Expression and Characterization of an Alginate Lyase and Its Thermostable Mutant in Pichia pastoris
Abstract
1. Introduction
2. Results and Discussion
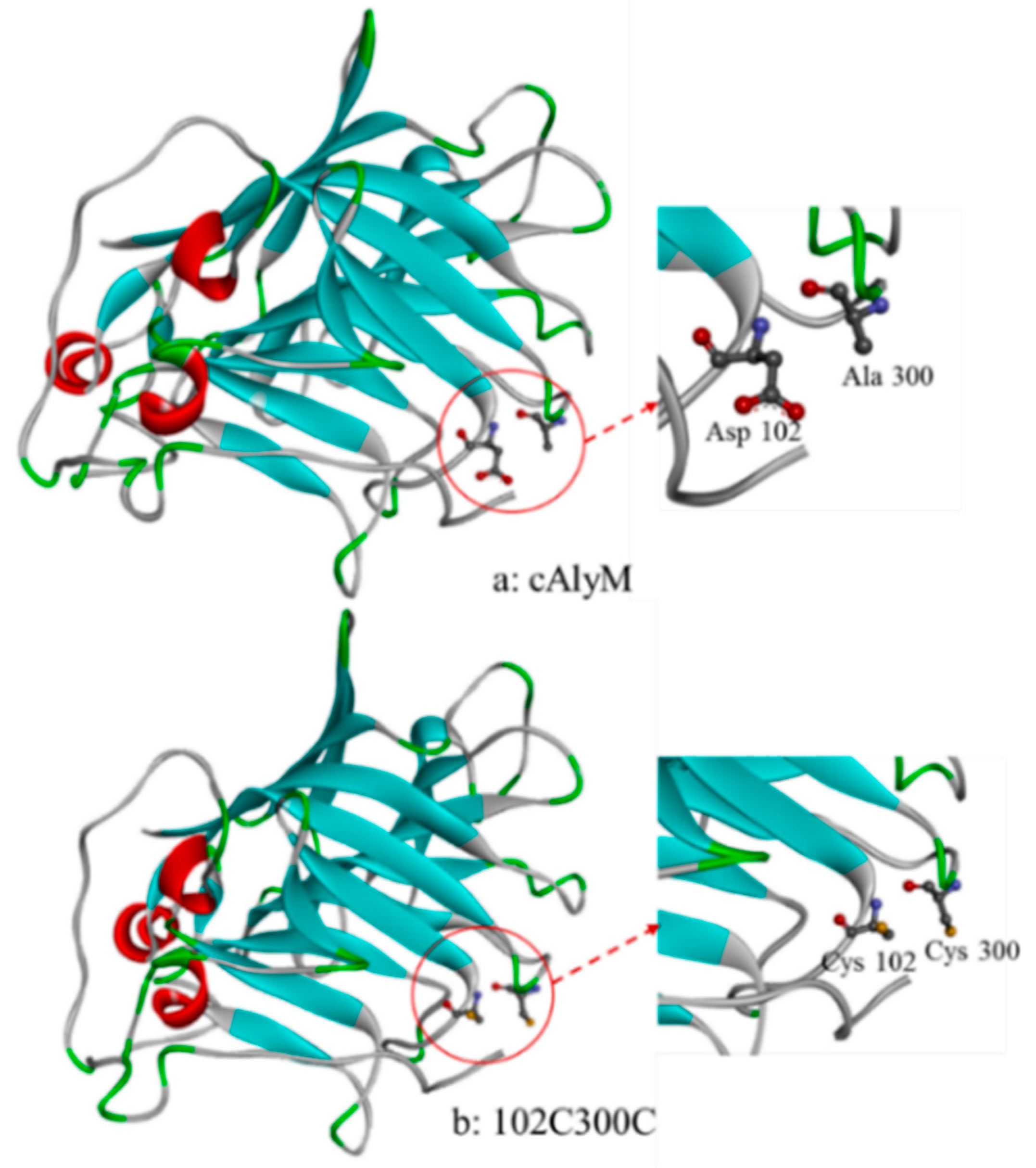
2.1. Recombinant Expression of cAlyM and 102C300C
2.2. Purification of Recombinant cAlyM and 102C300C
2.3. Kinetic Parameters of cAlyM and 102C300C
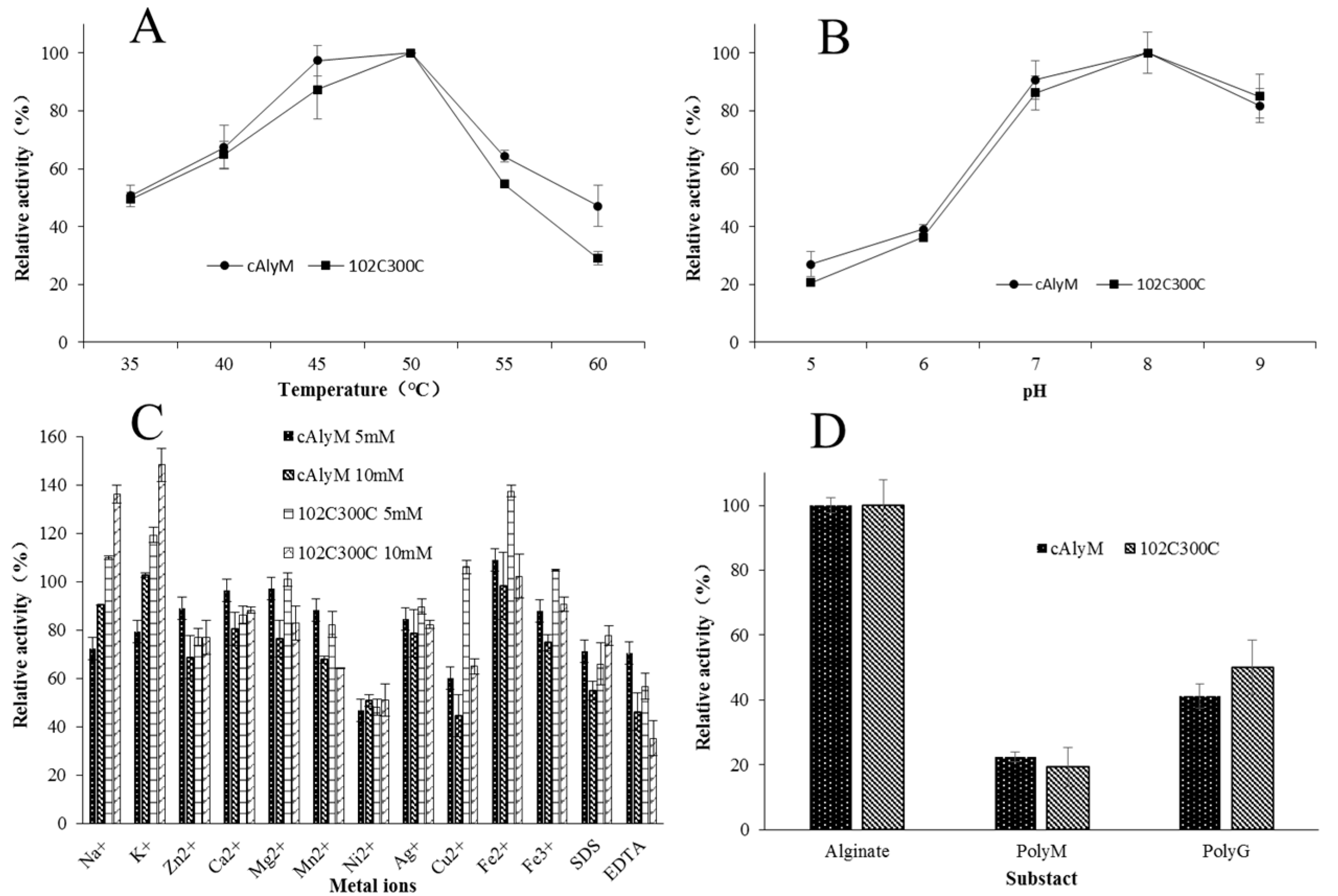
2.4. Enzymatic Properties of cAlyM and 102C300C
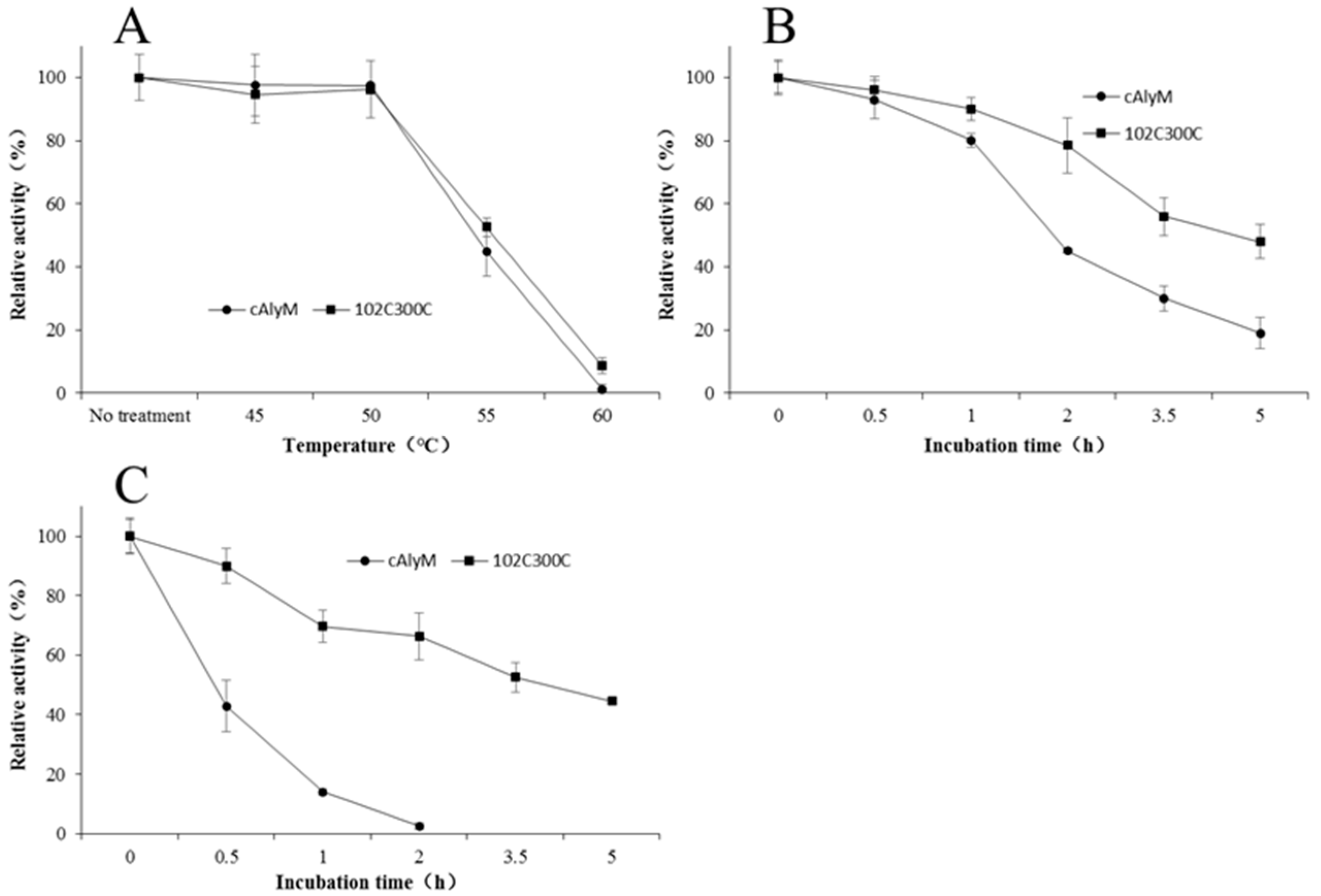
2.5. Thermal Stability of cAlyM and 102C300C
2.6. ESI-MS and HPLC Analyses of Degradation Products
3. Materials and Methods
3.1. Strains, Plasmids, and Reagents
3.2. Construction of Alginate Lyase and Mutant Recombinant Plasmid
3.3. Transformation and Colony Screening of P. pastoris
3.4. Purification of Recombinant cAlyM and 102C300C
3.5. Enzymatic Activity Assay
3.6. Kinetic parameters of cAlyM and 102C300C
3.7. Enzymatic Properties of cAlyM and 102C300C
3.8. Thermal Stability of cAlyM and 102C300C
3.9. ESI-MS and HPLC Analyses of Degradation Products
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhu, B.; Yin, H. Alginate lyase: Review of major sources and classification, properties, structure-function analysis and applications. Bioengineered 2015, 6, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Smidsrod, O.; Haug, A.; Larsen, B.R. The influence of pH on the rate of hydrolysis of acidic polysaccharides. Acta Chem. Scand. 1966, 20, 1026–1034. [Google Scholar] [CrossRef] [PubMed]
- Reddy, B.H.N.; Rauta, P.R.; Lakshmi, V.V.; Sreenivasa, S. Development, formulation, and evaluation of sodium alginate-g-poly (acryl amide-co-acrylic acid/cloiste-30b)/agnps hydrogel composites and their applications in paclitaxel drug delivery and anticancer activity. Int. J. Appl. Pharm. 2018, 10, 141–150. [Google Scholar]
- Draget, K.I.; Skjåk-Bræk, G.; Stokke, B.T. Similarities and differences between alginic acid gels and ionically crosslinked alginate gels. Food Hydrocoll. 2006, 20, 170–175. [Google Scholar] [CrossRef]
