Enzyme Bioprospection of Marine-Derived Actinobacteria from the Chilean Coast and New Insight in the Mechanism of Keratin Degradation in Streptomyces sp. G11C
Abstract
1. Introduction
2. Results
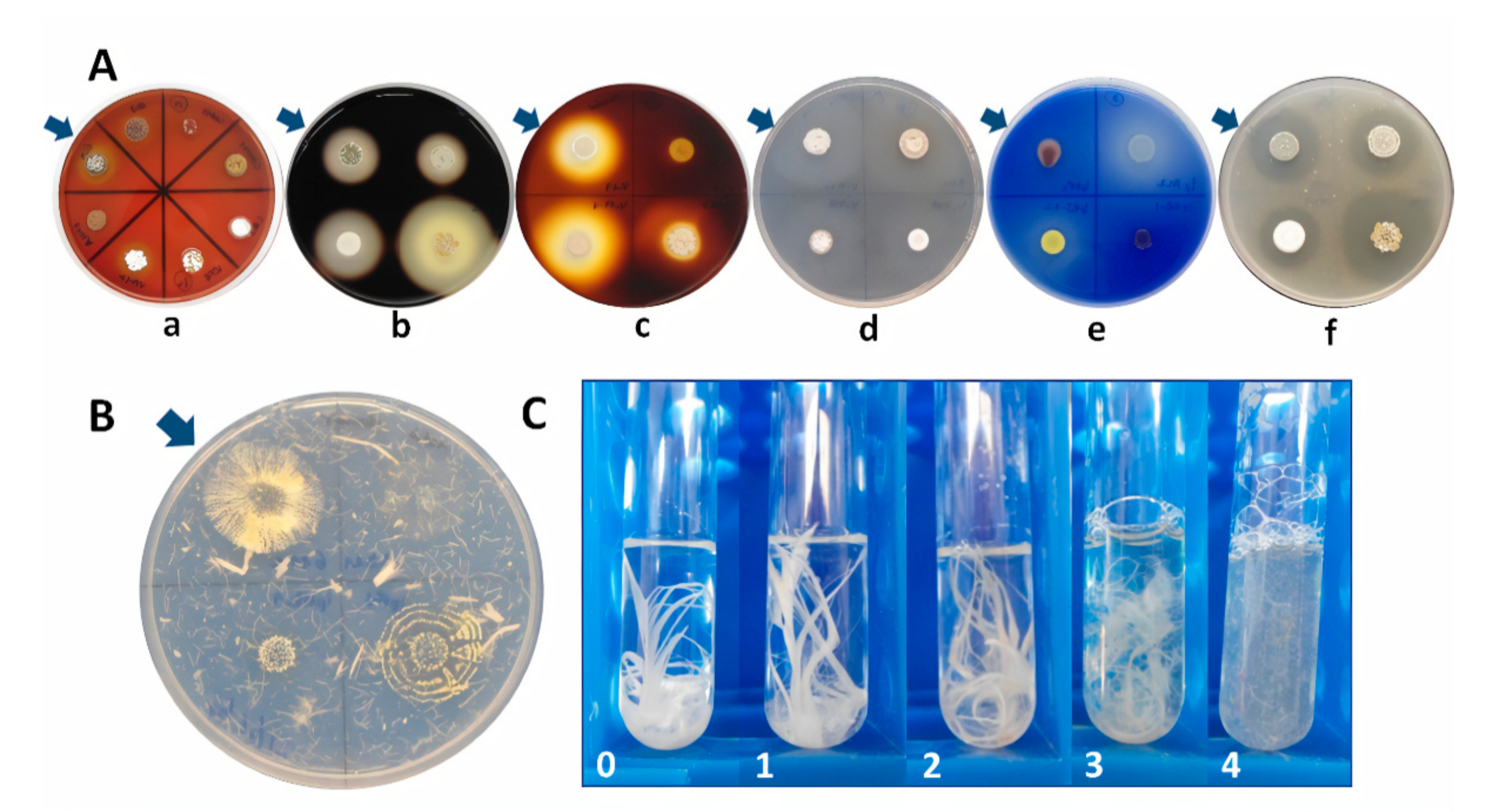
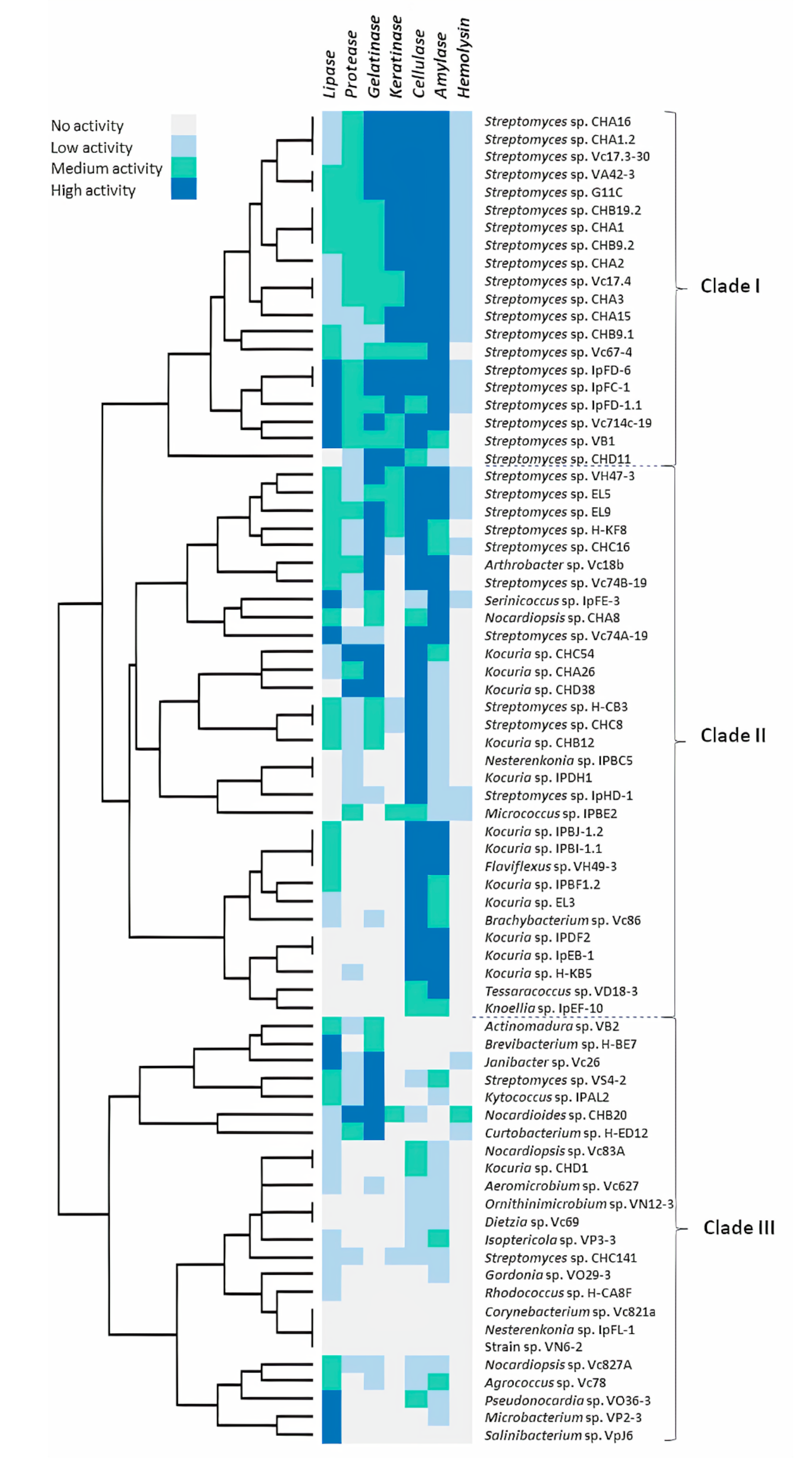
2.1. Hydrolytic Enzyme Screening
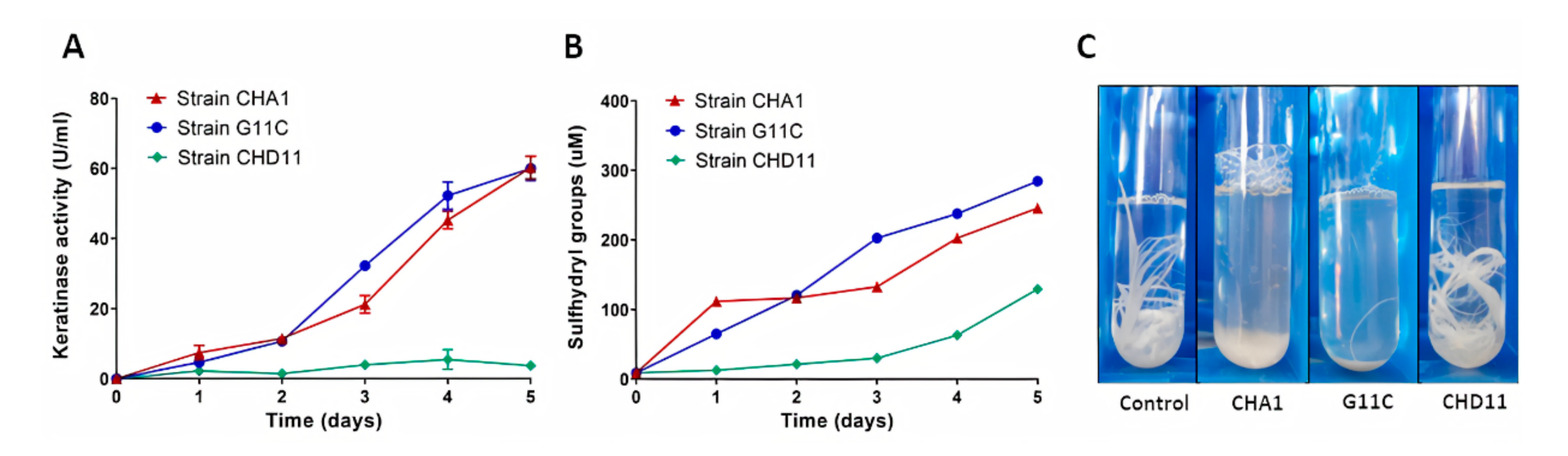
2.2. Screening of Keratinolytic Actinobacteria in Liquid Medium
2.3. Production of Keratinolytic Enzymes by Streptomyces sp. CHA1 and Streptomyces sp. G11C: Effect of Feather Concentration, Temperature, and Inoculum Percentage
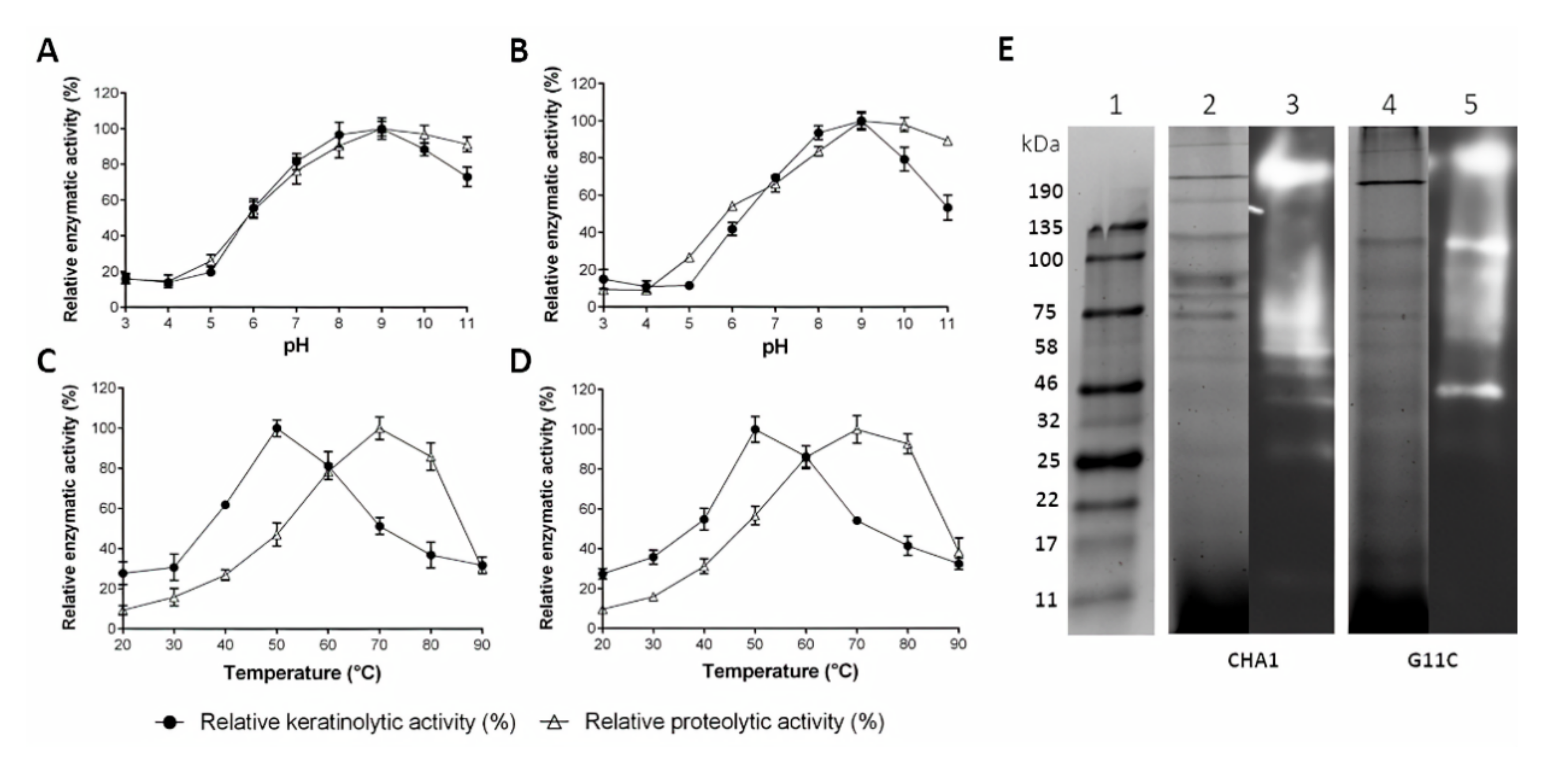
2.4. Partial Characterization of Cell-Free Culture Supernatant Containing Crude Enzymes
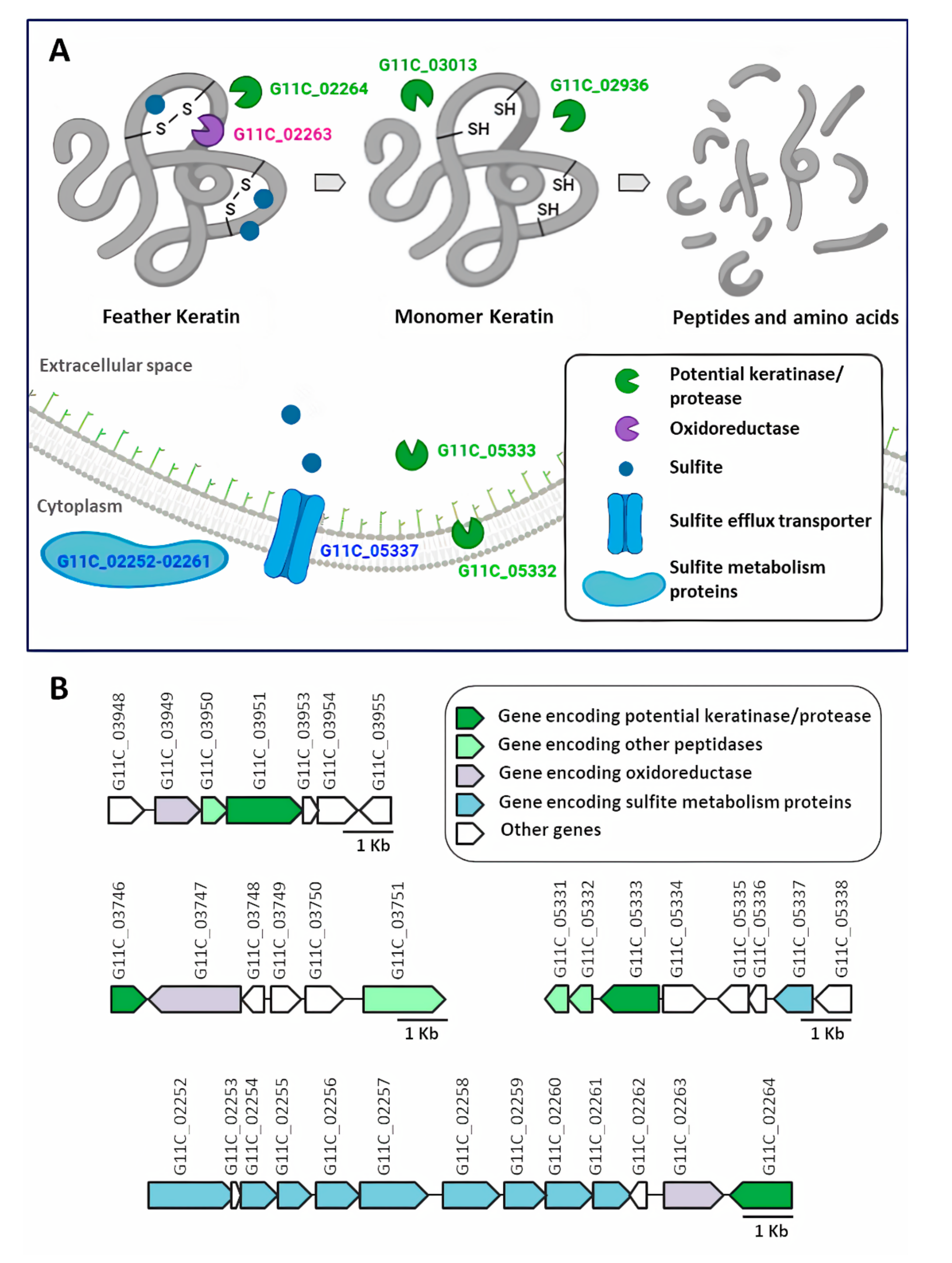
2.5. Secretome Analysis of Streptomyces sp. G11C
3. Discussion
4. Materials and Methods
4.1. Actinobacterial Strains
4.2. Primary Screening of Extracellular Hydrolytic Enzymes on Solid Media
4.2.1. Amylase Activity
4.2.2. Cellulase Activity
4.2.3. Gelatinase Activity
4.2.4. Lipase Activity
4.2.5. Protease Activity
4.2.6. Hemolytic Activity
4.2.7. Keratinase Activity
4.3. Growth Conditions for Keratinolytic Activity in Liquid Medium
4.4. Quantification of Keratinolytic Activity
4.5. Quantification of Proteolytic Activity
4.6. Determination of Sulfhydryl Groups
4.7. Percentage of Feather Degradation
4.8. Protein Determination
