Probing the Therapeutic Potential of Marine Phyla by SPE Extraction
Abstract
:1. Introduction
2. Results and Discussion
2.1. Collection of Organisms
2.1.1. Cultured Microorganisms
2.1.2. Benthic Invertebrates and Algae
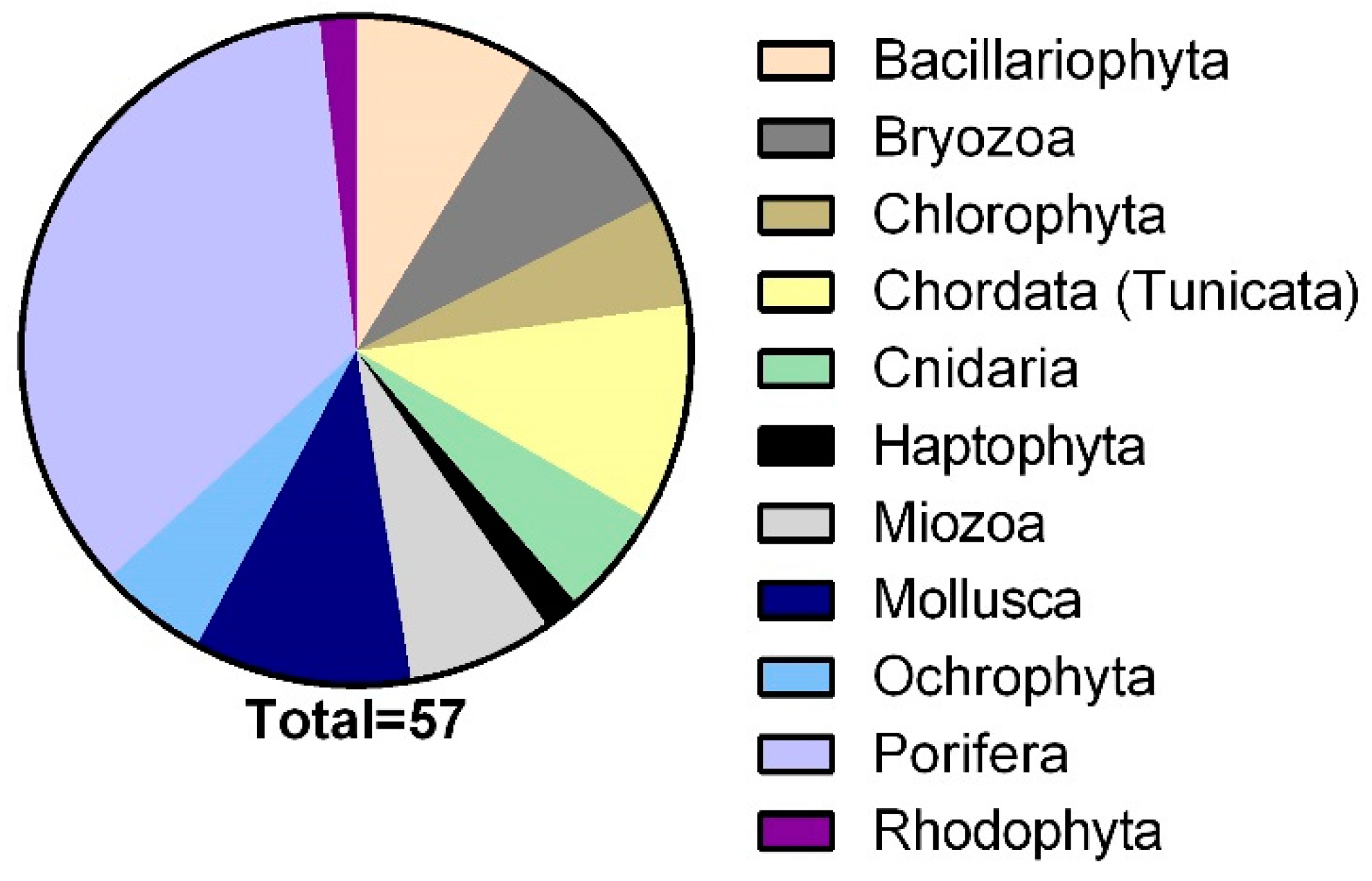
2.2. Library of Marine Natural Products Extracts
2.3. Bioassays
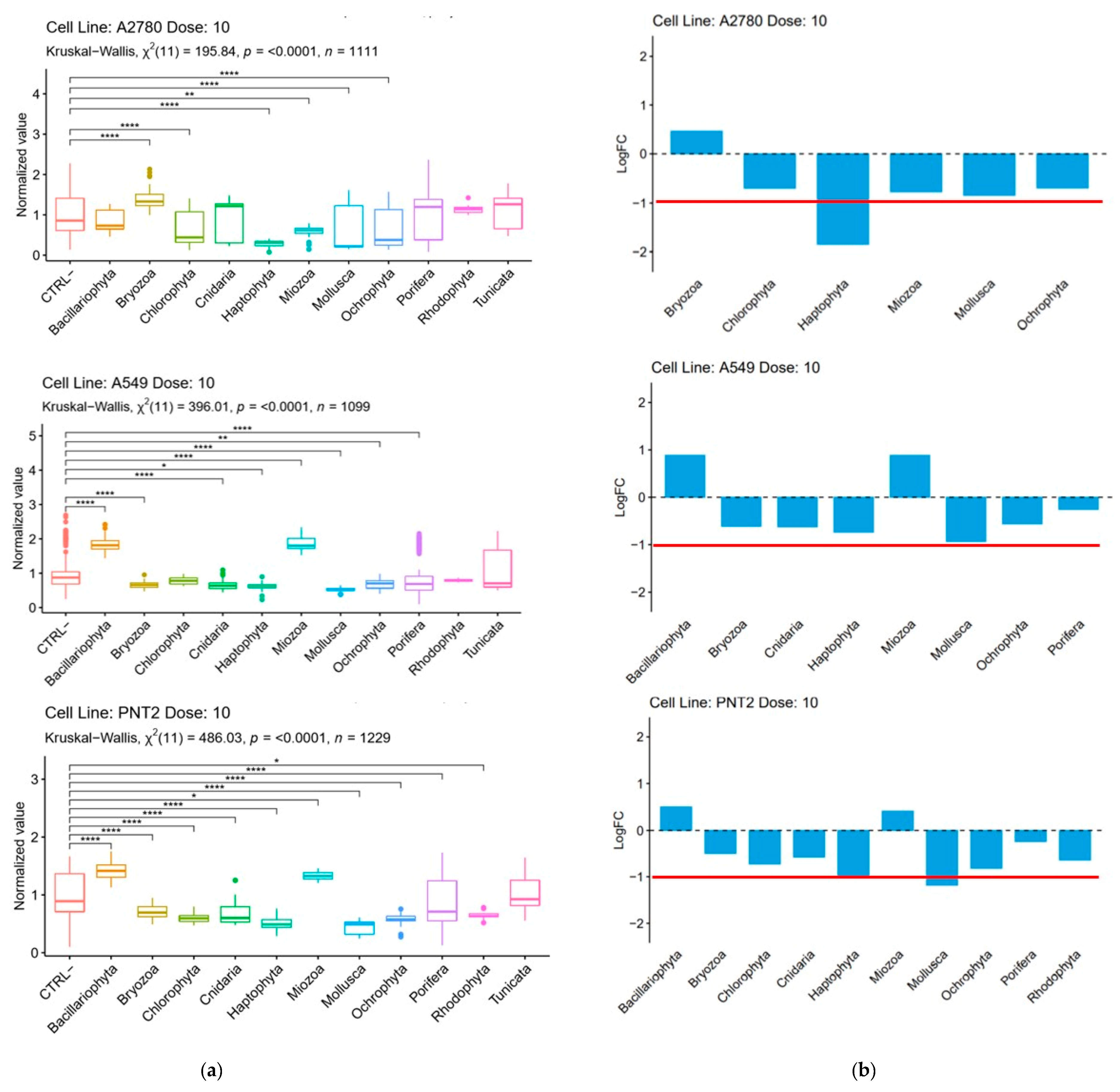
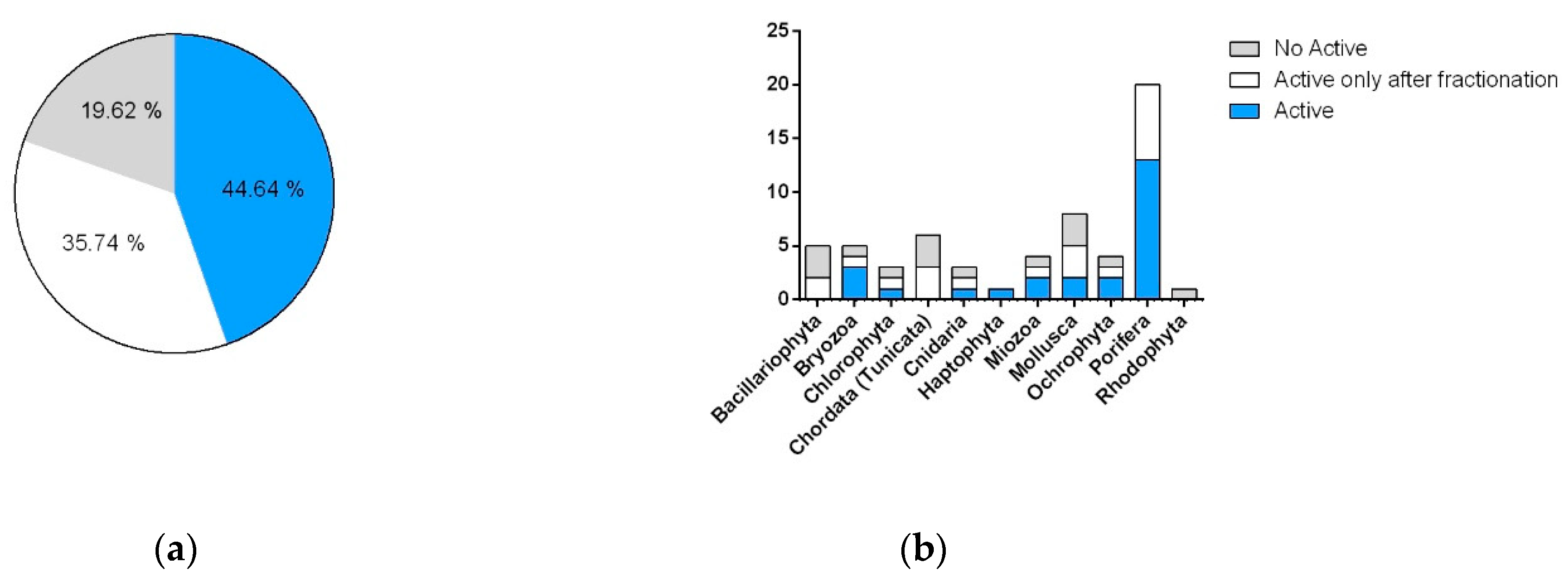
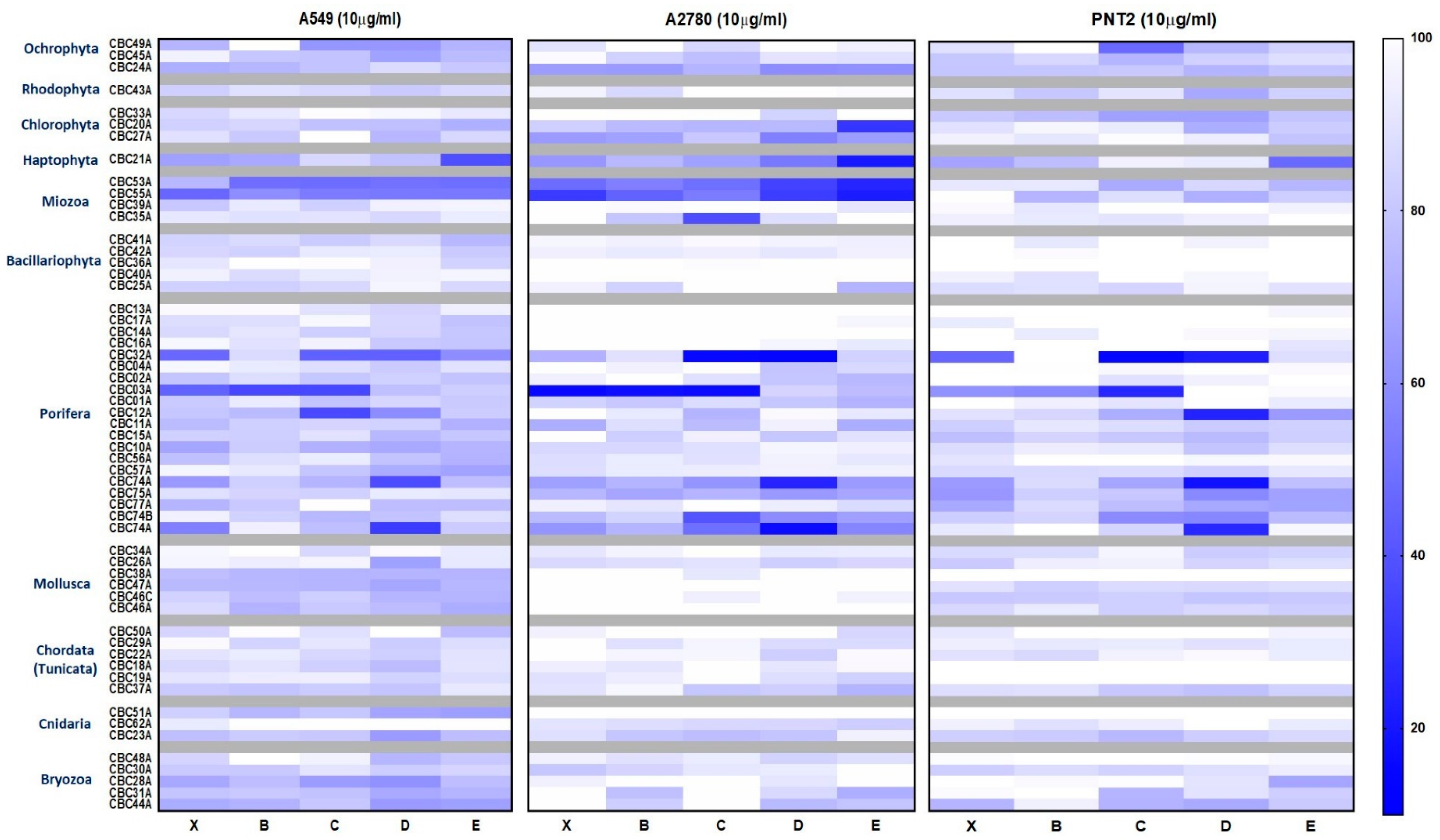
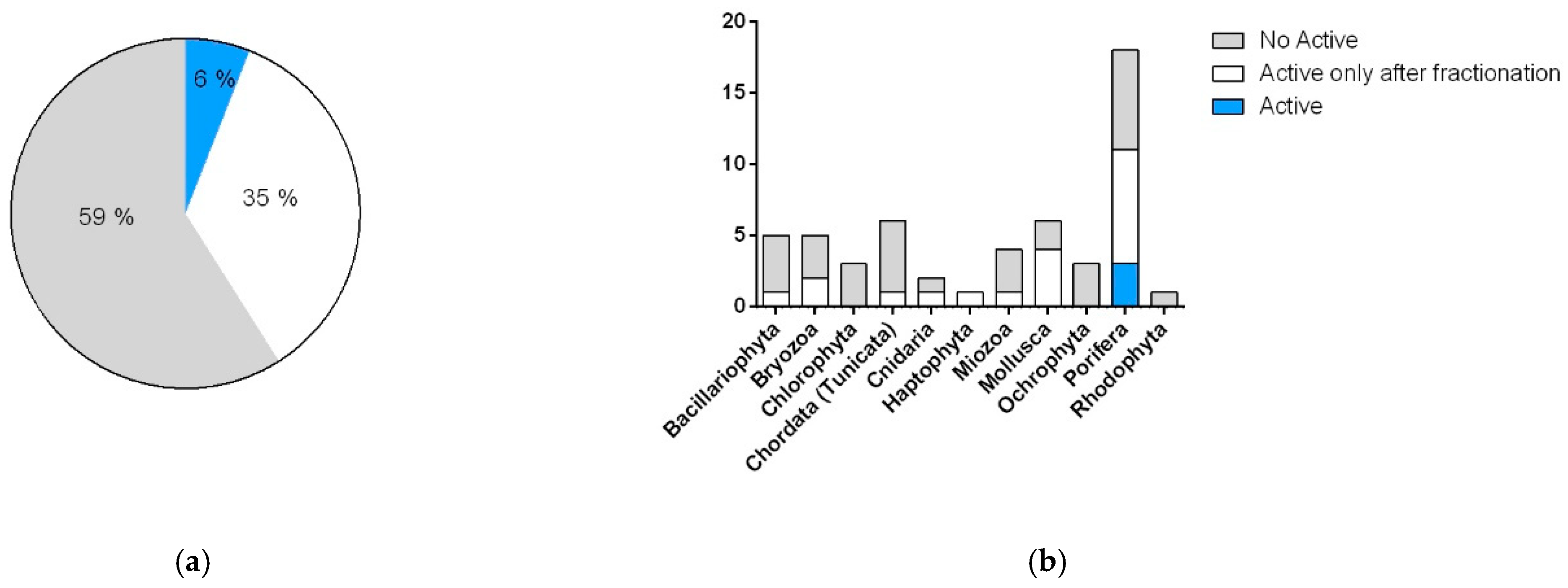
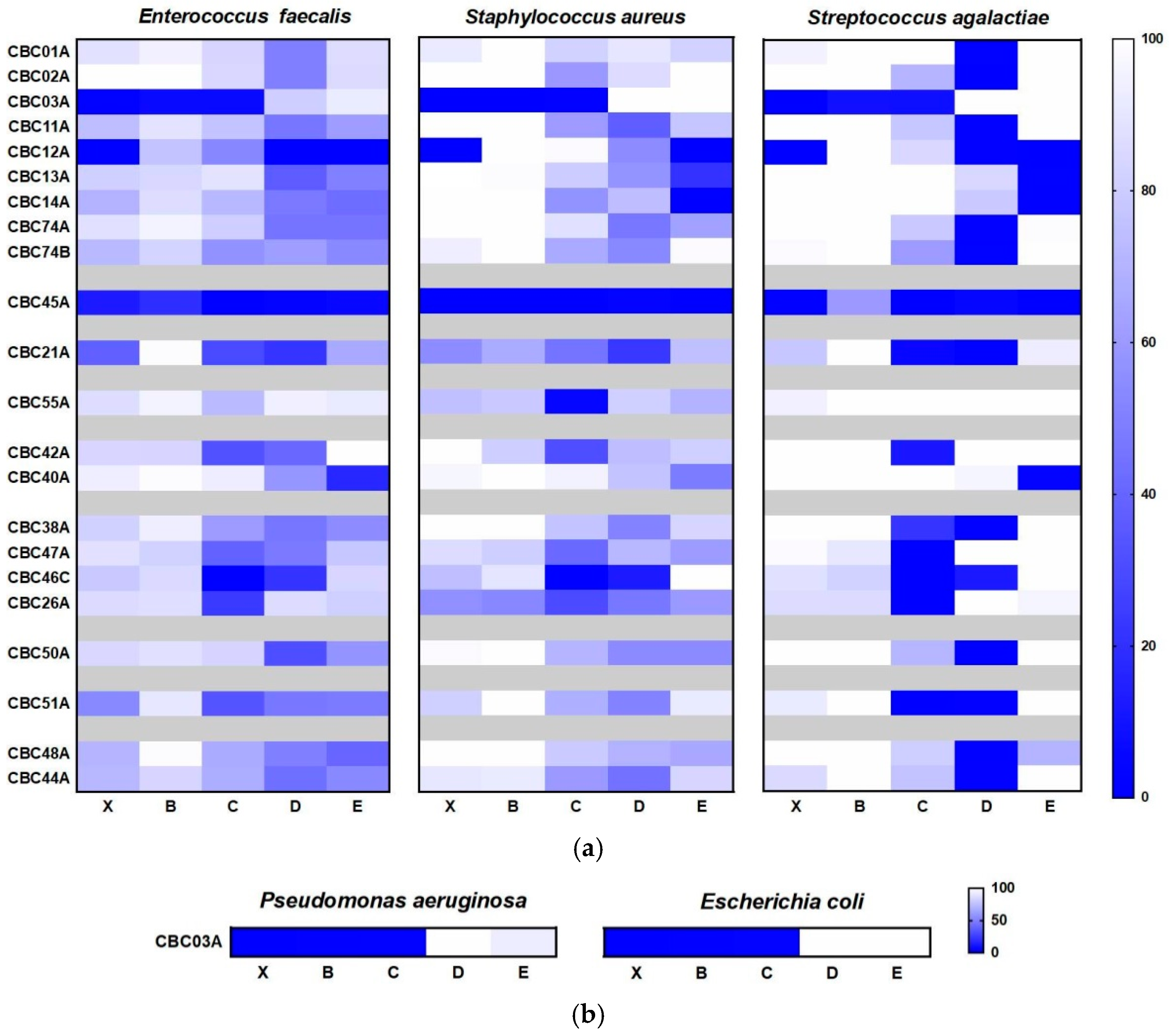
2.3.1. Cytotoxic and Antimicrobial Bioassays
2.3.2. Antimicrobial Bioassay
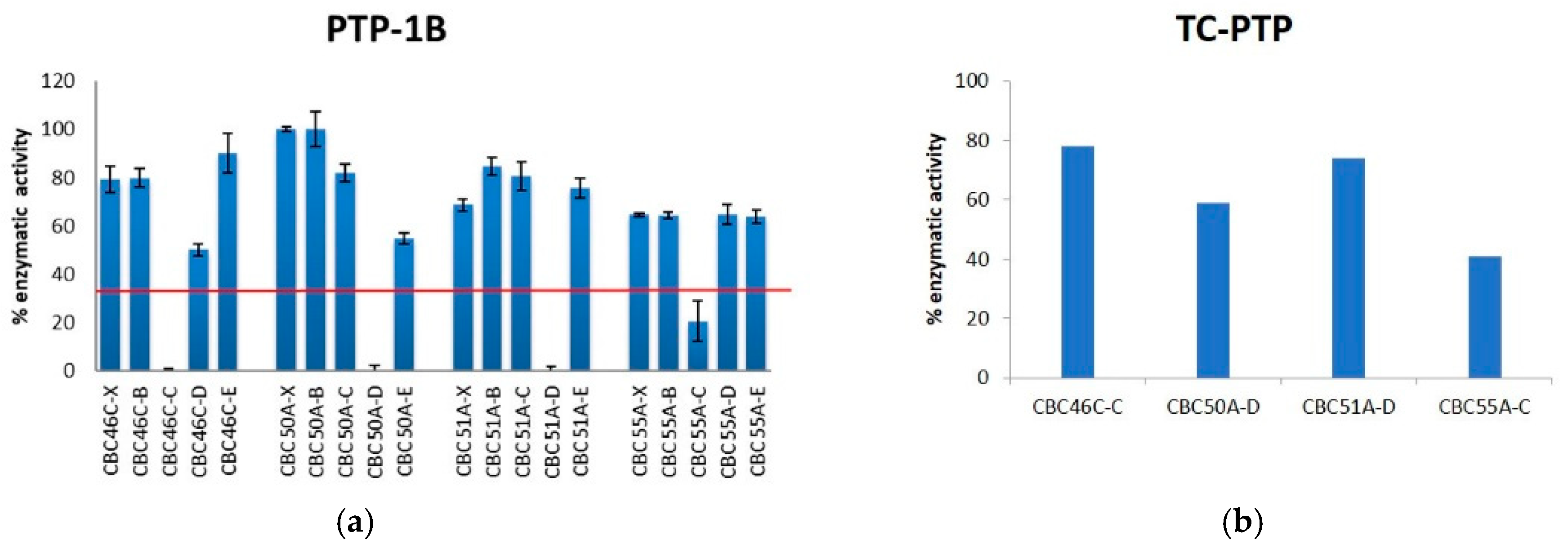
2.3.3. Antidiabetic Bioassay
3. Materials and Methods
3.1. General
3.2. Marine Organism Sources
3.2.1. Cultured Algae
3.2.2. Benthonic Invertebrates and Algae Sampled
3.3. Methanol Extraction of the Organisms
3.4. Solid Phase Extraction
3.5. Cytotoxicity Assays
3.6. Antibacterial Assays
3.7. Antidiabetic Assays
3.8. Statistical Analisys
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2020, 37, 175–223. [Google Scholar] [CrossRef] [PubMed]
- Sorokina, M.; Steinbeck, C. Review on natural products databases: Where to find data in 2020. J. Cheminform. 2020, 12, 20. [Google Scholar] [CrossRef] [Green Version]
- Kondo, J.; Inoue, M. Application of Cancer Organoid Model for Drug Screening and Personalized Therapy. Cells 2019, 8, 470. [Google Scholar] [CrossRef] [Green Version]
- Van Santen, J.A.; Jacob, G.; Singh, A.L.; Aniebok, V.; Balunas, M.J.; Bunsko, D.; Neto, F.C.; Castaño-Espriu, L.; Chang, C.; Clark, T.N.; et al. The Natural Products Atlas: An Open Access Knowledge Base for Microbial Natural Products Discovery. ACS Cent. Sci. 2019, 