De Novo Transcriptome Assembly and Gene Expression Profiling of the Copepod Calanus helgolandicus Feeding on the PUA-Producing Diatom Skeletonema marinoi
Abstract
1. Introduction
2. Results
2.1. Transcriptome Sequencing, De-Novo Assembly and Annotation
2.2. Differential Expression Analysis
2.3. RT-qPCR of Selected GOIs
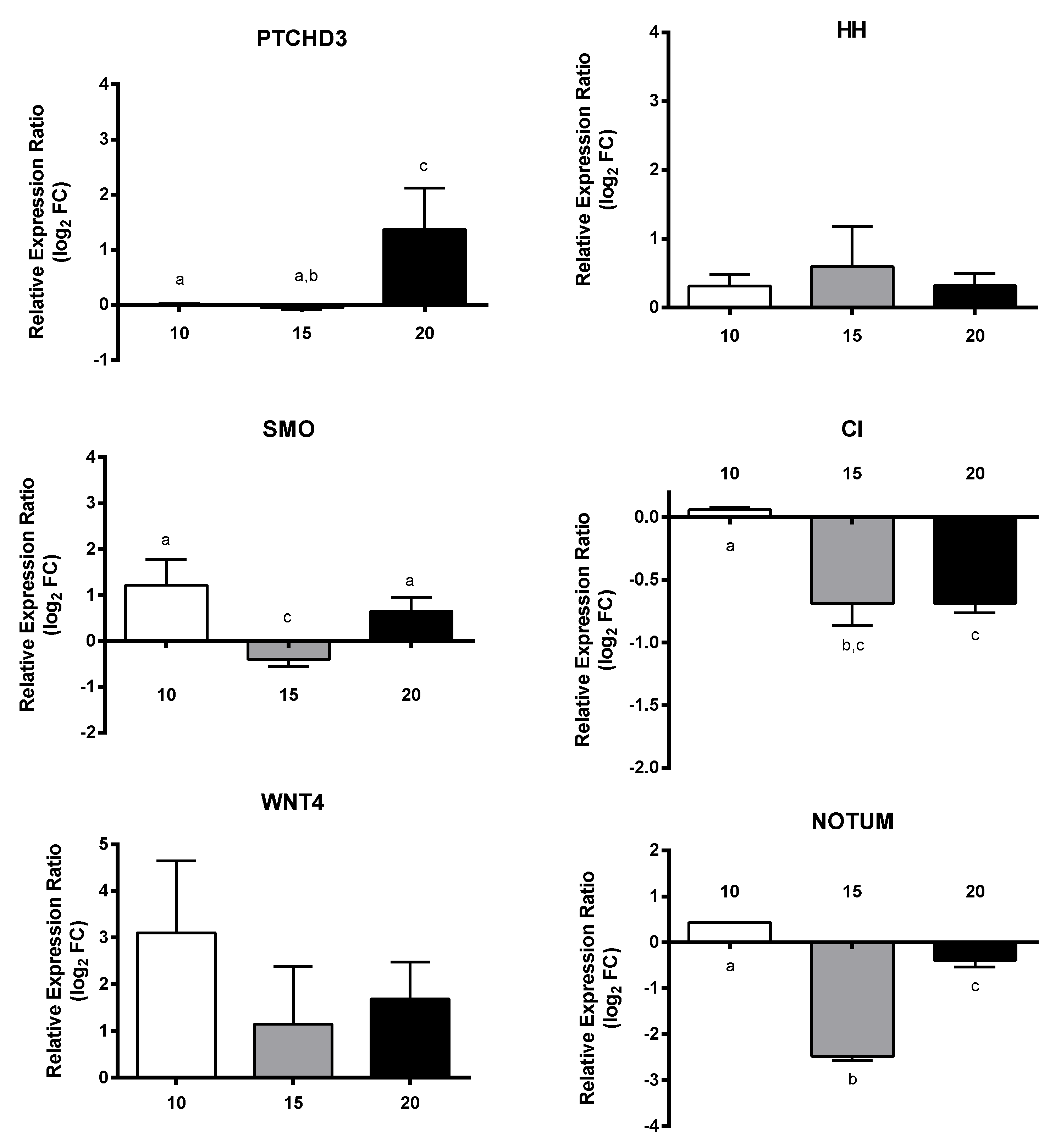
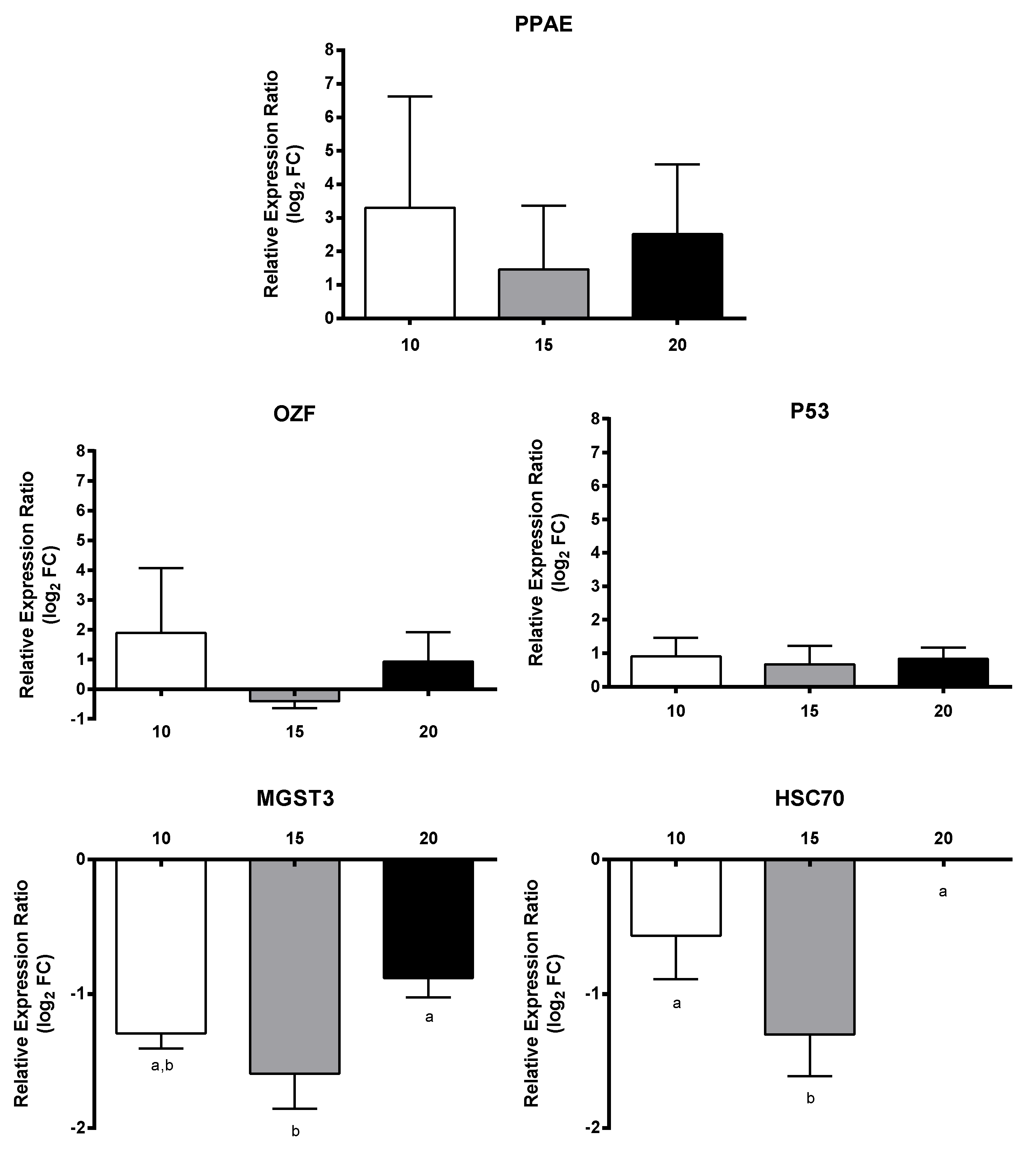
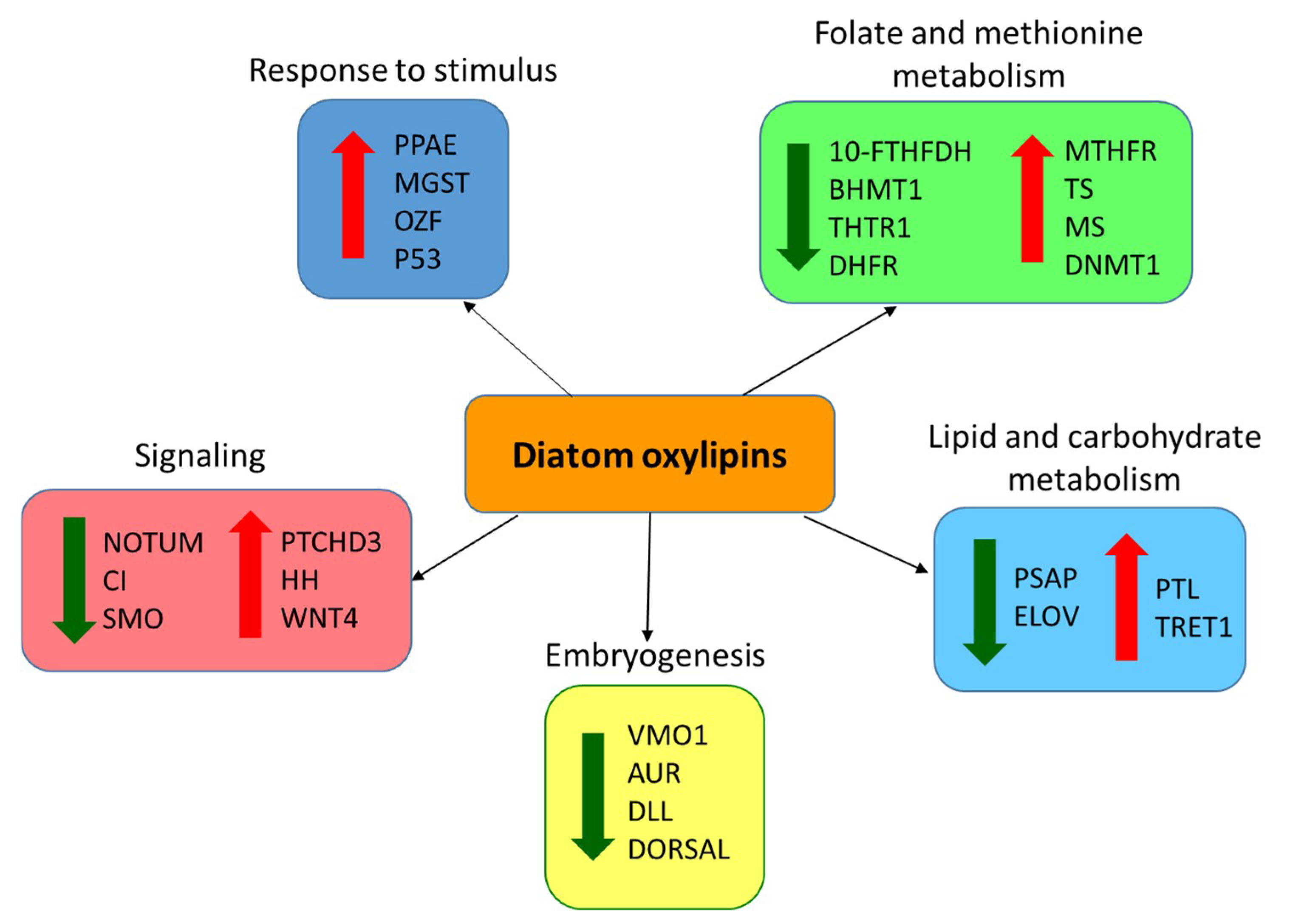
2.4. Effect of PUAs on Calanus helgolandicus Gene Expression
3. Discussion
4. Materials and Methods
4.1. Phytoplankton Culture
4.2. Copepod Collection and Feeding Experiments
4.3. Transcriptome Sequencing
4.4. De novo Transcriptome Assembly and Functional Annotation
4.5. Differential Gene Expression Analysis
4.6. RT-qPCR of Genes of Interest (GOIs)
4.7. PUAs Incubation Experiments and RT-qPCR of GOIs
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nelson, D.; Tréguer, P.; Brzezinski, M.; Leynaert, A.; Quéguiner, B.; Nelson, D.M.D.; Tréguer, P.; Brzezinski, M.A.; Leynaert, A.; Queguiner, B.; et al. Production and dissolution of biogenic silica in the ocean: Revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Glob. Biogeochem. Cycles 1995, 9, 359–732. [Google Scholar] [CrossRef]
- Ribalet, F.; Vidoudez, C.; Cassin, D.; Pohnert, G.; Ianora, A.; Miralto, A.; Casotti, R. High plasticity in the production of diatom derived polyunsaturated aldehydes under nutrient limitation: Physiological and ecological implications. Protist 2009, 160, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Vardi, A.; Bidle, K.; Kwityn, C.; Hirsh, D.J.; Thompson, S.M.; Callow, J.A.; Falkowski, P.; Bowler, C. A Diatom Gene Regulating Nitric-Oxide Signaling and Susceptibility to Diatom-Derived Aldehydes. Curr. Biol. 2008, 18, 895–899. [Google Scholar] [CrossRef] [PubMed]
- Ribalet, F.; Bastianini, M.; Vidoudez, C.; Acri, F.; Berges, J.; Ianora, A.; Miralto, A.; Pohnert, G.; Romano, G.; Wichard, T.; et al. Phytoplankton Cell Lysis Associated with Polyunsaturated Aldehyde Release in the Northern Adriatic Sea. PLoS ONE 2014, 9, e85947. [Google Scholar] [CrossRef] [PubMed]
- Russo, E.; d’Ippolito, G.; Fontana, A.; Sarno, D.; D’Alelio, D.; Busseni, G.; Ianora, A.; von Elert, E.; Carotenuto, Y. Density-dependent oxylipin production in natural diatom communities: Possible implications for plankton dynamics. ISME Journal 2020, 14, 164–177. [Google Scholar] [CrossRef] [PubMed]
- Ianora, A.; Bastianini, M.; Carotenuto, Y.; Casotti, R.; Roncalli, V.; Miralto, A.; Romano, G.; Gerecht, A.; Fontana, A.; Turner, J.T. Non-volatile oxylipins can render some diatom blooms more toxic for copepod reproduction. Harmful Algae 2015, 44, 1–7. [Google Scholar] [CrossRef]
- Ianora, A.; Miralto, A.; Poulet, S.A.; Carotenuto, Y.; Buttino, I.; Romano, G.; Casotti, R.; Pohnert, G.; Wichard, T.; Colucci-D’Amato, L.; et al. Aldehyde suppression of copepod recruitment in blooms of a ubiquitous planktonic diatom. Nature 2004, 429, 403–407. [Google Scholar] [CrossRef]
- Lauritano, C.; Romano, G.; Roncalli, V.; Amoresano, A.; Fontanarosa, C.; Bastianini, M.; Braga, F.; Carotenuto, Y.; Ianora, A. New oxylipins produced at the end of a diatom bloom and their effects on copepod reproductive success and gene expression levels. Harmful Algae 2016, 55, 221–229. [Google Scholar] [CrossRef]