- Xing, M.; Cao, Q.; Wang, Y.; Xiao, H.; Zhao, J.; Zhang, Q.; Ji, A.; Song, S. Advances in research on the bioactivity of alginate oligosaccharides. Mar. Drugs 2020, 18, 144. [Google Scholar] [CrossRef]
- Wilcox, M.D.; Brownlee, I.A.; Richardson, J.C.; Dettmar, P.W.; Pearson, J.P. The modulation of pancreatic lipase activity by alginates. Food Chem. 2014, 146, 479–484. [Google Scholar] [CrossRef]
- Fang, W.; Bi, D.; Zheng, R.; Cai, N.; Xu, H.; Zhou, R.; Lu, J.; Wan, M.; Xu, X. Identification and activation of TLR4-mediated signalling pathways by alginate-derived guluronate oligosaccharide in RAW264. 7 macrophages. Sci. Rep. 2017, 7, 1–13. [Google Scholar]
- Han, Y.; Zhang, L.; Yu, X.; Wang, S.; Xu, C.; Yin, H.; Wang, S. Alginate oligosaccharide attenuates α2, 6-sialylation modification to inhibit prostate cancer cell growth via the Hippo/YAP pathway. Cell Death Dis. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Xin, M.; Ren, L.; Sun, Y.; Li, H.H.; Guan, H.S.; He, X.X.; Li, C.X. Anticoagulant and antithrombotic activities of low-molecular-weight propylene glycol alginate sodium sulfate (PSS). Eur. J. Med. Chem. 2016, 114, 33–40. [Google Scholar] [CrossRef]
- Wang, X.; Sun, G.; Feng, T.; Zhang, J.; Huang, X.; Wang, T.; Xie, Z.; Chu, X.; Yang, J.; Wang, H. Sodium oligomannate therapeutically remodels gut microbiota and suppresses gut bacterial amino acids-shaped neuroinflammation to inhibit Alzheimer’s disease progression. Cell Res. 2019, 29, 787–803. [Google Scholar] [CrossRef]
- Zhu, X.; Li, X.; Shi, H.; Zhou, J.; Tan, Z.; Yuan, M.; Yao, P.; Liu, X. Characterization of a novel alginate lyase from marine bacterium Vibrio furnissii H1. Mar. Drugs 2018, 16, 30. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Wen, S.; Liao, S.; Wang, Q.; Huang, S. Characterization of a bifunctional alginate lyase as a new member of the polysaccharide lyase family 17 from a marine strain BP-2. Biotechnol. Lett. 2019, 41, 1187–1200. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Chen, P.; Zeng, Y.; Men, Y.; Mu, S.; Zhu, Y.; Chen, Y.; Sun, Y. The characterization and modification of a novel bifunctional and robust alginate lyase derived from Marinimicrobium sp. H1. Mar. Drugs 2019, 17, 545. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, X.; Bi, X.; Ren, Y.; Han, Q.; Zhou, Y.; Han, Y.; Yao, R.; Li, S. Characterization of an alkaline alginate lyase with pH-stable and thermo-tolerance property. Mar. Drugs 2019, 17, 308. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Ni, F.; Sun, Y.; Ning, L.; Yao, Z. Elucidation of degrading pattern and substrate recognition of a novel bifunctional alginate lyase from Flammeovirga sp. NJ-04 and its use for preparation alginate oligosaccharides. Biotechnol. Biofuels 2019, 12, 1–13. [Google Scholar] [CrossRef]
- Sun, M.; Sun, C.; Li, T.; Li, K.; Yan, S.; Yin, H. Characterization of a novel bifunctional mannuronan C-5 epimerase and alginate lyase from Pseudomonas mendocina sp. DICP-70. Int. J. Biol. Macromol. 2020, 150, 662–670. [Google Scholar] [CrossRef]
- Yang, M.; Li, N.; Yang, S.; Yu, Y.; Han, Z.; Li, L.; Mou, H. Study on expression and action mode of recombinant alginate lyases based on conserved domains reconstruction. Appl. Microbiol. Biotechnol. 2019, 103, 807–817. [Google Scholar] [CrossRef]
- Spohner, S.C.; Müller, H.; Quitmann, H.; Czermak, P. Expression of enzymes for the usage in food and feed industry with Pichia pastoris. J. Biotechnol. 2015, 202, 118–134. [Google Scholar] [CrossRef]
- Zhu, T.; Sun, H.; Wang, M.; Li, Y. Pichia pastoris as a versatile cell factory for the production of industrial enzymes and chemicals: Current status and future perspectives. Biotechnol. J. 2019, 14, 1800694. [Google Scholar] [CrossRef]
- Li, H.; Wang, S.; Zhang, Y.; Chen, L. High-level expression of a thermally stable alginate lyase using pichia pastoris, characterization and application in producing brown alginate oligosaccharide. Mar. Drugs 2018, 16, 158. [Google Scholar] [CrossRef]