5, 1824–1833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Appleton, D.R.; Buss, A.D.; Butler, M.S. A simple method for high-throughput extract prefractionation for biological screening. Chimia 2007, 61, 327–331. [Google Scholar] [CrossRef] [Green Version]
- Bugni, T.S.; Harper, M.K.; McCulloch, M.W.B.; Reppart, J.; Ireland, C.M. Fractionated marine invertebrate extract libraries for drug discovery. Molecules 2008, 13, 1372–1383. [Google Scholar] [CrossRef] [Green Version]
- Cutignano, A.; Nuzzo, G.; Ianora, A.; Luongo, E.; Romano, G.; Gallo, C.; Sansone, C.; Aprea, S.; Mancini, F.; D’Oro, U.; et al. Development and application of a novel SPE-method for bioassay-guided fractionation of marine extracts. Mar. Drugs 2015, 13, 5736–5749. [Google Scholar] [CrossRef] [Green Version]
- Lage, O.M.; Ramos, M.C.; Calisto, R.; Almeida, E.; Vasconcelos, V.; Vicente, F. Current screening methodologies in drug discovery for selected human diseases. Mar. Drugs 2018, 16, 279. [Google Scholar] [CrossRef] [Green Version]
- Lauritano, C.; Ianora, A. Marine organisms with anti-diabetes properties. Mar. Drugs 2016, 14, 220. [Google Scholar] [CrossRef]
- Thorpe, J.P.; Solé-Cava, A.M.; Watts, P.C. Exploited marine invertebrates: Genetics and fisheries. Hydrobiologia 2000, 420, 165–184. [Google Scholar] [CrossRef]
- Leal, M.C.; Madeira, C.; Brandão, C.A.; Puga, J.; Calado, R. Bioprospecting of marine invertebrates for new natural products—A chemical and zoogeographical perspective. Molecules 2012, 17, 9842–9854. [Google Scholar] [CrossRef] [PubMed]