4.9. Characterization of Cultural Conditions for Keratinolytic Activity
4.10. Partial Characterization of Culture Supernatants: Influence of Temperature and pH
4.11. Non-Reducing SDS-PAGE
4.12. Zymography
4.13. Proteomic Analysis
4.14. Analysis In Silico of Genetic Contexts of Some Selected Peptidases
4.15. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Choi, J.M.; Han, S.S.; Kim, H.S. Industrial applications of enzyme biocatalysis: Current status and future aspects. Biotechnol. Adv. 2015, 33, 1443–1454. [Google Scholar] [CrossRef] [PubMed]
- Prakash, D.; Nawani, N.; Prakash, M.; Bodas, M.; Mandal, A.; Khetmalas, M.; Kapadnis, B. Actinomycetes: A repertory of green catalysts with a potential revenue resource. Biomed. Res. Int. 2013, 2013, 264020. [Google Scholar] [CrossRef] [PubMed]
- Sarmiento, F.; Peralta, R.; Blamey, J.M. Cold and hot extremozymes: Industrial relevance and current trends. Front. Bioeng. Biotechnol. 2015, 3, 148. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Kim, S.K. Research and application of marine microbial enzymes: Status and prospects. Mar. Drugs 2010, 8, 1920–1934. [Google Scholar] [CrossRef]
- Bonugli-Santos, R.C.; Vasconcelos, R.D.S.; Passarini, M.R.Z.; Vieira, G.A.L.; Lopes, V.C.P.; Mainardi, P.H.; Santos, J.A.E.; Duarte, L.D.A.; Otero, I.V.R.; Yoshida, A.M.D.S.; et al. Marine-derived fungi: Diversity of enzymes and biotechnological applications. Front. Microbiol. 2015, 6, 269. [Google Scholar] [CrossRef]