- Kim, H.S.; Lee, C.-G.; Lee, E.Y. Alginate lyase: Structure, property, and application. Biotechnol. Bioprocess Eng. 2011, 16, 843–851. [Google Scholar] [CrossRef]
- Yang, M.; Yang, S.-X.; Liu, Z.-M.; Li, N.-N.; Li, L.; Mou, H.-J. Rational design of alginate lyase from Microbulbifer sp. Q7 to improve thermal stability. Mar. Drugs 2019, 17, 378. [Google Scholar] [CrossRef] [PubMed]
- Pei, X.; Chang, Y.; Shen, J. Cloning, expression and characterization of an endo-acting bifunctional alginate lyase of marine bacterium Wenyingzhuangia fucanilytica. Protein Expr. Purif. 2019, 154, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Adney, W.S.; Jeoh, T.; Beckham, G.T.; Chou, Y.-C.; Baker, J.O.; Michener, W.; Brunecky, R.; Himmel, M.E. Probing the role of N-linked glycans in the stability and activity of fungal cellobiohydrolases by mutational analysis. Cellulose 2009, 16, 699–709. [Google Scholar] [CrossRef]
- Skropeta, D. The effect of individual N-glycans on enzyme activity. Bioorg. Med. Chem. 2009, 17, 2645–2653. [Google Scholar] [CrossRef] [PubMed]
- Da, L.; Yang, X.; He, Q.; Pan, Y.; Li, Y. Glycosylation analysis of recombinant neutral protease I from Aspergillus oryzae expressed in Pichia pastoris. Biotechnol. Lett. 2013, 35, 2121–2127. [Google Scholar]
- Polizeli, M.d.L.T.d.M. Biochemical properties of glycosylation and characterization of a histidine acid phosphatase (phytase) expressed in Pichia pastoris. Protein Expr. Purif. 2014, 99, 43–49. [Google Scholar]
- Zhu, B.W.; Huang, L.-S.-X.; Tan, H.-D.; Qin, Y.-Q.; Du, Y.-G.; Yin, H. Characterization of a new endo-type polyM-specific alginate lyase from Pseudomonas sp. Biotechnol. Lett. 2015, 37, 409–415. [Google Scholar] [CrossRef]
- Iwamoto, Y.; Araki, R.; Iriyama, K.-I.; Oda, T.; Fukuda, H.; Hayashida, S.; Muramatsu, T. Purification and characterization of bifunctional alginate lyase from Alteromonas sp. strain no. 272 and its action on saturated oligomeric substrates. Biosci. Biotechnol. Biochem. 2001, 65, 133–142. [Google Scholar] [CrossRef]