- Mehbub, M.F.; Lei, J.; Franco, C.; Zhang, W. Marine sponge derived natural products between 2001 and 2010: Trends and opportunities for discovery of bioactives. Mar. Drugs 2014, 12, 4539–4577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayer, A.M.S.; Rodríguez, A.D.; Taglialatela-Scafati, O.; Fusetani, N. Marine pharmacology in 2012–2013: Marine compounds with antibacterial, antidiabetic, antifungal, anti-inflammatory, antiprotozoal, antituberculosis, and antiviral activities; affecting the immune and nervous systems, and other miscellaneous mechanisms of action. Mar. Drugs 2017, 15, 273. [Google Scholar] [CrossRef]
- Jiménez, C. Marine Natural Products in Medicinal Chemistry. ACS Med. Chem. Lett. 2018, 9, 959–961. [Google Scholar] [CrossRef] [Green Version]
- Gerlier, D.; Thomasset, N. Use of MTT colorimetric assay to measure cell activation. J. Immunol. Methods 1986, 94, 57–63. [Google Scholar] [CrossRef]
- Cafieri, F.; Fattorusso, E.; Mahajnah, Y.A.M. Complete 1H and 13C NMR assignment of ent-3β, 16α-atisanediol, a cytotoxic diterpene isolated from the sponge Spirastrella cunctatrix. Magn. Reson. Chem. 1995, 33, 286–289. [Google Scholar] [CrossRef]
- Pettit, G.R.; Herald, C.L.; Cichacz, Z.A.; Gao, F.; Schmidt, J.M.; Boyd, M.R.; Christie, N.D.; Boettner, F.E. Isolation and structure of the powerful human cancer cell growth inhibitors spongistatins 4 and 5 from an African Spirastrella spinispirulifera (porifera). J. Chem. Soc. Chem. Commun. 1995, 24, 2473–2670. [Google Scholar] [CrossRef]
- Elizabeth, A.J.; Ryuichi, S.; Kenneth, L.R. Crambescidins: New antiviral and cytotoxic compounds from the sponge Crambe crambe. J. Org. Chem. 1991, 56, 5712–5715. [Google Scholar] [CrossRef]
- Fontana, A.; Scognamiglio, G.; Cimino, G. Dendrinolide, a New Degraded Diterpenoid from the Antarctic Sponge Dendrilla membranosa. J. Nat. Prod. 1997, 60, 455–457. [Google Scholar] [CrossRef]