- Chater, K.F.; Biró, S.; Lee, K.J.; Palmer, T.; Schrempf, H. The complex extracellular biology of Streptomyces. FEMS Microbiol. Rev. 2010, 34, 171–198. [Google Scholar] [CrossRef]
- Ramesh, S.; Rajesh, M.; Mathivanan, N. Characterization of a thermostable alkaline protease produced by marine Streptomyces fungicidicus MML1614. Bioprocess Biosyst. Eng. 2009, 32, 791–800. [Google Scholar] [CrossRef]
- El-Sersy, N.A.; Abd-Elnaby, H.; Abou-Elela, G.M.; Ibrahim, H.A.; El-Toukhy, N.M. Optimization, economization and characterization of cellulase produced by marine Streptomyces ruber. Afr. J. Biotechnol. 2010, 9, 6355–6364. [Google Scholar] [CrossRef]
- Simkhada, J.R.; Yoo, H.Y.; Cho, S.S.; Choi, Y.H.; Kim, S.W.; Park, D.H.; Yoo, J.C. A novel cold-adapted lipase, LP28, from a mesophilic Streptomyces strain. Bioprocess Biosyst. Eng. 2012, 35, 217–225. [Google Scholar] [CrossRef]
- Habbeche, A.; Saoudi, B.; Jaouadi, B.; Haberra, S.; Kerouaz, B.; Boudelaa, M.; Badis, A.; Ladjama, A. Purification and biochemical characterization of a detergent-stable keratinase from a newly thermophilic actinomycete Actinomadura keratinilytica strain Cpt29 isolated from poultry compost. J. Biosci. Bioeng. 2014, 117, 413–421. [Google Scholar] [CrossRef]
