Novel Fredericamycin Variant Overproduced by a Streptomycin-Resistant Streptomyces albus subsp. chlorinus Strain
Abstract
1. Introduction
2. Results
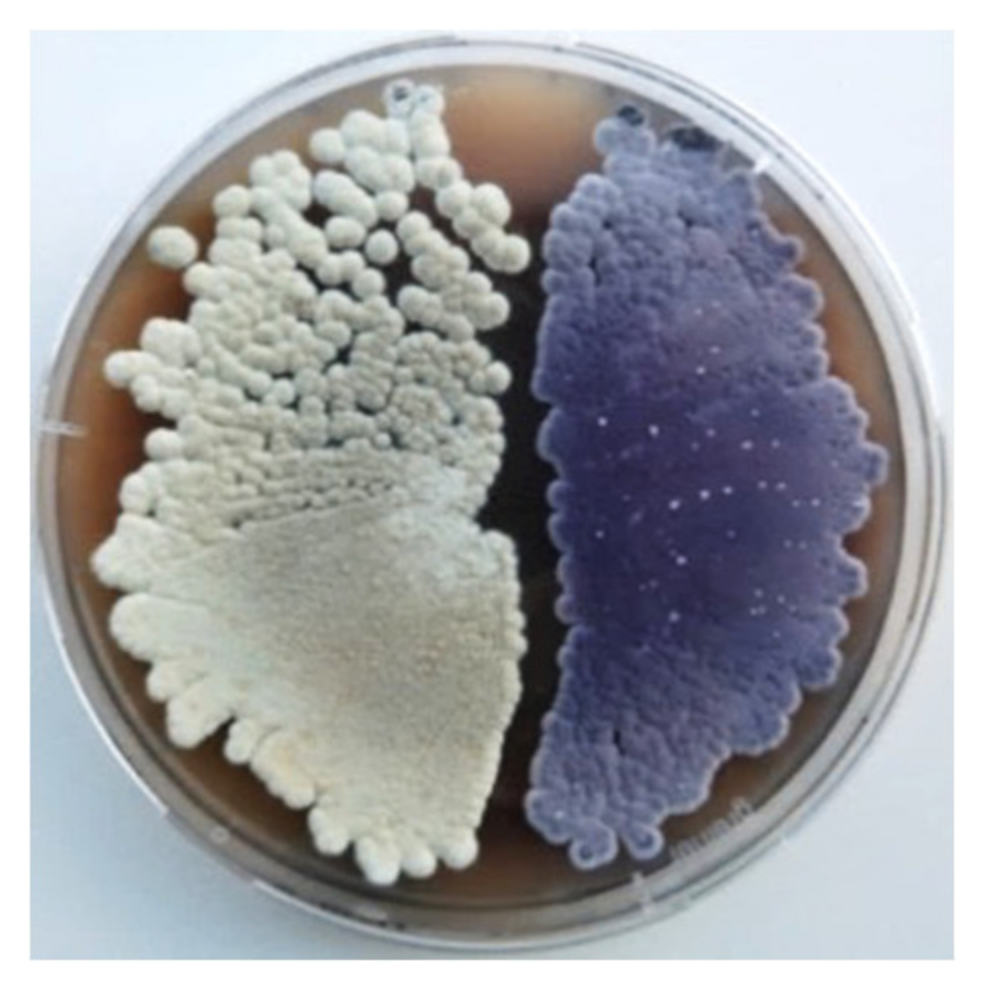
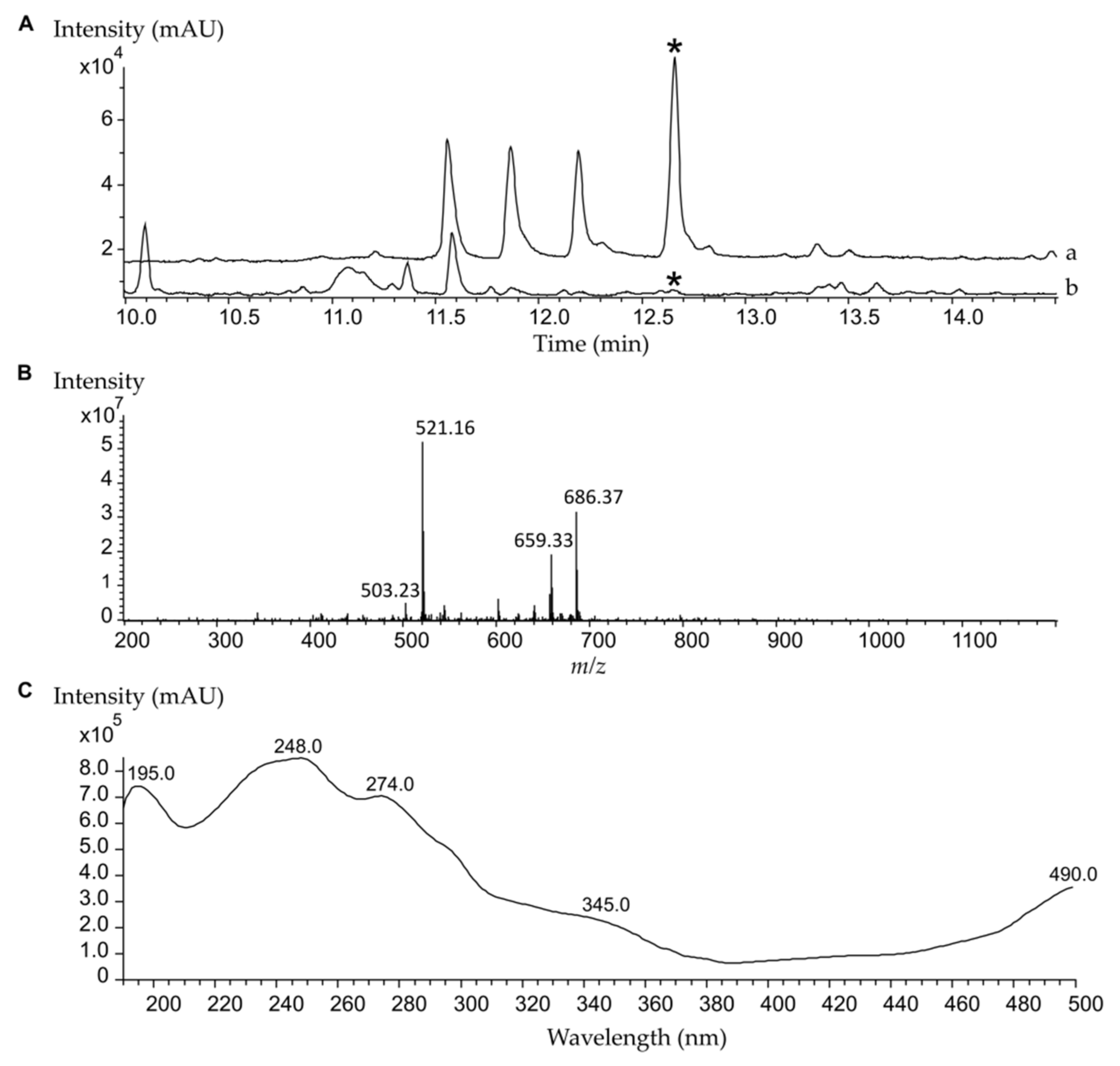
2.1. High-Level Streptomycin-Resistant Mutant S. albus subsp. chlorinus Overproduces the Novel Compound Fredericamycin C2
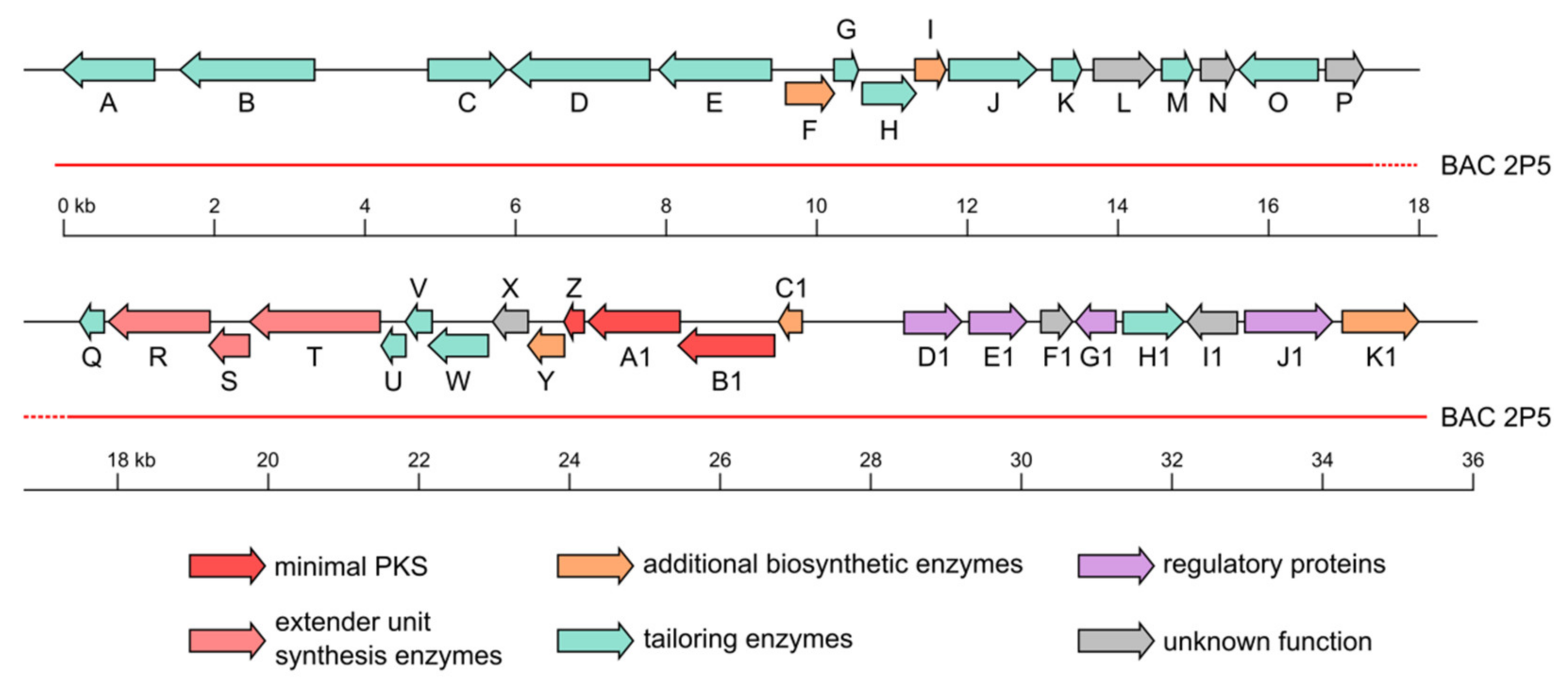
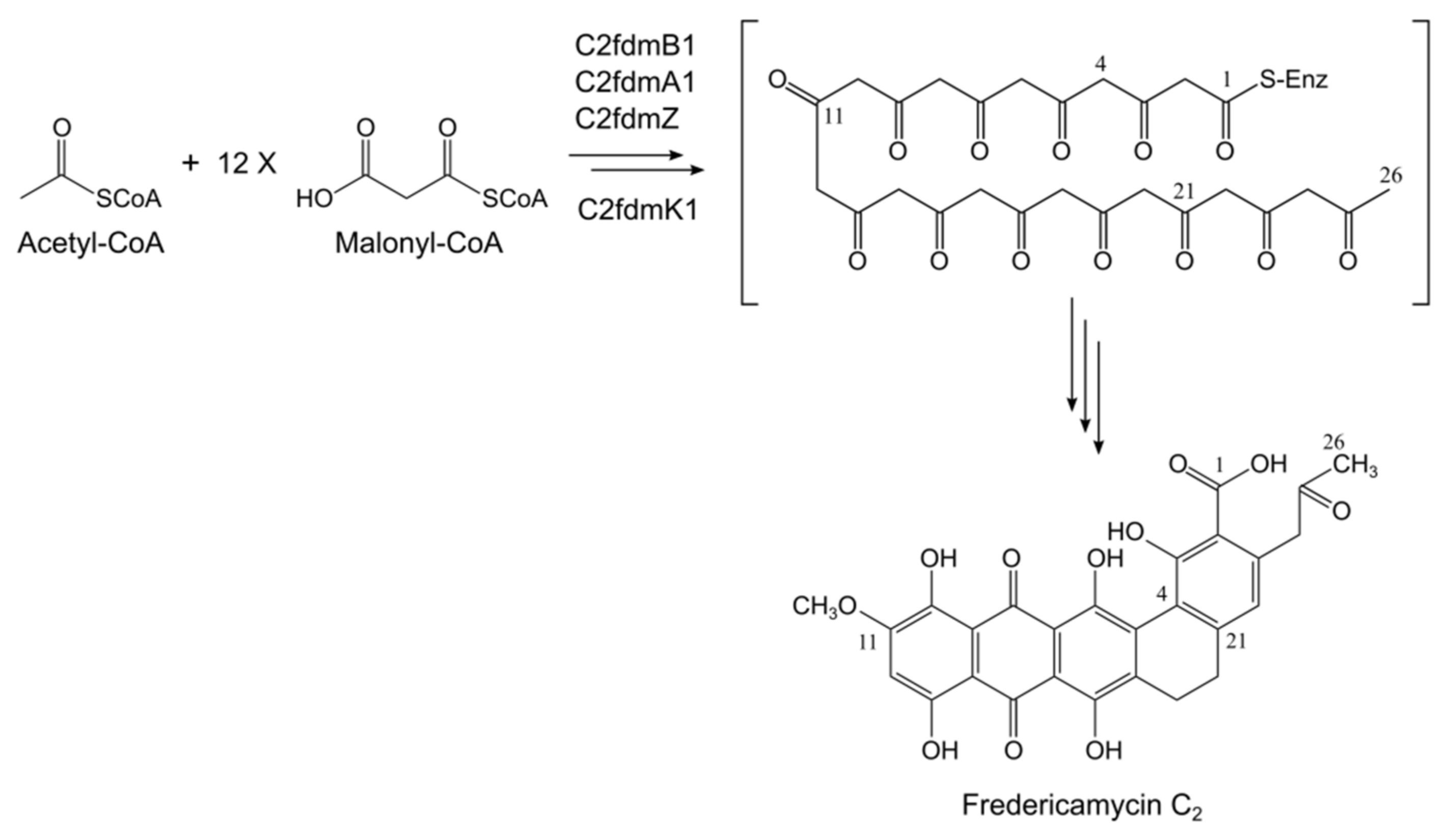
2.2. Fredericamycin C2 Is Biosynthesized by a Type II PKS Gene Cluster
2.3. Screening for the Mutation Causing Fredericamycin C2 Overproduction and Streptomycin Resistance in S. albus subsp. chlorinus JR1
2.4. Biological Activity of Fredericamycin C2
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Isolation and Manipulation of DNA
4.3. Strain Generation, Metabolite Extraction, and Analysis
4.4. Fredericamycin C2 Purification and Quantification
4.5. H-NMR Spectroscopy
4.6. Antimicrobial Susceptibility Test
4.7. Genome Sequencing, Genome Assembly, and Analysis
4.8. Genome Mining and Bioinformatics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Protasov, E.S.; Axenov-Gribanov, D.V.; Rebets, Y.V.; Voytsekhovskaya, I.V.; Tokovenko, B.T.; Shatilina, Z.M.; Luzhetskyy, A.N.; Timofeyev, M.A. The diversity and antibiotic properties of actinobacteria associated with endemic deepwater amphipods of Lake Baikal. Antonie Van Leeuwenhoek 2017, 110, 1593–1611. [Google Scholar] [CrossRef]
- Axenov-Gribanov, D.V.; Voytsekhovskaya, I.V.; Tokovenko, B.T.; Protasov, E.S.; Gamaiunov, S.V.; Rebets, Y.V.; Luzhetskyy, A.N.; Timofeyev, M.A. Actinobacteria isolated from an underground lake and moonmilk speleothem from the biggest conglomeratic karstic cave in Siberia as sources of novel biologically active compounds. PLoS ONE 2016, 11, e0152957. [Google Scholar] [CrossRef] [PubMed]
- Shima, J.; Hesketh, A.; Okamoto, S.; Kawamoto, S.; Ochi, K. Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J. Bacteriol. 1996, 178, 7276–7284. [Google Scholar] [CrossRef] [PubMed]