- Ankisetty, S.; Amsler, C.D.; McClintock, J.B.; Bill, J.B. Further Membranolide Diterpenes from the Antarctic Sponge Dendrilla membranosa. J. Nat. Prod. 2004, 67, 1172–1174. [Google Scholar] [CrossRef] [PubMed]
- Piddock, L.J.V. Understanding drug resistance will improve the treatment of bacterial infections. Nat. Rev. Microbiol. 2017, 15, 639–640. [Google Scholar] [CrossRef]
- Burkholder, P.R.; Ruetzler, K. Antimicrobial Activity of some Marine Sponge. Nature 1969, 222, 983–984. [Google Scholar] [CrossRef]
- Manjusha, W.A.; Ansusha, W.A.; George, M.J. Antimicrobial screening and GC-MS analysis of marine sponge Crambe crambe. Asian J. Microbiol. Biotechnol. Environ. Sci. 2011, 13, 633–638. [Google Scholar]
- Saltiel, A.R.; Kahn, C.R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 2001, 414, 799–806. [Google Scholar] [CrossRef]
- Verma, M.; Gupta, S.J.; Chaudhary, A.; Garg, V.K. Protein tyrosine phosphatase 1B inhibitors as antidiabetic agents—A brief review. Bioorg. Chem. 2017, 70, 267–283. [Google Scholar] [CrossRef]
- Harris, L.K.; Frumm, S.M.; Bishop, A.C. A general assay for monitoring the activities of protein tyrosine phosphatases in living eukaryotic cells. Anal. Biochem. 2013, 435, 99–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keller, D.K.; Selvin, R.C.; Claus, W.; Guillard, R.R.L. Media for the culture of oceanic ultraphytoplankton. J. Phycol. 1987, 23, 633–638. [Google Scholar] [CrossRef]
- Guillard, R.R.L. Culture of Phytoplankton for Feeding Marine Invertebrates. In Culture of Marine Invertebrate Animals; Smith, W.L., Chanley, M.H., Eds.; Springer: Boston, MA, USA, 1975. [Google Scholar] [CrossRef]