- Gaber, Y.; Mekasha, S.; Vaaje-Kolstad, G.; Eijsink, V.G.H.; Fraaije, M.W. Characterization of a chitinase from the cellulolytic actinomycete Thermobifida fusca. Biochim. Biophys. Acta-Proteins Proteom. 2016, 1864, 1253–1259. [Google Scholar] [CrossRef] [PubMed]
- Holkar, C.R.; Jain, S.S.; Jadhav, A.J.; Pinjari, D.V. Valorization of keratin based waste. Process Saf. Environ. Prot. 2018, 115, 85–98. [Google Scholar] [CrossRef]
- Vidmar, B.; Vodovnik, M. Microbial keratinases: Enzymes with promising biotechnological applications. Food Technol. Biotechnol. 2018, 56, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Jaouadi, B.; Abdelmalek, B.; Fodil, D.; Ferradji, F.Z.; Rekik, H.; Zaraî, N.; Bejar, S. Purification and characterization of a thermostable keratinolytic serine alkaline proteinase from Streptomyces sp. strain AB1 with high stability in organic solvents. Bioresour. Technol. 2010, 101, 8361–8369. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Singh, H.; Anwar, S.; Chattopadhyay, A.; Tiwari, K.K.; Kaur, S.; Dhilon, G.S. Microbial keratinases: Industrial enzymes with waste management potential. Crit. Rev. Biotechnol. 2017, 37, 476–491. [Google Scholar] [CrossRef]
- Gopinath, S.C.B.; Anbu, P.; Lakshmipriya, T.; Tang, T.H.; Chen, Y.; Hashim, U.; Ruslinda, A.R.; Arshad, M.K.M. Biotechnological aspects and perspective of microbial Keratinase production. Biomed. Res. Int. 2015, 2015, 140726. [Google Scholar] [CrossRef]