- Lee, S.; Choi, S.H.; Lee, E.Y.; Kim, H.S. Molecular cloning, purification, and characterization of a novel polyMG-specific alginate lyase responsible for alginate MG block degradation in Stenotrophomas maltophilia KJ-2. Appl. Microbiol. Biotechnol. 2012, 95, 1643–1653. [Google Scholar] [CrossRef]
- Yagi, H.; Fujise, A.; Itabashi, N.; Ohshiro, T. Purification and characterization of a novel alginate lyase from the marine bacterium Cobetia sp. NAP1 isolated from brown algae. Biosci. Biotechnol. Biochem. 2016, 80, 2338–2346. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, M.; Obata, J.; Iwamoto, Y.; Oda, T.; Muramatsu, T. Calcium-sensitive extracellular poly (α-L-guluronate) lyase from a marine bacterium Pseudomonas sp. strain F6: Purification and some properties. Fish. Sci. 2001, 67, 956–964. [Google Scholar] [CrossRef]
- Zhang, Y.-H.; Shao, Y.; Jiao, C.; Yang, Q.-M.; Weng, H.-F.; Xiao, A.-F. Characterization and application of an alginate lyase, aly1281 from marine bacterium Pseudoalteromonas carrageenovora ASY5. Mar. Drugs 2020, 18, 95. [Google Scholar] [CrossRef]
- Yang, M.; Yu, Y.; Yang, S.; Shi, X.; Mou, H.; Li, L. Expression and characterization of a new polyG-specific alginate lyase from marine bacterium Microbulbifer sp. Q7. Front. Microbiol. 2018, 9, 2894. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Shi, X.; Gao, Y.; Cai, N.; Jiang, Z.; Xu, X. Anti-inflammatory activity of guluronate oligosaccharides obtained by oxidative degradation from alginate in lipopolysaccharide-activated murine macrophage RAW 264.7 cells. J. Agric. Food Chem. 2015, 63, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Powell, L.C.; Pritchard, M.F.; Emanuel, C.; Onsoyen, E.; Rye, P.D.; Wright, C.J.; Thomas, D.W. A nanoscale characterization of the interaction of a novel alginate oligomer with the cell surface and motility of Pseudomonas aeruginosa. Am. J. Respir. Cell Mol. Biol. 2014, 50, 483–492. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]