| Phylum | Taxon | Ident Code | Origin |
|---|---|---|---|
| Bacillariophyta | Cyclotella cryptica | CBC25A | CCMP331 (NCMA) |
| Bacillariophyta | Thalassiosira pseudonana | CBC36A | CCMP1335 (NCMA) |
| Bacillariophyta | Skeletonema marinoi | CBC40A | CCMP2092 (NCMA) |
| Bacillariophyta | Thalassiosira rotula | CBC41A | New isolate (Gulf of Naples, Western Mediterranean Sea) |
| Bacillariophyta | Phaeodactylum tricornutum | CBC42A | New isolate (Gulf of Naples, Western Mediterranean Sea) |
| Chlorophyta | Tetraselmis suecica | CBC20A | Neoalgae |
| Chlorophyta | Tetraselmis sp. | CBC27A | Phytobloom |
| Haptophyta | Isochrysis galbana | CBC21A | Phytobloom |
| Ochrophyta | Nannochloropsis sp. | CBC24A | Phytobloom |
| Miozoa | Amphidinium massartii | CBC39A | New isolate (Gulf of Naples, Western Mediterranean Sea) |
| Miozoa | Amphidinium carterae | CBC35A | New isolate (Fusaro lake, brakish lagoon) |
| Miozoa | Heterocapsa sp. | CBC53A | New isolate (Gulf of Naples, Western Mediterranean Sea) |
| Miozoa | Scrippsiella sp. | CBC55A | New isolate (Gulf of Naples, Western Mediterranean Sea) |
| Phylum | Taxonomy/Voucher Code | Identification Code | Collection Site |
|---|---|---|---|
| Bryozoa | Bugula neritina complex | CBC28A | Gulf of Naples, Western Mediterranean Sea |
| Bryozoa | Amathia verticillata | CBC30A | Gulf of Naples, Western Mediterranean Sea |
| Bryozoa | Schizoporella errata | CBC31A | Gulf of Naples, Western Mediterranean Sea |
| Bryozoa | Lepraliellidae ind. | CBC44A | Gulf of Naples, Western Mediterranean Sea |
| Bryozoa | ITA-AM 15 12/13 | CBC48A | Antarctica |
| Chlorophyta | Codium bursa | CBC33A | Gulf of Naples, Western Mediterranean Sea |