- Bernal, C.; Cairó, J.; Coello, N. Purification and characterization of a novel exocellular keratinase from Kocuria rosea. Enzym. Microb. Technol. 2006, 38, 49–54. [Google Scholar] [CrossRef]
- Longshaw, C.M.; Wright, J.D.; Farrell, A.M.; Holland, K.T. Kytococcus sedentarius, the organism associated with pitted keratolysis, produces two keratin-degrading enzymes. J. Appl. Microbiol. 2002, 93, 810–816. [Google Scholar] [CrossRef]
- Thys, R.C.S.; Lucas, F.S.; Riffel, A.; Heeb, P.; Brandelli, A. Characterization of a protease of a feather-degrading Microbacterium species. Lett. Appl. Microbiol. 2004, 39, 181–186. [Google Scholar] [CrossRef]
- Mitsuiki, S.; Sakai, M.; Moriyama, Y.; Goto, M.; Furukawa, K. Purification and Some Properties of a Keratinolytic Enzyme from an Alkaliphilic Nocardiopsis sp. TOA-1. Biosci. Biotechnol. Biochem. 2002, 66, 164–167. [Google Scholar] [CrossRef][Green Version]
- Nickerson, W.J.; Noval, J.J.; Robison, R.S. Keratinase, Properties of the enzyme conjugate elaborated by Streptomyces fradiae. Biochim. Biophys. Acta 1963, 77, 73–86. [Google Scholar] [CrossRef]
- Böckle, B.; Galunsky, B.; Muller, R. Characterization of a Keratinolytic Serine Proteinase from Streptomyces Pactum DSM 40530. Appl. Environ. Microbiol. 1995, 61, 3705–3710. [Google Scholar] [CrossRef] [PubMed]
- Chitte, R.R.; Nalawade, V.K.; Dey, S. Keratinolytic activity from the broth of a feather-degrading thermophilic Streptomyces thermoviolaceus strain SD8. Lett. Appl. Microbiol. 1999, 28, 131–136. [Google Scholar] [CrossRef]