- Hosoya, Y.; Okamoto, S.; Muramatsu, H.; Ochi, K. Acquisition of certain streptomycin-resistant (str) mutations enhances antibiotic production in bacteria. Antimicrob. Agents Chemother. 1998, 42, 2041–2047. [Google Scholar] [CrossRef] [PubMed]
- Hesketh, A.; Ochi, K. A novel method for improving Streptomyces coelicolor A3(2) for production of actinorhodin by introduction of rpsL (encoding ribosomal protein S12) mutations conferring resistance to streptomycin. J. Antibiot. 1997, 50, 532–535. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Ochi, K. Novel approach for improving the productivity of antibiotic-producing strains by inducing combined resistant mutations. Appl. Environ. Microbiol. 2001, 67, 1885–1892. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.Y.Z.; Liu, Q.; Huang, Y.; Hu, C.; Liao, G. Improvement of A21978C production in Streptomyces roseosporus by reporter-guided rpsL mutation selection. J. Appl. Microbiol. 2012, 112, 1095–1101. [Google Scholar] [CrossRef]
- Cashel, M.; Rudd, K.E. The stringent response. In Escherichia Coli and Salmonella Typhimurium: Cellular and Molecular Biology; Neidhart, F.C., Ed.; American Society for Microbiology: Washington, DC, USA, 1987. [Google Scholar]
- Ochi, K. Occurrence of the stringent response in Streptomyces sp. and its significance for the initiation of morphological and physiological differentiation. J. Gen. Microbiol. 1986, 132, 2621–2631. [Google Scholar] [CrossRef]
- Strauch, E.; Takano, E.; Baylis, H.A.; Bibb, M.J. The stringent response in Streptomyces coelicolor A3(2). Mol. Microbiol. 1991, 5, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Sands, M.K.; Roberts, R.B. The effects of a tryptophan-histidine deficiency in a mutant of Escherichia coli. J. Bacteriol. 1952, 63, 505–511. [Google Scholar] [CrossRef]