| Chordata (Tunicata) | Ciona robusta | CBC18A | Gulf of Naples, Western Mediterranean Sea |
| Chordata (Tunicata) | Halocynthia papillosa | CBC19A | Gulf of Naples, Western Mediterranean Sea |
| Chordata (Tunicata) | Styela plicata | CBC22A | Gulf of Naples, Western Mediterranean Sea |
| Chordata (Tunicata) | Ascidiella aspersa | CBC29A | Off the coast of Crotone, Ionian Sea |
| Chordata (Tunicata) | Botryllus schlosseri complex | CBC37A | Gulf of Naples, Western Mediterranean Sea |
| Chordata (Tunicata) | ITA-AM 12 12/13 | CBC50A | Antarctica |
| Cnidaria | Leptogorgia sarmentosa | CBC23A | Gulf of Naples, Western Mediterranean Sea |
| Cnidaria | ITA-AM 13 12/13 | CBC51A | Antarctica |
| Cnidaria | ITA-AM 14 12/13 | CBC62A | Antarctica |
| Mollusca | Philine quadripartita | CBC26A | Off the coast of Crotone, Ionian Sea |
| Mollusca | Hexaplex trunculus complex | CBC34A | Gulf of Naples, Western Mediterranean Sea |
| Mollusca | Euthria cornea | CBC38A | Gulf of Naples, Western Mediterranean Sea |
| Mollusca | Gasteropteron rubrum | CBC46C | Off the coast of Crotone, Ionian Sea |
| Mollusca | Gasteropteron rubrum | CBC46A | Western Mediterranean Sea |
| Mollusca | Scaphander lignarius complex | CBC47A | Off the coast of Crotone, Ionian Sea |
| Ochrophyta | Cystoseira dubia | CBC49A | Off the coast of Crotone, Ionian Sea |
| Ochrophyta | Dictyota sp. | CBC45A | Gulf of Naples, Western Mediterranean Sea |
| Porifera | Haliclona mediterranea | CBC1A | Gulf of Naples, Western Mediterranean Sea |
| Porifera | Chondrosia reniformis | CBC2A | Gulf of Naples, Western Mediterranean Sea |
| Porifera | Crambe crambe | CBC3A | Gulf of Naples, Western Mediterranean Sea |