- Poulet, S.A.; Laabir, M.; Ianora, A.; Miralto, A. Reproductive response of Calanus helgolandicus.1. abnormal embryonic and naupliar development. Mar. Ecol. Progr. Ser. 1995, 129, 85–95. [Google Scholar] [CrossRef]
- Ask, J.; Reinikainen, M.; Bamstedt, U. Variation in hatching success and egg production of Eurytemora affinis (Calanoida, Copepoda) from the Gulf of Bothnia, Baltic Sea, in relation to abundance and clonal differences of diatoms. J. Plankton Res. 2006, 28, 683–694. [Google Scholar] [CrossRef]
- Halsband-Lenka, C.; Piersona, J.J.; Leisingb, A.W. Reproduction of Pseudocalanus newmani (Copepoda: Calanoida) is deleteriously affected by diatom blooms—A field study. Progr. Oceanogr. 2005, 67, 332–348. [Google Scholar] [CrossRef]
- Poulet, S.A.; Escribano, R.; Hidalgo, P.; Cueff, A.; Wichard, T.; Aguilera, V.; Vargas, C.A.; Pohnert, G. Collapse of Calanus chilensis reproduction in a marine environment with high diatom concentration. J. Exp. Mar. Biol. Ecol. 2007, 352, 187–199. [Google Scholar] [CrossRef]
- Fontana, A.; d’Ippolito, G.; Cutignano, A.; Romano, G.; Lamari, N.; Massa Gallucci, A.; Cimino, G.; Miralto, A.; Ianora, A. LOX-Induced Lipid Peroxidation Mechanism Responsible for the Detrimental Effect of Marine Diatoms on Zooplankton Grazers. ChemBioChem. 2007, 8, 1810–1818. [Google Scholar] [CrossRef] [PubMed]
- Barreiro, A.; Carotenuto, Y.; Lamari, N.; Esposito, F.; D’Ippolito, G.; Fontana, A.; Romano, G.; Ianora, A.; Miralto, A.; Guisande, C. Diatom induction of reproductive failure in copepods: The effect of PUAs versus non-volatile oxylipins. J. Exp. Mar. Biol. Ecol. 2011, 401, 13–19. [Google Scholar] [CrossRef]
- Russo, E.; Ianora, A.; Carotenuto, Y. Re-shaping marine plankton communities: Effects of diatom oxylipins on copepods and beyond. Mar. Biol. 2019, 166, 9. [Google Scholar] [CrossRef]
- Ceballos, S.; Ianora, A. Different diatoms induce contrasting effects on the reproductive success of the copepod Temora stylifera. J. Exp. Mar. Biol. Ecol. 2003, 294, 189–202. [Google Scholar] [CrossRef]
- Ianora, A.; Romano, G.; Carotenuto, Y.; Esposito, F.; Roncalli, V.; Buttino, I.; Miralto, A. Impact of the diatom oxylipin 15S-HEPE on the reproductive success of the copepod Temora stylifera. Hydrobiologia 2011, 666, 265–275. [Google Scholar] [CrossRef]
- Ruocco, N.; Costantini, S.; Zupo, V.; Lauritano, C.; Caramiello, D.; Ianora, A.; Budillon, A.; Romano, G.; Nuzzo, G.; d’Ippolito, G.; et al. Toxigenic effects of two benthic diatoms upon grazing activity of the sea urchin: Morphological, metabolomic and de novo transcriptomic analysis. Sci. Rep. 2018, 8, 5622. [Google Scholar] [CrossRef]
- Sansone, C.; Braca, A.; Ercolesi, E.; Romano, G.; Palumbo, A.; Casotti, R.; Francone, M.; Ianora, A. Diatom-Derived Polyunsaturated Aldehydes Activate Cell Death in Human Cancer Cell Lines but Not Normal Cells. PLoS ONE 2014, 9, e101220. [Google Scholar] [CrossRef]
- Bonnet, D.; Richardson, A.; Harris, R.; Hirst, A.; Beaugrand, G.; Edwards, M.; Ceballos, S.; Diekman, R.; Lopez-Urrutia, A.; Valdes, L.; et al. An overview of Calanus helgolandicus ecology in European waters. Prog. Oceanogr. 2005, 65, 1–53. [Google Scholar] [CrossRef]
- Mauchline, J. The biology of Calanoid Copepods. In Advances in Marine Biology; Blaxter, J.H.S., Southward, A.J., Tyler, P.A., Eds.; Academic Press: Cambridge, MA, USA, 1998; p. 710. [Google Scholar]