- Syed, D.G.; Lee, J.C.; Li, W.J.; Kim, C.J.; Agasar, D. Production, characterization and application of keratinase from Streptomyces gulbargensis. Bioresour. Technol. 2009, 100, 1868–1871. [Google Scholar] [CrossRef] [PubMed]
- Gousterova, A.; Braikova, D.; Goshev, I.; Christov, P.; Tishinov, K.; Vasileva-Tonkova, E.; Haertlé, T.; Nedkov, P. Degradation of keratin and collagen containinq wastes by newly isolated thermoactinomycetes or by alkaline hydrolysis. Lett. Appl. Microbiol. 2005, 40, 335–340. [Google Scholar] [CrossRef]
- Verma, A.; Singh, H.; Anwar, M.S.; Kumar, S.; Ansari, M.W.; Agrawal, S. Production of thermostable organic solvent tolerant keratinolytic protease from Thermoactinomyces sp. RM4: IAA production and plant growth promotion. Front. Microbiol. 2016, 7, 1–13. [Google Scholar] [CrossRef]
- Gupta, R.; Ramnani, P. Microbial keratinases and their prospective applications: An overview. Appl. Microbiol. Biotechnol. 2006, 7, 1189. [Google Scholar] [CrossRef]
- Brandelli, A.; Daroit, D.J.; Riffel, A. Biochemical features of microbial keratinases and their production and applications. Appl. Microbiol. Biotechnol. 2010, 85, 1735–1750. [Google Scholar] [CrossRef]
- Lin, X.; Kelemen, D.W.; Miller, E.S.; Shih, J.C.H. Nucleotide sequence and expression of kerA, the gene encoding a keratinolytic protease of Bacillus licheniformis PWD-1. Appl. Environ. Microbiol. 1995, 61, 1469–1474. [Google Scholar] [CrossRef]