| Porifera | Chondrilla nucula | CBC4A | Gulf of Naples, Western Mediterranean Sea |
| Porifera | ITA-AM 09 12/13 | CBC10A | Antarctica |
| Porifera | Spongia sp. (ITA-AM 06 16) | CBC12A | Off the coast of Crotone, Ionian Sea |
| Porifera | ITA-AM 07 02 | CBC11A | Antarctica |
| Porifera | ITA-AM 08 02 | CBC15A | Antarctica |
| Porifera | ITA-AM 02 02 | CBC13A | Antarctica |
| Porifera | ITA-AM 04 02 | CBC14A | Antarctica |
| Porifera | ITA-AM 05 02 | CBC16A | Antarctica |
| Porifera | ITA-AM 03 02 | CBC17A | Antarctica |
| Porifera | Spirastrella cunctatrix | CBC32A | Gulf of Naples, Western Mediterranean Sea |
| Porifera | ITA-AM 10 12/13 | CBC56A | Antarctica |
| Porifera | ITA-AM 11 12/13 | CBC57A | Antarctica |
| Porifera | Dendrilla sp. (GAN-38) | CBC74A-1 | Antarctica |
| Porifera | Dendrilla sp. (GAN-54) | CBC74B | Antarctica |
| Porifera | Dendrilla sp. (GAN-53) | CBC74A-3 | Antarctica |
| Porifera | GAN-3 | CBC75A | Antarctica |
| Porifera | GAN-37 | CBC77A | Antarctica |
| Rhodophyta | Pterocladiella capillacea | CBC43A | Gulf of Naples, Western Mediterranean Sea |
| Phylum | Frac. A % | Frac. B % | Frac. C % | Frac. D % | Frac. E% | Total OCs % |
|---|---|---|---|---|---|---|
| Bacillariophyta | 39.8 ± 9.3 | 11.7 ± 6.4 | 11.4 ± 3 | 10.4 ± 4.2 | 15.2 ± 10.4 | 48.7 |
| Bryozoa | 71.3 ± 8.2 | 3.2 ± 2.0 | 3.6 ± 1.2 | 2.1 ± 0.7 | 5.3 ± 2.0 | 14.2 |
| Chlorophyta | 52.6 ± 18.8 | 5.0 ± 6.1 | 8.0 ± 6.7 | 5.8 ± 4.3 | 6.6 ± 5.0 | 25.4 |
| Chordata (Tunicata) | 72.3 ± 9.7 | 2.6 ± 0.8 | 3.3 ± 1.7 | 3.1 ± 1.8 | 3.9 ± 1.8 | 12.9 |
| Cnidaria | 44.1 ± 12.9 | 3.6 ± 1.0 | 4.1 ± 1.4 | 5.8 ± 1.5 | 7.0 ± 3.6 | 20.6 |
| Haptophyta | 49.4 | 8.4 | 0.9 | 16.9 | 7.8 | 34.0 |