- Kaartvedt, S. Life history of Calanus finmarchicus in the Norwegian Sea in relation to planktivorous fish. ICES J. Mar. Sci. 2000, 57, 1819–1824. [Google Scholar] [CrossRef]
- Frangoulis, C.; Christou, E.D.; Hecq, J.H. Comparison of marine copepod outfluxes: Nature, rate, fate and role in the carbon and nitrogen cycles. Adv. Mar. Biol. 2004, 47, 254. [Google Scholar]
- Mazzocchi, M.G.; Licandro, P.; Dubroca, L.; Di Capua, I.; Saggiomo, V. Zooplankton associations in a Mediterranean long-term time-series. J. Plankton Res. 2011, 33, 1163–1181. [Google Scholar] [CrossRef]
- Yebra, L.; Bonnet, D.; Harris, R.P.; Lindeque, P.K.; Peijnenburg, K.T.C.A. Barriers in the pelagic: Population structuring of Calanus helgolandicus and C. euxinus in European waters. Mar. Ecol. Progr. Ser. 2011, 428, 135–149. [Google Scholar] [CrossRef]
- Carotenuto, Y.; Esposito, F.; Pisano, F.; Lauritano, C.; Perna, M.; Miralto, A.; Ianora, A. Multi-generation cultivation of the copepod Calanus helgolandicus in a re-circulating system. J. Exp. Mar. Biol. Ecol. 2012, 418, 46–58. [Google Scholar] [CrossRef]
- Lauritano, C.; Borra, M.; Carotenuto, Y.; Biffali, E.; Miralto, A.; Procaccini, G.; Ianora, A. First molecular evidence of diatom effects in the copepod Calanus helgolandicus. J. Exp. Mar. Biol. Ecol. 2011, 404, 79–86. [Google Scholar] [CrossRef]
- Lauritano, C.; Borra, M.; Carotenuto, Y.; Biffali, E.; Miralto, A.; Procaccini, G.; Ianora, A. Molecular evidence of the toxic effects of diatom diets on gene expression patterns in copepods. PLoS ONE 2011, 6, e26850. [Google Scholar] [CrossRef]
- Lauritano, C.; Carotenuto, Y.; Miralto, A.; Procaccini, G.; Ianora, A. Copepod population-specific response to a toxic diatom diet. PLoS ONE 2012, e47262. [Google Scholar] [CrossRef]
- Carotenuto, Y.; Dattolo, E.; Lauritano, C.; Pisano, F.; Sanges, R.; Miralto, A.; Procaccini, G. Insights into the transcriptome of the marine copepod Calanus helgolandicus feeding on the oxylipin-producing diatom Skeletonema marinoi. Harmful Algae 2014, 31, 153–162. [Google Scholar] [CrossRef]
- Amato, A.; Carotenuto, Y. Planktonic calanoids embark into the “omics” era. In Trends in Copepod Studies—Distribution, Biology and Ecology; Uttieri, M., Ed.; Nova Science Publishers Inc.: New York, NY, USA, 2018; pp. 287–314. ISBN 978-1 53612-593-1. [Google Scholar]
- Lenz, P.H.; Roncalli, V.; Hassett, R.P.; Wu, L.-S.; Cieslak, M.C.; Hartline, D.K.; Christie, A.E. De Novo Assembly of a Transcriptome for Calanus finmarchicus (Crustacea, Copepoda)—The Dominant Zooplankter of the North Atlantic Ocean. PLoS ONE 2014, 9, e88589. [Google Scholar] [CrossRef]
- Ning, J.; Wang, M.; Li, C.; Sun, S. Transcriptome Sequencing and De Novo Analysis of the Copepod Calanus sinicus Using 454 GS FLX. PLoS ONE 2013, 8, e63741. [Google Scholar] [CrossRef] [PubMed]
- Roncalli, V.; Sommer, S.A.; Cieslak, M.C.; Clarke, C.; Hopcroft, R.R.; Lenz, P.H. Physiological characterization of the emergence from diapause: A transcriptomics approach. Sci. Rep. 2018, 8, 12577. [Google Scholar] [CrossRef] [PubMed]
- Semmouri, I.; Asselman, J.; Van Nieuwerburgh, F.; Deforce, D.; Janssen, C.R.; De Schamphelaere, K.A.C. The transcriptome of the marine calanoid copepod Temora longicornis under heat stress and recovery. Mar. Environ. Res. 2019, 143, 10–23. [Google Scholar] [CrossRef]