- Gralla, J.D. Escherichia coli ribosomal RNA transcription: Regulatory roles for ppGpp, NTPs, architectural proteins and a polymerase-binding protein. Mol. Microbiol. 2005, 55, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Myronovskyi, M.; Rosenkranzer, B.; Stierhof, M.; Petzke, L.; Seiser, T.; Luzhetskyy, A. Identification and Heterologous Expression of the Albucidin Gene Cluster from the Marine Strain Streptomyces Albus Subsp. Chlorinus NRRL B-24108. Microorganisms 2020, 8, 237. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez Estevez, M.; Myronovskyi, M.; Gummerlich, N.; Nadmid, S.; Luzhetskyy, A. Heterologous expression of the nybomycin gene cluster from the marine strain Streptomyces albus subsp. chlorinus NRRL B-24108. Mar. Drugs 2018, 16, 435. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.C.; Toussaint, M.W.; Stroshane, R.M.; Kalita, C.C.; Aszalos, A.A.; Garretson, A.L.; Wei, T.T.; Byrne, K.M.; Geoghegan, R.F., Jr.; White, R.J. Fredericamycin A, a new antitumor antibiotic. I. Production, isolation and physicochemical properties. J. Antibiot. 1981, 34, 1389–1401. [Google Scholar] [CrossRef]
- Misra, R.; Pandey, R.C.; Hilton, B.D.; Roller, P.P.; Silverton, J.V. Structure of fredericamycin A, an antitumor antibiotic of a novel skeletal type; spectroscopic and mass spectral characterization. J. Antibiot. 1987, 40, 786–802. [Google Scholar] [CrossRef]
- Sontag, B.; Muller, J.G.; Hansske, F.G. Fredericamycin B and C from Streptomyces griseus: Structure elucidation after 23 years. J. Antibiot. 2004, 57, 823–828. [Google Scholar] [CrossRef][Green Version]
- Chen, Y.; Luo, Y.; Ju, J.; Wendt-Pienkowski, E.; Rajski, S.R.; Shen, B. Identification of fredericamycin E from Streptomyces griseus: Insights into fredericamycin A biosynthesis highlighting carbaspirocycle formation. J. Nat. Prod. 2008, 71, 431–437. [Google Scholar] [CrossRef]
- Warnick-Pickle, D.J.; Byrne, K.M.; Pandey, R.C.; White, R.J. Fredericamycin A, a new antitumor antibiotic. II. Biological properties. J. Antibiot. 1981, 34, 1402–1407. [Google Scholar] [CrossRef]
- Lu, K.P.; Fischer, G. Methods of Inhibiting Pin1-Associated States Using a Fredericamycin A Compound. WO/2004/002429. 2004. Available online: https://patentscope2.wipo.int/search/en/detail.jsf?docId=WO2004002429 (accessed on 2 March 2020).