| Miozoa | 44.7 ± 8.1 | 3.9 ± 2.3 | 6.8 ± 4.1 | 8.4 ± 4.5 | 9.0 ± 5.1 | 28.0 |
| Mollusca | 63.7 ± 7.2 | 3.9 ± 0.9 | 3.9 ±1.4 | 2.9 ± 1.3 | 5.8 ± 2.4 | 16.5 |
| Ochrophyta | 56.1 ± 10.6 | 1.8 ± 2.4 | 4.1 ± 3.6 | 4.2 ± 4.7 | 6.2 ± 6.3 | 16.4 |
| Porifera | 62.0 ± 17.4 | 4.7 ± 2.1 | 3.7 ± 2.4 | 3.8 ± 2.7 | 5.8 ± 3.0 | 18.1 |
| Rhodophyta | 71.0 | 1.1 | 2.9 | 0.7 | 1.2 | 5.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreiras-Figueruelo, A.; Nuzzo, G.; Galasso, C.; Sansone, C.; Crocetta, F.; Mazzella, V.; Gallo, C.; Barra, G.; Sardo, A.; Iuliano, A.; et al. Probing the Therapeutic Potential of Marine Phyla by SPE Extraction. Mar. Drugs 2021, 19, 640. https://doi.org/10.3390/md19110640
Moreiras-Figueruelo A, Nuzzo G, Galasso C, Sansone C, Crocetta F, Mazzella V, Gallo C, Barra G, Sardo A, Iuliano A, et al. Probing the Therapeutic Potential of Marine Phyla by SPE Extraction. Marine Drugs. 2021; 19(11):640. https://doi.org/10.3390/md19110640
Chicago/Turabian StyleMoreiras-Figueruelo, Alejandro, Genoveffa Nuzzo, Christian Galasso, Clementina Sansone, Fabio Crocetta, Valerio Mazzella, Carmela Gallo, Giusi Barra, Angela Sardo, Antonella Iuliano, and et al. 2021. "Probing the Therapeutic Potential of Marine Phyla by SPE Extraction" Marine Drugs 19, no. 11: 640. https://doi.org/10.3390/md19110640
APA StyleMoreiras-Figueruelo, A., Nuzzo, G., Galasso, C., Sansone, C., Crocetta, F., Mazzella, V., Gallo, C., Barra, G., Sardo, A., Iuliano, A., Manzo, E., d’Ippolito, G., Albrigtsen, M., Andersen, J. H., Ianora, A., & Fontana, A. (2021). Probing the Therapeutic Potential of Marine Phyla by SPE Extraction. Marine Drugs, 19(11), 640. https://doi.org/10.3390/md19110640