| Enzymes | Step | Total Activity (U) | Total Protein (mg) | Specific Activity (U/mg) | Recovery Rate (%) | Fold | Yield (U/mL) |
|---|---|---|---|---|---|---|---|
| cAlyM | Fermentation medium | 507.4 ± 8.5 | 3.00 ± 0.06 | 169.1 | 100 | 1 | 33.8 ± 0.57 |
| Magnetic beads | 188.4 ± 4.6 | 0.68 ± 0.01 | 277.1 | 37.13 | 1.64 | 26.9 ± 0.66 | |
| 102C300C | Fermentation medium | 617.6 ± 6.4 | 3.82 ± 0.05 | 161.7 | 100 | 1 | 41.2 ± 0.43 |
| Magnetic beads | 237.1 ± 5.1 | 0.95 ± 0.01 | 249.6 | 38.39 | 1.54 | 33.9 ± 0.73 |
| Enzyme | Vmax (U/mg) | Km (mg/mL) | kcat (s−1) | kcat/Km (mL/s/mg) |
|---|---|---|---|---|
| cAlyM | 344.8 ± 5.6 | 1.31 ± 0.21 | 189.1 ± 5.3 | 144.3 |
| 102C300C | 312.5 ± 10.1 | 1.43 ± 0.20 | 171.3 ± 4.3 | 119.2 |
| Enzyme | Origin | Source | Substrate Preference | Optimal Catalytic Temperature | Thermalstability | Specific activity (U/mg) | Reference |
|---|---|---|---|---|---|---|---|
| Aly08 | Vibrio sp. SY01 | E. coli | Poly G | 45 °C | Retained 48.7% activity at 20 °C for 1 h | 841 | [14] |
| Aly7B_Wf | Wenyingzhuangia fucanilytica | E. coli | Poly M | 40 °C | Retained 75% activity at 35 °C for 24 h | 23.24 * | [23] |
| AlyH1 | Vibrio furnissii H1 | Native | Poly G | 40 °C | Retained 60% activity at 40 °C for 30 min | 2.40 * | [11] |
| FsAlgB | Flammeovirga sp. NJ-04 | E. coli | Poly M and alginate | 40 °C | Retained 80% activity at 40 °C for 30 min | 1760.8 | [15] |
| AlgH | Marinimicrobium sp. H1 | E. coli | Poly G | 45 °C | Retained 80% activity at 40 °C for 2 h | 5510 | [13] |
| PmC5A | Pseudomonas mendocina DICP-70 | E. coli | Poly G and Poly M | 40 °C | Retained 80% activity at 45 °C for 1 h | N.D. | [16] |
| Aly1281 | Pseudoalteromonas carrageenovora ASY5 | E. coli | Poly G | 50 °C | Stable at temperatures lower than 55 °C | 1.15 * | [33] |
| Alg17B | BP-2 | Native | Poly M | 45 °C | Retained 10% activity at 45 °C for 1 h | 4036 | [12] |
| KJ-2 | Stenotrophomas maltophilia KJ-2 | E. coli | Poly MG | 40 °C | Inactivated at higher than 40 °C for 30 min | 848.3 | [30] |