- Yasuzawa, T.; Yoshida, M.; Shirahata, K.; Sano, H. Structure of a novel Ca2+ and calmodulin-dependent cyclic nucleotide phosphodiesterase inhibitor KS-619-1. J. Antibiot. 1987, 40, 1111–1114. [Google Scholar] [CrossRef]
- Wendt-Pienkowski, E.; Huang, Y.; Zhang, J.; Li, B.; Jiang, H.; Kwon, H.; Hutchinson, C.R.; Shen, B. Cloning, sequencing, analysis, and heterologous expression of the fredericamycin biosynthetic gene cluster from Streptomyces griseus. J. Am. Chem. Soc. 2005, 127, 16442–16452. [Google Scholar] [CrossRef]
- Zhang, Q.; Pang, B.; Ding, W.; Liu, W. Aromatic Polyketides Produced by Bacterial Iterative Type I Polyketide Synthases. ACS Catal. 2013, 3, 1439–1447. [Google Scholar] [CrossRef]
- Kudo, F.; Kasama, Y.; Hirayama, T.; Eguchi, T. Cloning of the pactamycin biosynthetic gene cluster and characterization of a crucial glycosyltransferase prior to a unique cyclopentane ring formation. J. Antibiot. 2007, 60, 492–503. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhao, Q.; He, Q.; Ding, W.; Tang, M.; Kang, Q.; Yu, Y.; Deng, W.; Zhang, Q.; Fang, J.; Tang, G.; et al. Characterization of the azinomycin B biosynthetic gene cluster revealing a different iterative type I polyketide synthase for naphthoate biosynthesis. Chem. Biol. 2008, 15, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Li, S.; Niu, S.; Ma, L.; Zhang, G.; Zhang, H.; Zhang, G.; Ju, J.; Zhang, C. Characterization of tiacumicin B biosynthetic gene cluster affording diversified tiacumicin analogues and revealing a tailoring dihalogenase. J. Am. Chem. Soc. 2011, 133, 1092–1105. [Google Scholar] [CrossRef]
- Hertweck, C.; Luzhetskyy, A.; Rebets, Y.; Bechthold, A. Type II polyketide synthases: Gaining a deeper insight into enzymatic teamwork. Nat. Prod. Rep. 2007, 24, 162–190. [Google Scholar] [CrossRef]
- Funatsu, G.; Wittmann, H.G. Ribosomal proteins. 33. Location of amino-acid replacements in protein S12 isolated from Escherichia coli mutants resistant to streptomycin. J. Mol. Biol. 1972, 68, 547–550. [Google Scholar] [CrossRef]