- Zhou, C.; Carotenuto, Y.; Vitiello, V.; Wu, C.; Zhang, J.; Buttino, I. De novo transcriptome assembly and differential gene expression analysis of the calanoid copepod Acartia tonsa exposed to nickel nanoparticles. Chemosphere 2018, 209, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Roncalli, V.; Cieslak, M.C.; Lenz, P.H. Transcriptomic responses of the calanoid copepod Calanus finmarchicus to the saxitoxin producing dinoflagellate Alexandrium fundyense. PLoS ONE 2016, 6, 25708. [Google Scholar] [CrossRef] [PubMed]
- Smolina, I.; Kollias, S.; Møller, E.F.; Lindeque, P.; Sundaram, A.Y.M.; Fernandes, J.M.O.; Hoarau, G. Contrasting transcriptome response to thermal stress in two key zooplankton species, Calanus finmarchicus and C. glacialis. Mar. Ecol. Progr. Ser. 2015, 534, 79–93. [Google Scholar] [CrossRef]
- Tarrant, A.; Baumgartner, M.; Hansen, B.; Altin, D.; Nordtug, T.; Olsen, A. Transcriptional profiling of reproductive development, lipid storage and molting throughout the last juvenile stage of the marine copepod Calanus finmarchicus. Front. Zool. 2014, 11, 91. [Google Scholar] [CrossRef]
- Yang, Q.; Sun, F.; Yang, Z.; Li, H. Comprehensive Transcriptome Study to Develop Molecular Resources of the Copepod Calanus sinicus for Their Potential Ecological Applications. BioMed Res. Int. 2014, 2014, 12. [Google Scholar] [CrossRef]
- Aubry, F.B.; Berton, A.; Bastianini, M.; Socal, G.; Acri, F. Phytoplankton succession in a coastal area of the NW Adriatic, over a 10-year sampling period (1990–1999). Cont. Shelf Res. 2004, 24, 97–115. [Google Scholar] [CrossRef]
- Li, C.; Weng, S.; Chen, Y.; Yu, X.; Lü, L.; Zhang, H.; He, J.; Xu, X. Analysis of Litopenaeus vannamei Transcriptome Using the Next-Generation DNA Sequencing Technique. PLoS ONE 2012, 7, e47442. [Google Scholar] [CrossRef]
- Gerecht, A.; Carotenuto, Y.; Ianora, A.; Romano, G.; Fontana, A.; d’Ippolito, G.; Jakobsen, H.H.; Nejstgaard, J. Oxylipin production during a mesocosm bloom of Skeletonema marinoi. J. Exp. Mar. Biol. Ecol. 2013, 446, 159–165. [Google Scholar] [CrossRef]
- Ayala, A.; Muñoz, M.F.; Argüelles, S. Lipid Peroxidation: Production, Metabolism, and Signaling Mechanisms of Malondialdehyde and 4-Hydroxy-2-Nonenal. Oxidative Med. Cell. Longev. 2014, 2014, 360438. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, J.; Lerman, L.O.; Lerman, A. Ubiquitin and ubiquitin-like proteins in protein regulation. Circ. Res. 2007, 100, 1276–1291. [Google Scholar] [CrossRef] [PubMed]
- Hansen, B.H.; Altin, D.; Booth, A.; Vang, S.-H.; Frenzel, M.; Sørheim, K.R.; Brakstad, O.G.; Størseth, T.R. Molecular effects of diethanolamine exposure on Calanus finmarchicus (Crustacea: Copepoda). Aquat. Toxicol. 2010, 99, 212–222. [Google Scholar] [CrossRef]
- Vazquez, L.; Alpuche, J.; Maldonado, G.; Agundis, C.; Pereyra-Morales, A.; Zenteno, E. Review: Immunity mechanisms in crustaceans. Innate Immun. 2009, 15, 179–188. [Google Scholar] [CrossRef]
- Li, S.; Zhang, X.; Sun, Z.; Li, F.; Xiang, J. Transcriptome analysis on Chinese shrimp Fenneropenaeus chinensis during WSSV acute infection. PLoS ONE 2013, 8, e58627. [Google Scholar] [CrossRef]
- Guo, H.; Ye, C.-X.; Wang, A.-L.; Xian, J.-A.; Liao, S.-A.; Miao, Y.-T.; Zhang, S.-P. Trascriptome analysis of the Pacific white shrimp Litopenaeus vannamei exposed to nitrite by RNA-seq. Fish Shellfish Immun. 2013, 35, 2008–2016. [Google Scholar] [CrossRef]
