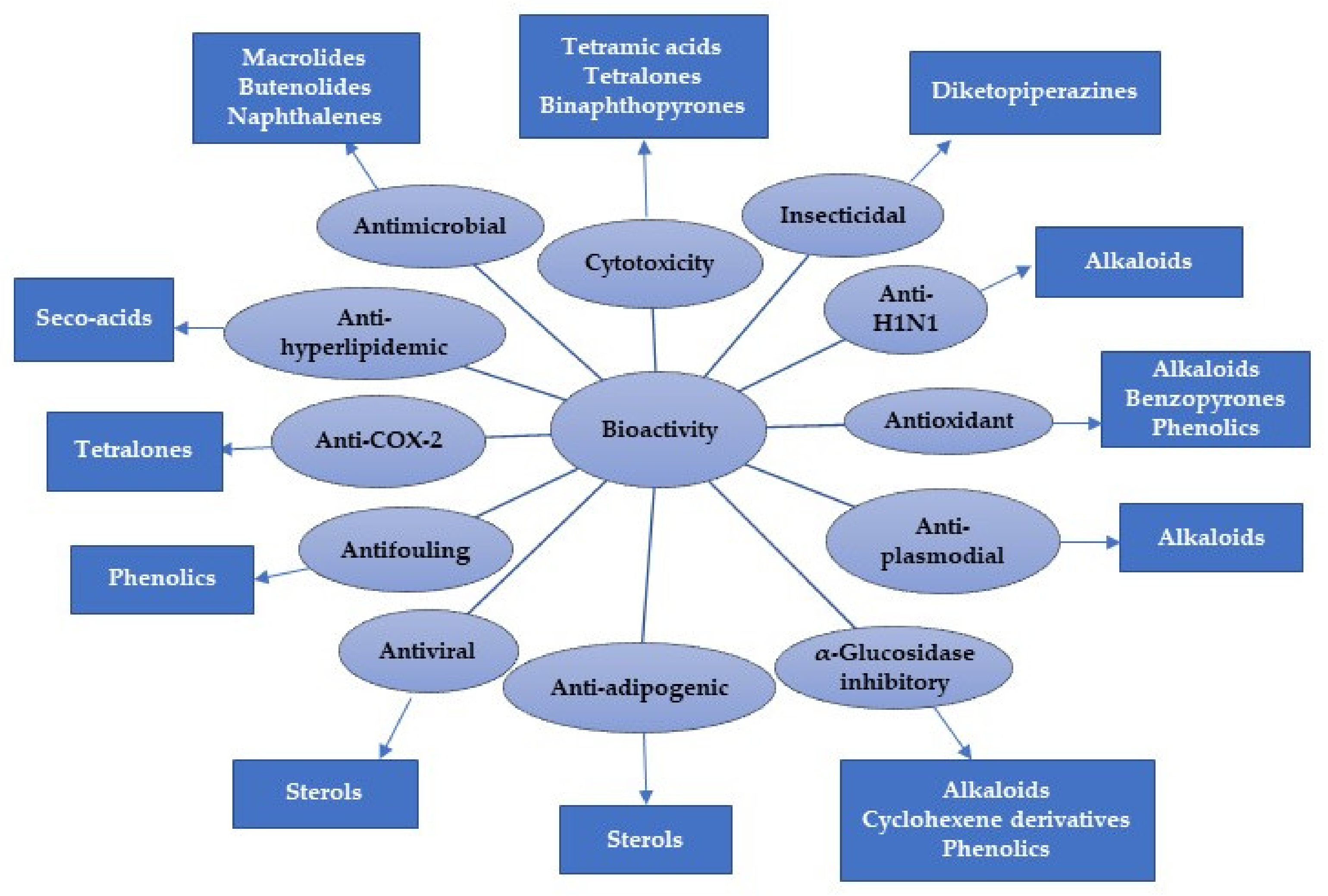
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohamed, G.A.; Ibrahim, S.R.M. Untapped Potential of Marine-Associated Cladosporium Species: An Overview on Secondary Metabolites, Biotechnological Relevance, and Biological Activities. Mar. Drugs 2021, 19, 645. https://doi.org/10.3390/md19110645
Mohamed GA, Ibrahim SRM. Untapped Potential of Marine-Associated Cladosporium Species: An Overview on Secondary Metabolites, Biotechnological Relevance, and Biological Activities. Marine Drugs. 2021; 19(11):645. https://doi.org/10.3390/md19110645
Chicago/Turabian StyleMohamed, Gamal A., and Sabrin R. M. Ibrahim. 2021. "Untapped Potential of Marine-Associated Cladosporium Species: An Overview on Secondary Metabolites, Biotechnological Relevance, and Biological Activities" Marine Drugs 19, no. 11: 645. https://doi.org/10.3390/md19110645
APA StyleMohamed, G. A., & Ibrahim, S. R. M. (2021). Untapped Potential of Marine-Associated Cladosporium Species: An Overview on Secondary Metabolites, Biotechnological Relevance, and Biological Activities. Marine Drugs, 19(11), 645. https://doi.org/10.3390/md19110645