- Finken, M.; Kirschner, P.; Meier, A.; Wrede, A.; Bottger, E.C. Molecular basis of streptomycin resistance in Mycobacterium tuberculosis: Alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol. Microbiol. 1993, 9, 1239–1246. [Google Scholar] [CrossRef]
- Meier, A.; Kirschner, P.; Bange, F.C.; Vogel, U.; Bottger, E.C. Genetic alterations in streptomycin-resistant Mycobacterium tuberculosis: Mapping of mutations conferring resistance. Antimicrob. Agents Chemother. 1994, 38, 228–233. [Google Scholar] [CrossRef]
- Tanaka, Y.; Tokuyama, S.; Ochi, K. Activation of secondary metabolite-biosynthetic gene clusters by generating rsmG mutations in Streptomyces griseus. J. Antibiot. 2009, 62, 669–673. [Google Scholar] [CrossRef]
- Mikheil, D.M.; Shippy, D.C.; Eakley, N.M.; Okwumabua, O.E.; Fadl, A.A. Deletion of gene encoding methyltransferase (gidB) confers high-level antimicrobial resistance in Salmonella. J. Antibiot. 2012, 65, 185–192. [Google Scholar] [CrossRef]
- Ulrych, A.; Holeckova, N.; Goldova, J.; Doubravova, L.; Benada, O.; Kofroňová, O.; Halada, P.; Branny, P. Characterization of pneumococcal Ser/Thr protein phosphatase phpP mutant and identification of a novel PhpP substrate, putative RNA binding protein Jag. BMC Microbiol. 2016, 16, 247. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, K.; Hosaka, T.; Tokuyama, S.; Okamoto, S.; Ochi, K. Mutations in rsmG, encoding a 16S rRNA methyltransferase, result in low-level streptomycin resistance and antibiotic overproduction in Streptomyces coelicolor A3(2). J. Bacteriol. 2007, 189, 3876–3883. [Google Scholar] [CrossRef] [PubMed]
- Lopatniuk, M.; Myronovskyi, M.; Nottebrock, A.; Busche, T.; Kalinowski, J.; Ostash, B.; Fedorenko, V.; Luzhetskyy, A. Effect of “ribosome engineering” on the transcription level and production of S. albus indigenous secondary metabolites. Appl. Microbiol. Biotechnol. 2019, 103, 7097–7110. [Google Scholar] [CrossRef] [PubMed]
- Carter, A.P.; Clemons, W.M.; Brodersen, D.E.; Morgan-Warren, R.J.; Wimberly, B.T.; Ramakrishnan, V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 2000, 407, 340–348. [Google Scholar] [CrossRef]