- Hansen, D.K.; Billings, R.E. Phenytoin teratogenicity and effects on embryonic and maternal folate metabolism [published errtum appears in Teratology 1986 Dec;34(3):487]. Teratology 1985, 31, 363–371. [Google Scholar] [CrossRef]
- Lee, M.S.; Bonner, J.R.; Bernard, D.J.; Sanchez, E.L.; Sause, E.T.; Prentice, R.R.; Burgess, S.M.; Brody, L.C. Disruption of the folate pathway in zebrafish causes developmental defects. BMC Dev. Biol. 2012, 12. [Google Scholar] [CrossRef]
- Padmanabhan, N.; Jia, D.; Geary-Joo, C.; Wu, X.; Ferguson-Smith, A.C.; Fung, E.; Bieda, M.C.; Snyder, F.F.; Gravel, R.A.; Cross, J.C.; et al. Mutation in folate metabolism causes epigenetic instability and transgenerational effects on development. Cell 2013, 155, 81–93. [Google Scholar] [CrossRef]
- Affleck, J.G.; Neumann, K.; Wong, L.; Walker, V.K. The Effects of Methotrexate on Drosophila Development, Female Fecundity, and Gene Expression. Toxicol. Sci. 2006, 89, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Hernandorena, A. The patterning function of purines in the brine shrimp Artemia. Comptes Rendus De L Acad. Des. Sci. Ser. III Sci. De La Vie-Life Sci. 1999, 322, 289–301. [Google Scholar] [CrossRef]
- Affleck, J.G.; Walker, V.K. Transgenic rescue of methotrexate-induced teratogenicity in Drosophila melanogaster. Toxicol. Sci. 2007, 99, 522–531. [Google Scholar] [CrossRef]
- Arvizu-Flores, A.A.; Aispuro-Hernandez, E.; Garcia-Orozco, K.D.; Varela-Romero, A.; Valenzuela-Soto, E.; Velazquez-Contreras, E.F.; Rojo-Domínguez, A.; Yepiz-Plascencia, G.; Maley, F.; Sotelo-Mundo, R.R. Functional identity of the active sites of crustacean and viral thymidylate synthases. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2009, 150, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Greene, N.D.; Stanier, P.; Copp, A.J. Genetics of human neural tube defects. Hum. Mol. Genet. 2009, 18, R113–R129. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.N.; Lee, G.H.; Kao, T.T.; Lin, C.Y.; Hsiao, T.H.; Tsai, J.N.; Chen, B.H.; Chen, Y.H.; Wu, H.R.; Tsai, H.J.; et al. Knocking down 10-Formyltetrahydrofolate dehydrogenase increased oxidative stress and impeded zebrafish embryogenesis by obstructing morphogenetic movement. Biochim. Biophys. Acta 2014, 1840, 2340–2350. [Google Scholar] [CrossRef]
- Hazra, A.; Selhub, J.; Chao, W.; Ueland, P.M.; Hunter, D.J.; Baron, J.A. Uracil misincorporation into DNA and folic acid supplementation. Am. J Clin. Nutr. 2010, 91, 160–165. [Google Scholar] [CrossRef]
- Roy, M.; Leclerc, D.; Wu, Q.; Gupta, S.; Kruger, W.D.; Rozen, R. Valproic acid increases expression of methylenetetrahydrofolate reductase (MTHFR) and induces lower teratogenicity in MTHFR deficiency. J. Cell. Biochem. 2008, 105, 467–476. [Google Scholar] [CrossRef]
- Hirasawa, R.; Chiba, H.; Kaneda1, M.; Tajima, S.; Li, E.; Jaenisch, R.; Sasaki, H. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Develop. 2008, 22, 1607–1616. [Google Scholar] [CrossRef]
- Aşkın, H.; Uysal, H.; Altun, D. Preventive role of folic acid on the developmental toxicity of phenol in Drosophila melanogaster. Toxicol. Ind. Health 2007, 23, 591–598. [Google Scholar] [CrossRef]
- Wolfram, S.; Nejstgaard, J.C.; Pohnert, G. Accumulation of Polyunsaturated Aldehydes in the Gonads of the Copepod Acartia tonsa Revealed by Tailored Fluorescent Probes. PLoS ONE 2014, 9, e112522. [Google Scholar] [CrossRef] [PubMed]