| algA | Pseudomonas sp. E03 | E. coli | Poly M | 30 °C | Retained 50% activity at 50 °C for 30 min | 222 | [28] |
| SAGL | Flavobacterium sp. H63 | P. pastoris | Poly M and alginate | 45 °C | Retained 49.0% activity at 50 °C for 72 h | 4044 * | [20] |
| AlgC-PL7 | Cobetia sp. NAP1 | E. coli | Poly G and Poly M | 45 °C | Retained 80% activity at 70 °C for 1 h | 30 | [31] |
| GLyase | Pseudomonas sp. F6 | Native | Poly G | N.D. | Retained 60% activity at 80 °C for 15 min | 222.8 | [32] |
| cAlyM | Microbulbifer sp. Q7 | P. pastoris | Poly G and alginate | 50 °C | Retained 50% activity at 45 °C for 2 h | 277.1 * | This study |
| 102C300C | Microbulbifer sp. Q7 | P. pastoris | Poly G and alginate | 50 °C | Retained 50% activity at 45 °C for 5.2 h | 249.6 * | This study |
| Sample | Peak Area Percentage of G | Peak Area Percentage of M | M/G |
|---|---|---|---|
| 1-fold | 15.05% | 84.95% | 5.61 |
| 3-fold | 31.17% | 68.83% | 2.08 |
| 5-fold | 72.91% | 27.09% | 0.37 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Liu, Z.; Fu, X.; Zhu, C.; Kong, Q.; Yang, M.; Mou, H. Expression and Characterization of an Alginate Lyase and Its Thermostable Mutant in Pichia pastoris. Mar. Drugs 2020, 18, 305. https://doi.org/10.3390/md18060305
Yang S, Liu Z, Fu X, Zhu C, Kong Q, Yang M, Mou H. Expression and Characterization of an Alginate Lyase and Its Thermostable Mutant in Pichia pastoris. Marine Drugs. 2020; 18(6):305. https://doi.org/10.3390/md18060305
Chicago/Turabian StyleYang, Suxiao, Zhemin Liu, Xiaodan Fu, Changliang Zhu, Qing Kong, Min Yang, and Haijin Mou. 2020. "Expression and Characterization of an Alginate Lyase and Its Thermostable Mutant in Pichia pastoris" Marine Drugs 18, no. 6: 305. https://doi.org/10.3390/md18060305
APA StyleYang, S., Liu, Z., Fu, X., Zhu, C., Kong, Q., Yang, M., & Mou, H. (2020). Expression and Characterization of an Alginate Lyase and Its Thermostable Mutant in Pichia pastoris. Marine Drugs, 18(6), 305. https://doi.org/10.3390/md18060305