- Davies, J.; Gorini, L.; Davis, B.D. Misreading of RNA codewords induced by aminoglycoside antibiotics. Mol. Pharmacol. 1965, 1, 93–106. [Google Scholar]
- Das, A.; Szu, P.H.; Fitzgerald, J.T.; Khosla, C. Mechanism and engineering of polyketide chain initiation in fredericamycin biosynthesis. J. Am. Chem. Soc. 2010, 132, 8831–8833. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2012. [Google Scholar]
- Kieser, T.B.M.J.; Buttner, M.J.; Charter, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Rebets, Y.; Kormanec, J.; Lutzhetskyy, A.; Bernaerts, K.; Anne, J. Cloning and expression of metagenomic DNA in Streptomyces lividans and subsequent fermentation for optimized production. In Metagenomics. Methods in Molecular Biology; Streit, W., Daniel, R., Eds.; Humana Press: New York, NY, USA, 2017. [Google Scholar]
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Muller, R.; Wohlleben, W.; et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
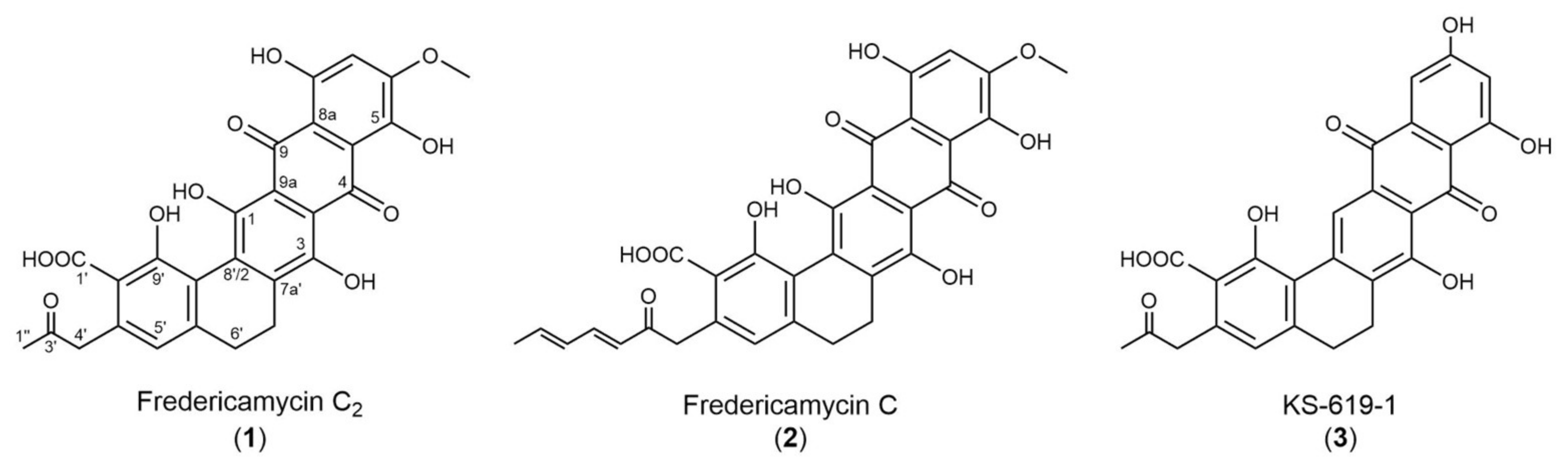
| Pos. a | δC | δH (J Hz) | COSY b | HMBC b | ROESY b |
|---|---|---|---|---|---|
| 9’ | 164.3 | 5’-H | |||
| 9a’ | 115.3 | 4’-H2, 5’-H | |||
| 3’ | 140.7 | 4’-H2, (5’-H) | |||
| 5’ | 120.4 | 6.52 s | (4‘-H2) | 1J, 4’-H2, (5’-H), 6’-H2 | 4’-H2, 6’-H2, (7’-H2) |
| 5a’ | 144.3 | 5‘-H, 6‘-H2, 7‘-H2 | |||
| 6’ | 28.8 | 2.63 br t (7Hz) | 7’-H2 | 5’-H, 7’-H2 | 5’-H, 7’-H2 |
| 7’ | 21.2 | 2.79 br t (7Hz) | 6’-H2 | 6’-H2 | |
| 7a’ | 137.3 | 6’-H2, 7’-H2 | |||
| 3 | 152.5 | 7’-H2 | |||
| 3a | 112.2 | ||||
| 4 | 186.9 | ||||
| 4a | 113.2 | (7-H) | |||
| 5 | 148.8 c | 7-H | |||
| 6 | 157.6 | 6-OCH3, 7-H | |||
| 6-OCH3 | 56.4 | 3.94 s | 6-H | 1J | 7-H |
| 7 | 105.8 | 6.88 s | 6-OCH3 | 6-OCH3 | 6-OCH3 |
| 8 | 158.6 | 7-H | |||
| 8a | 105.8 | 7-H | |||
| 9 | 187.6 | 7-H | |||
| 9a | 114.4 | ||||
| 1 | 156.3 | ||||
| 2 = 8’ | 136.3 | (5’-H), 7’-H2 | |||
| 8a’ | 121.0 | 5’-H | |||
| 1’ | 169.3 | 5’-H | |||
| 4’ | 49.5 | 4.06 s | (5’-H, 1’’-H3) | 5’-H, (1’’-H3) | 5’-H, 1’’-H3 |
| 3’ | (204.1) d | ||||
| 1’’ | 29.7 | 2.17 s | 4’-H2 | ||