- Buttino, I.; De Rosa, G.; Carotenuto, Y.; Mazzaella, M.; Ianora, A.; Esposito, F.; Vitiello, V.; Quaglia, F.; La Rotonda, M.I.; Miralto, A. Aldehyde-encapsulating liposomes impair marine grazer survivorship. J. Exp. Biol. 2008, 211, 1426–1433. [Google Scholar] [CrossRef] [PubMed]
- Byrne, B.M.; Gruber, M.; Ab, G. The evolution of egg yolk proteins. Progr. Biophys. Mol. Biol. 1989, 53, 33–69. [Google Scholar] [CrossRef]
- Elalayli, M.; Hall, J.D.; Fakhouri, M.; Neiswender, H.; Ellison, T.T.; Han, Z.; Roon, P.; LeMosy, E.K. Palisade is required in the Drosophila ovary for assembly and function of the protective vitelline membrane. Develop. Biol. 2008, 319, 359–369. [Google Scholar] [CrossRef][Green Version]
- Stevens, L.M.; Beuchle, D.; Jurcsak, J.; Tong, X.; Stein, D. The Drosophila embryonic patterning determinant torsolike is a component of the eggshell. Curr. Biol. 2003, 13, 1058–1063. [Google Scholar] [CrossRef]
- Heckmann, L.H.; Sibly, R.M.; Connon, R.; Hooper, H.L.; Hutchinson, T.H.; Maund, S.J.; Hill, C.J.; Bouetard, A.; Callaghan, A. Systems biology meets stress ecology: Linking molecular and organismal stress responses in Daphnia magna. Genome Biol. 2008, 9, R40. [Google Scholar] [CrossRef]
- Poulet, S.A.; Cueff, A.; Wichard, T.; Marchetti, J.; Dancie, C.; Pohnert, G. Influence of diatoms on copepod reproduction. III. Consequences of abnormal oocyte maturation on reproductive factors in Calanus helgolandicus. Mar. Biol. 2007, 152, 415–428. [Google Scholar] [CrossRef]
- Farley, B.M.; Ryder, S.P. Regulation of maternal mRNAs in early development. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 135–162. [Google Scholar] [CrossRef]
- Yao, L.-J.; Zhong, Z.-S.; Zhang, L.-S.; Chen, D.-Y.; Schatten, H.; Sun, Q.-Y. Aurora-A Is a Critical Regulator of Microtubule Assembly and Nuclear Activity in Mouse Oocytes, Fertilized Eggs, and Early Embryos. Biol. Reprod. 2004, 70, 1392–1399. [Google Scholar] [CrossRef][Green Version]
- Glover, D.M.; Leibowitz, M.H.; McLean, D.A.; Parry, H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell 1995, 81, 32–44. [Google Scholar] [CrossRef]
- Panganiban, G.; Rubenstein, J.L.R. Developmental functions of the Distal-less/Dlx homeobox genes. Development 2002, 129, 4371–4386. [Google Scholar] [PubMed]
- Panganiban, G.; Irvine, S.M.; Lowe, C.; Roehl, H.; Corley, L.S.; Sherbon, B.; Grenier, J.K.; Fallon, J.F.; Kimble, J.; Walker, M.; et al. The origin and evolution of animal appendages. Proc. Natl. Acad. Sci. USA 1997, 94, 5162–5166. [Google Scholar] [CrossRef] [PubMed]
- Steward, R.; Zusman, S.B.; Huang, L.H.; Schedl, P. The dorsal protein is distributed in a gradient in early Drosophila embryos. Cell 1988, 55, 487–495. [Google Scholar] [CrossRef]
- Perrimon, N.; Pitsouli, C.; Shilo, B.Z. Signaling mechanisms controlling cell fate and embryonic patterning. Cold Spring Harb. Perspect. Biol. 2012, 4, a005975. [Google Scholar] [CrossRef]
- Jiang, J.; Struhl, G. Protein kinase A and hedgehog signaling in Drosophila limb development. Cell 1995, 80, 563–572. [Google Scholar] [CrossRef]
- Cohen, S.M.; Brönner, G.; Küttner, F.; Jürgens, G.; Jäckle, H. Distal-less encodes a homoeodomain protein required for limb development in Drosophila. Nature 1989, 338, 432–434. [Google Scholar] [CrossRef]