| OH | 13.22 br s 12.80 br s 5.5 br s |
| Gene | Size (aa) | Proposed Function | GenBank Homologue 1 | Identity/Similarity (%) | Fredericamycin A Gene Cluster Homologue | Identity/Similarity (%) |
|---|---|---|---|---|---|---|
| c2fdmA | 406 | Cytochrome P450 oxygenase | WP_017596471.1 | 66/73 | - | - |
| c2fdmB | 594 | Monooxygenase | WP_017596470.1 | 67/75 | - | - |
| c2fdmC | 337 | O-methyltransferase | WP_043504920.1 | 42/55 | - | - |
| c2fdmD | 620 | Asparagine synthase | WP_017596467.1 | 79/88 | - | - |
| c2fdmE | 497 | Monooxygenase | WP_081620749.1 | 59/70 | - | - |
| c2fdmF | 216 | Polyketide cyclase | REH43750.1 | 43/54 | - | - |
| c2fdmG | 107 | Monooxygenase | WP_027732672.1 | 44/64 | - | - |
| c2fdmH | 237 | 3-ketoacyl-ACP reductase | WP_017596481.1 | 72/83 | - | - |
| c2fdmI | 134 | Polyketide cyclase | WP_109361109.1 | 72/83 | - | - |
| c2fdmJ | 389 | O-methyltransferase | WP_061257536.1 | 67/79 | fdmN | 56/70 |
| c2fdmK | 138 | Oxidoreductase | WP_020573863.1 | 56/68 | - | - |
| c2fdmL | 263 | Unknown | WP_017596478.1 | 56/71 | - | - |
| c2fdmM | 138 | Oxidoreductase | WP_017596477.1 | 70/85 | - | - |
| c2fdmN | 150 | Unknown | WP_017596476.1 | 76/89 | - | - |
| c2fdmO | 358 | O-methyltransferase | WP_043504920.1 | 40/54 | - | - |
| c2fdmP | 169 | Unknown | WP_017596474.1 | 62/75 | - | - |
| c2fdmQ | 113 | Monooxygenase | WP_017596454.1 | 65/76 | - | - |
| c2fdmR | 454 | Biotin carboxylase | WP_017596455.1 | 80/87 | - | - |
| c2fdmS | 175 | Biotin carboxyl carrier protein | WP_026120848.1 | 60/71 | - | - |
| c2fdmT | 585 | Carboxyl transferase | WP_017596457.1 | 75/80 | - | - |
| c2fdmU | 111 | Monooxygenase | WP_017596458.1 | 70/78 | fdmQ | 50/66 |
| c2fdmV | 113 | Monooxygenase | WP_017596459.1 | 76/83 | fdmP | 51/70 |
| c2fdmW | 248 | 3-ketoacyl-ACP reductase | WP_017596460.1 | 83/92 | fdmO | 55/71 |
| c2fdmX | 153 | Unknown | WP_015621174.1 | 61/74 | - | - |
| c2fdmY | 156 | Polyketide cyclase | WP_075740187.1 | 67/82 | fdmI | 56/74 |
| c2fdmZ | 87 | ACP | WP_017596463.1 | 46/71 | - | - |
| c2fdmA1 | 409 | KSβ | WP_017596464.1 | 77/86 | fdmG | 61/72 |
| c2fdmB1 | 422 | KSα | WP_017596465.1 | 83/90 | fdmF | 63/77 |
| c2fdmC1 | 112 | Polyketide cyclase | WP_017596466.1 | 83/90 | fdmD | 64/76 |
| c2fdmD1 | 254 | Transcriptional regulator | WP_116247593.1 | 58/78 | fdmR1 | 46/64 |
| c2fdmE1 | 256 | Transcriptional regulator | WP_081620746.1 | 67/80 | - | - |
| c2fdmF1 | 144 | Unknown | WP_017596472.1 | 78/89 | fdmE | 59/72 |
| c2fdmG1 | 179 | Transcriptional regulator | KPC87453.1 | 80/84 | - | - |
| c2fdmH1 | 271 | Serine hydrolase | WP_099880484.1 | 90/93 | - | - |
| c2fdmI1 | 225 | Unknown | WP_099880487.1 | 81/86 | - | - |
| c2fdmJ1 | 394 | Transcriptional regulator | WP_055497612.1 | 98/98 | - | - |
| c2fdmK1 | 315 | ACP S-malonyltransferase | WP_099880491.1 | 87/92 | - | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez Estévez, M.; Myronovskyi, M.; Rosenkränzer, B.; Paululat, T.; Petzke, L.; Ristau, J.; Luzhetskyy, A. Novel Fredericamycin Variant Overproduced by a Streptomycin-Resistant Streptomyces albus subsp. chlorinus Strain. Mar. Drugs 2020, 18, 284. https://doi.org/10.3390/md18060284
Rodríguez Estévez M, Myronovskyi M, Rosenkränzer B, Paululat T, Petzke L, Ristau J, Luzhetskyy A. Novel Fredericamycin Variant Overproduced by a Streptomycin-Resistant Streptomyces albus subsp. chlorinus Strain. Marine Drugs. 2020; 18(6):284. https://doi.org/10.3390/md18060284
Chicago/Turabian StyleRodríguez Estévez, Marta, Maksym Myronovskyi, Birgit Rosenkränzer, Thomas Paululat, Lutz Petzke, Jeanette Ristau, and Andriy Luzhetskyy. 2020. "Novel Fredericamycin Variant Overproduced by a Streptomycin-Resistant Streptomyces albus subsp. chlorinus Strain" Marine Drugs 18, no. 6: 284. https://doi.org/10.3390/md18060284
APA StyleRodríguez Estévez, M., Myronovskyi, M., Rosenkränzer, B., Paululat, T., Petzke, L., Ristau, J., & Luzhetskyy, A. (2020). Novel Fredericamycin Variant Overproduced by a Streptomycin-Resistant Streptomyces albus subsp. chlorinus Strain. Marine Drugs, 18(6), 284. https://doi.org/10.3390/md18060284