- Liubicich, D.M.; Serano, J.M.; Pavlopoulos, A.; Kontarakis, Z.; Protas, M.E.; Kwan, E.; Chatterjee, S.; Tran, K.D.; Averof, M.; Patel, N.H. Knockdown of Parhyale Ultrabithorax recapitulates evolutionary changes in crustacean appendage morphology. Proc. Natl. Acad. Sci. USA 2009, 106, 13892–13896. [Google Scholar] [CrossRef]
- Kato, Y.; Shiga, Y.; Kobayashi, K.; Tokishita, S.; Yamagata, H.; Iguchi, T.; Watanabe, H. Development of an RNA interference method in the cladoceran crustacean Daphnia magna. Dev. Genes Evol. 2011, 220, 337–345. [Google Scholar] [CrossRef]
- Hiruta, C.; Toyota, K.; Miyakawa, H.; Ogino, Y.; Miyagawa, S.; Tatarazako, N.; Shaw, J.R.; Iguchi, T. Development of a microinjection system for RNA interference in the water flea Daphnia pulex. BMC Biotech. 2013, 13, 96. [Google Scholar] [CrossRef]
- Asai, S.; Ianora, A.; Lauritano, C.; Lindeque, P.K.; Carotenuto, Y. High-quality RNA extraction from copepods for Next Generation Sequencing: A comparative study. Mar. Genom. 2015, 24, 115–118. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Bolger, A.M.; Nagel, A.; Fernie, A.R.; Lunn, J.E.; Stitt, M.; Usadel, B. RobiNA: A user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 2012, 40, W622–W627. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotech. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Roncalli, V.; Christie, A.E.; Sommer, S.A.; Cieslak, M.C.; Hartline, D.K.; Lenz, P.H. A deep transcriptomic resource for the copepod crustacean Labidocera madurae: A potential indicator species for assessing near shore ecosystem health. PLoS ONE 2017, 12, e0186794. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef] [PubMed]
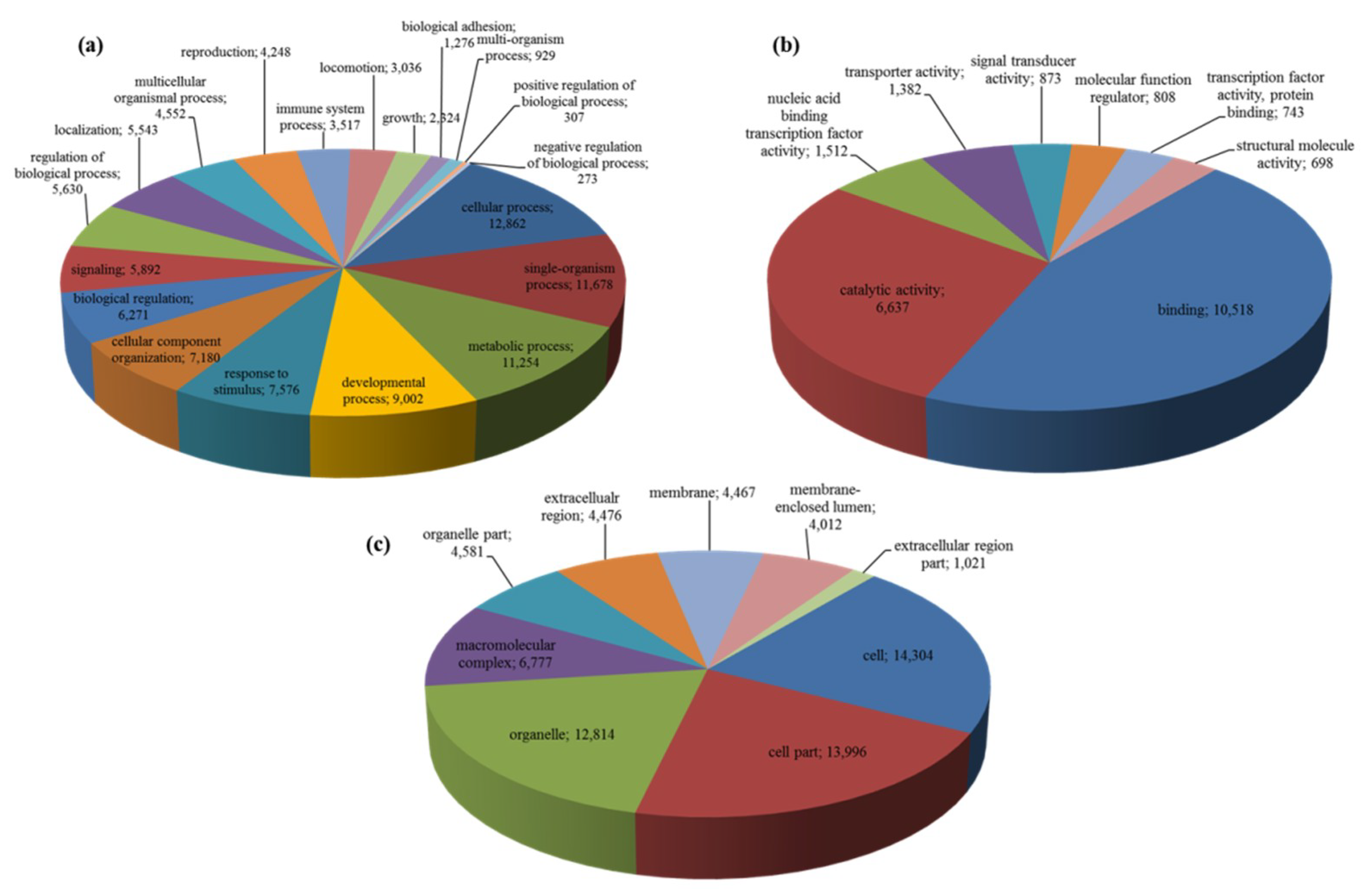

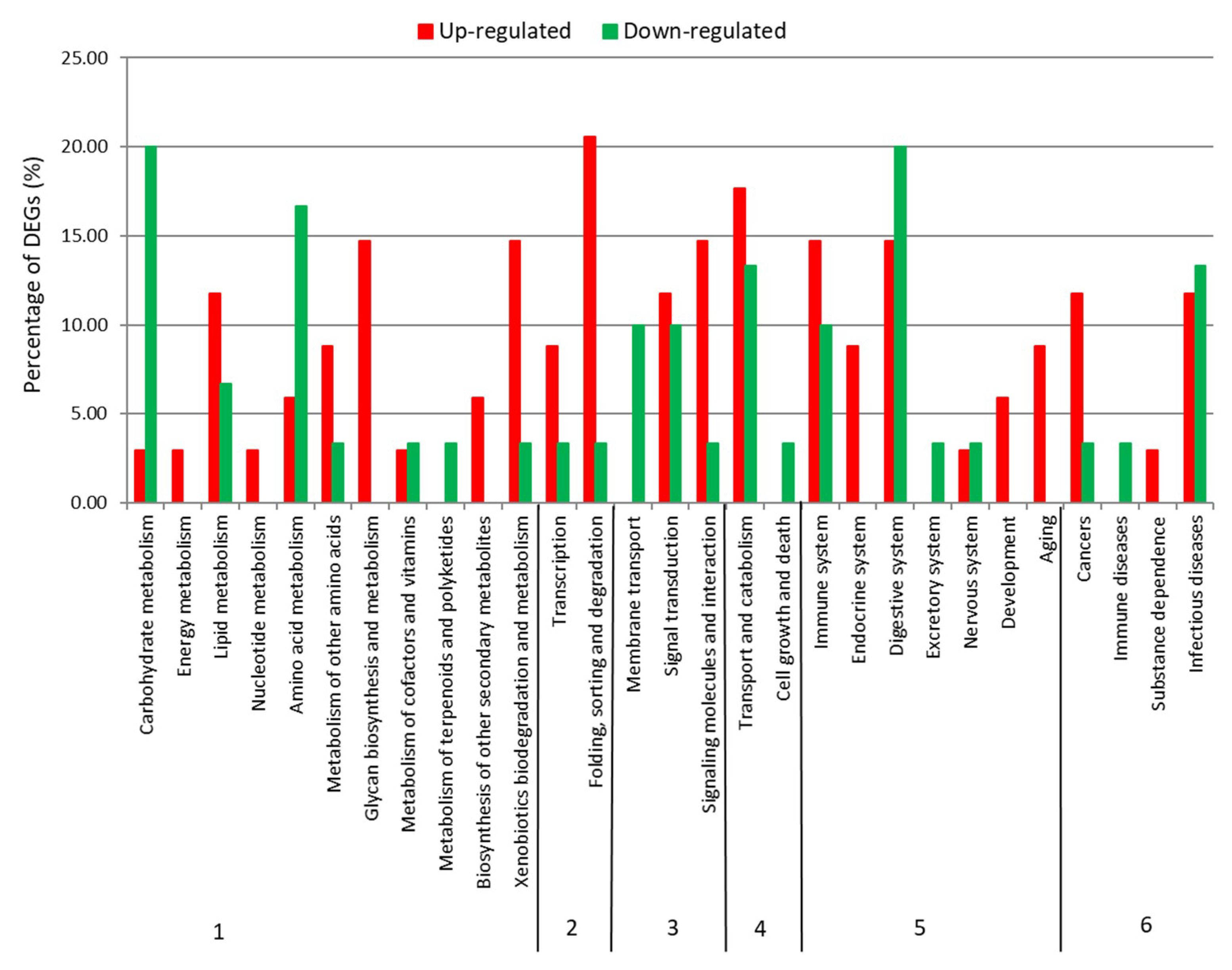
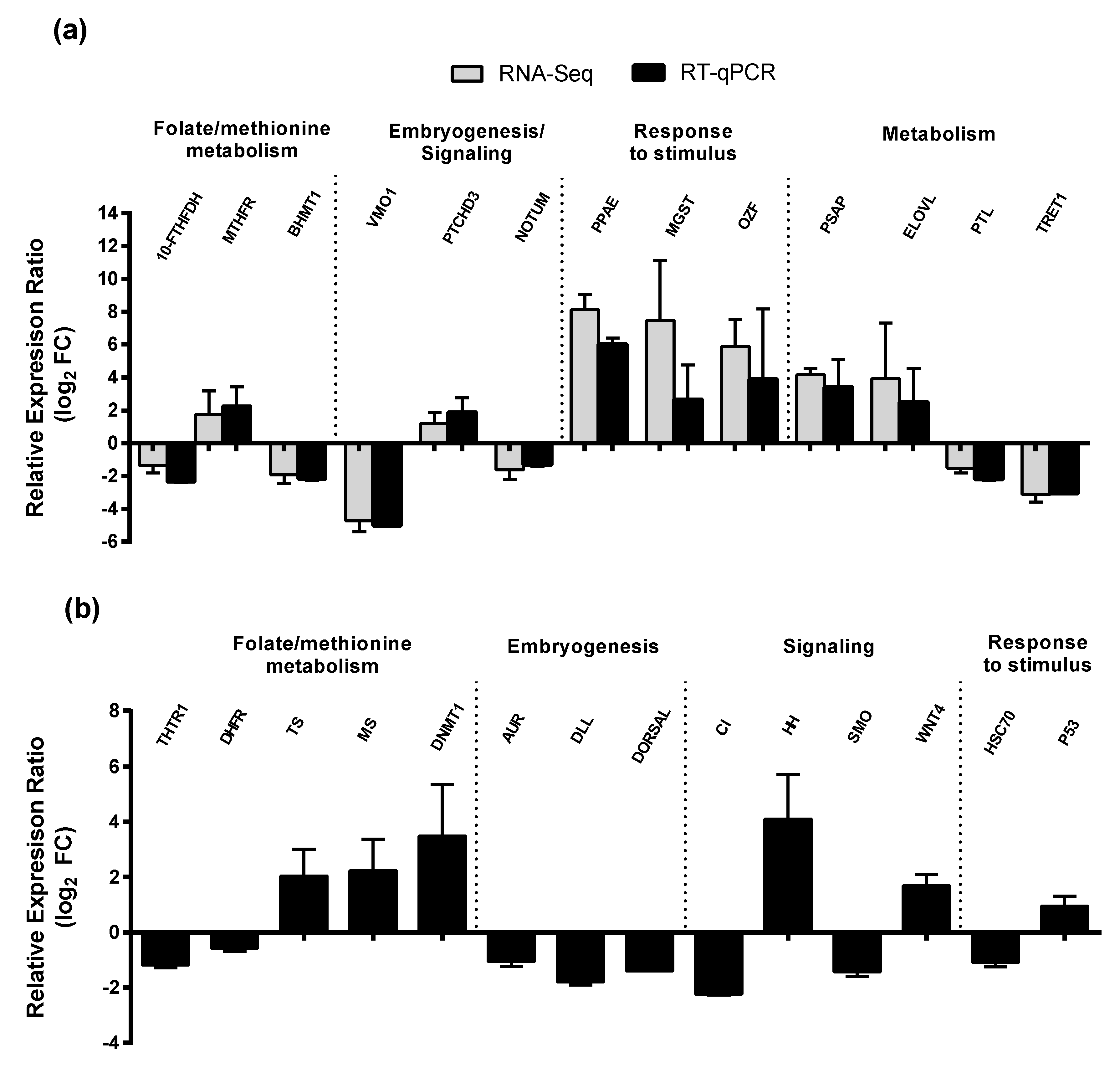

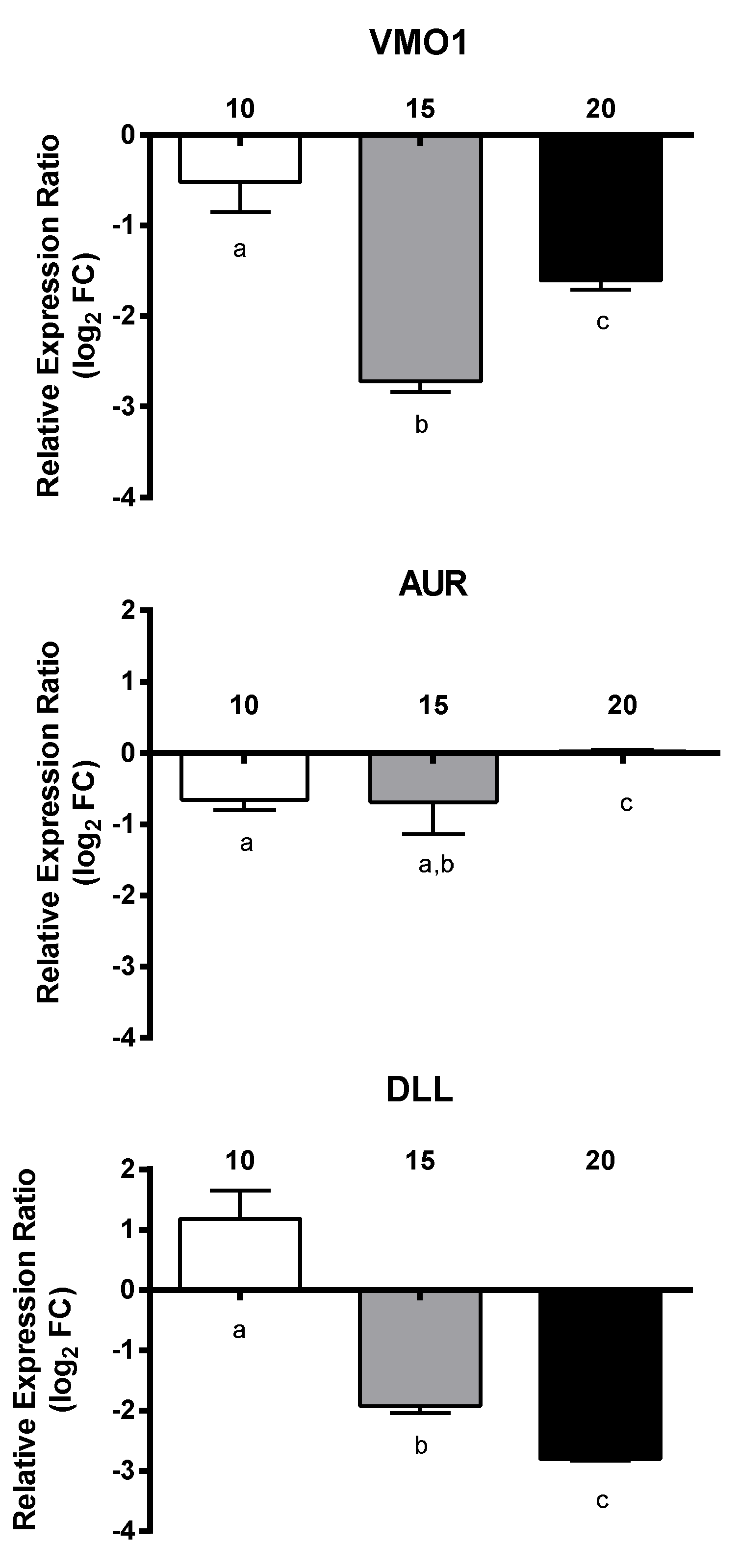

| Category | Number/Length |
|---|---|
| Reads from raw data | 726,490,334 |
| Average read length (bp) | 50 |
| Reads after trimming | 605,681,621 |
| Percentage retained | 83.4% |
| Unigenes | 30,339 |
| Average length (bp) | 1427 |
| N50 (bp) | 1784 |
| Database | Number | % |
|---|---|---|
| All unigenes | 30,339 | 100 |
| Nr | 21,337 | 70.3 |
| Swissprot | 19,386 | 63.9 |
| GO | 18,167 | 59.9 |
| KEGG | 6361 | 21.0 |
| Name | Primer Sequence 5′–3′ | As (bp) | E (%) | |
|---|---|---|---|---|
| 10-Formyltetrahydrofolate dehydrogenase | 10-FTHFDH | CTTGCCAGGAACAGGAAGAG | 170 | 111 |
| AGATCAGCGGAGACTTTCCA | ||||
| Betaine homocysteine s-methyltransferase 1 | BHMT1 | TCGTGCTGGAGCTGATATTG | 143 | 102 |
| GGGTGTGATATGCCAGAGGT | ||||
| Methylenetetrahydrofolate reductase | MTHFR | TATCCACCAGGCAACACAGA | 172 | 102 |
| GGCAGGATCAGCAGAAAGTC | ||||
| Vitelline membrane outer layer protein 1 | VMO1 | CTGGCATGAGGAACACCTTT | 148 | 117 |
| AGCAGCATCCAGGTCAGTTT | ||||
| Patched domain-containing protein 3 | PTCHD3 | TGGAGGAATATCGGACTTGC | 148 | 118 |
| TGGTGATGTCCCAGAAGTGA | ||||
| Palmitoleoyl-protein carboxylesterase NOTUM | NOTUM | TTGTACACAGGCACCAGGAA | 142 | 117 |
| CACCAATGAGCACAAATTGC | ||||
| Elongation of very long-chain fatty acids protein | ELOVL | GCCCAAGATTTATTGGTGGA | 192 | 107 |
| GCTGGATAGCGTGGAAGAAA | ||||
| Prosaposin | PSAP | AGACTTGGACAATTGGCTGGT | 112 | 101 |
| GCACATTGTTTCCAGGTCCTC | ||||
| Pancreatic triacylglycerol lipase | PTL | CTGGCTTGAGGCTATTCCTG | 179 | 103 |
| CTGAGCCTCCACTTGGGTAG | ||||
| Facilitated trehalose transporter 1 | TRET1 | TTTGGCTGAAAGGATTGGTC | 110 | 92 |
| ACATCATCAAGGACGGGAAC | ||||
| Prophenoloxidase activating enzyme | PPAE | ATCTGCTGCCGAGTGTAACC | 127 | 126 |
| TCCCCCATTATCTGCATAGC | ||||
| Microsomal glutathione s-transferase 3 | MGST3 | CCAGAGAGCACACCAGAACA | 156 | 106 |
| GGCTCGCCTGTGTAATATCC | ||||
| Zinc finger protein OZF | OZF | TGTTTGGCTGTGAAGTTTGC | 153 | 101 |
| TCAATGTGTGGGTCTTCAGG | ||||
| Name | Primer Sequence 5′–3′ | As (bp) | E (%) | |
|---|---|---|---|---|
| Thymidylate synthase | TS | CCGAATACACCAACATGCAC | 184 | 109 |
| TCTGCCACGTAGAACTGCAC | ||||
| Methionine synthase | MS | GGGACCTTTGATGAGTGGAA | 152 | 95 |
| ACAGTGCGGCTTGTCTTTCT | ||||
| DNA (cytosine-5)-methyltransferase 1 | DNMT1 | ACAACAACTGGGCTGGTCTC | 188 | 100 |
| GGGTGTGCCGTAGAACTTGT | ||||
| Thiamine transporter 1 | THTR1 | CCCGAACCAACTGTTCAAAT | 189 | 88 |
| ATGGGCTGGCTTTATCTCCT | ||||
| Dihydrofolate reductase | DHFR | GATCAAGTCTGAGCTGGCGT | 154 | 112 |
| CCTGGAGAGCACGATGTTGA | ||||
| Aurora kinase B | AUR | CTCAAGGAGAGCCACCATGT | 193 | 122 |
| CCTCAGGTCCACCCTTGTAA | ||||
| Homeotic protein distal-less | DLL | AGTTCCCATTCCCAGGAGGT | 199 | 97 |
| GGCAGAGCTAGGTACTGGGT | ||||
| Embryonic polarity protein dorsal | DORSAL | CAGCCAGCACCCAAGAGAAT | 143 | 104 |
| GCATCCTTCCTTCCCAACCA | ||||
| Heat shock cognate protein 70 | HSC70 | TCGGAATTGATCTTGGAACC | 149 | 103 |
| TGCAGCATCTCCAACAAGTC | ||||
| Sonic hedgehog protein | HH | TCTGATCTCGGACTGGTTGA | 189 | 116 |
| CTGGCAGGGTAGAGAGCAAC | ||||
| Transcriptional activator cubitus interruptus | CI | TGCACGTTTGAAGGCTGTTG | 153 | 100 |
| ATTCTGGTGTTTCGCCCTGT | ||||
| Protein smoothened | SMO | AATGAGGTGGAGGAGTGTGG | 184 | 100 |
| AGAAGATTGCCAGAGCAGGA | ||||
| Protein Wnt-4 | WNT4 | GACGCACAAGACAGACGAAA | 123 | 107 |
| GCACTTGCATTCAACCTTCA | ||||
| Cellular tumor antigen p53 | P53 | AGACCCTTCCAACAGAGCAA | 186 | 129 |
| CAAGACCCGAGACACATGAA | ||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asai, S.; Sanges, R.; Lauritano, C.; Lindeque, P.K.; Esposito, F.; Ianora, A.; Carotenuto, Y. De Novo Transcriptome Assembly and Gene Expression Profiling of the Copepod Calanus helgolandicus Feeding on the PUA-Producing Diatom Skeletonema marinoi. Mar. Drugs 2020, 18, 392. https://doi.org/10.3390/md18080392
Asai S, Sanges R, Lauritano C, Lindeque PK, Esposito F, Ianora A, Carotenuto Y. De Novo Transcriptome Assembly and Gene Expression Profiling of the Copepod Calanus helgolandicus Feeding on the PUA-Producing Diatom Skeletonema marinoi. Marine Drugs. 2020; 18(8):392. https://doi.org/10.3390/md18080392
Chicago/Turabian StyleAsai, Sneha, Remo Sanges, Chiara Lauritano, Penelope K. Lindeque, Francesco Esposito, Adrianna Ianora, and Ylenia Carotenuto. 2020. "De Novo Transcriptome Assembly and Gene Expression Profiling of the Copepod Calanus helgolandicus Feeding on the PUA-Producing Diatom Skeletonema marinoi" Marine Drugs 18, no. 8: 392. https://doi.org/10.3390/md18080392
APA StyleAsai, S., Sanges, R., Lauritano, C., Lindeque, P. K., Esposito, F., Ianora, A., & Carotenuto, Y. (2020). De Novo Transcriptome Assembly and Gene Expression Profiling of the Copepod Calanus helgolandicus Feeding on the PUA-Producing Diatom Skeletonema marinoi. Marine Drugs, 18(8), 392. https://doi.org/10.3390/md18080392