- Riffel, A.; Brandelli, A.; Bellato, C.D.M.; Souza, G.H.M.F.; Eberlin, M.N.; Tavares, F.C.A. Purification and characterization of a keratinolytic metalloprotease from Chryseobacterium sp. kr6. J. Biotechnol. 2007, 128, 693–703. [Google Scholar] [CrossRef]
- Lange, L.; Huang, Y.; Busk, P.K. Microbial decomposition of keratin in nature—a new hypothesis of industrial relevance. Appl. Microbiol. Biotechnol. 2016, 100, 2083–2096. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Sharma, R.; Beg, Q.K. Revisiting microbial keratinases: Next generation proteases for sustainable biotechnology. Crit. Rev. Biotechnol. 2012, 33, 216–228. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, S.; Morita, Y.; Hasan, Q.; Yokoyama, K.; Tamiya, E. Keratin degradation: A cooperative action of two enzymes from Stenotrophomonas sp. Biochem. Biophys. Res. Commun. 2002, 294, 1138–1143. [Google Scholar] [CrossRef]
- Ramnani, P.; Singh, R.; Gupta, R. Keratinolytic potential of Bacillus licheniformis RG1: Structural and biochemical mechanism of feather degradation. Can. J. Microbiol. 2005, 51, 191–196. [Google Scholar] [CrossRef]
- Böckle, B.; Muller, R. Reduction of Disulfite Bonds by Streptomyces Pactum During Growth on Chiken Feathers. Appl. Environ. Microbiol. 1997, 63, 790–792. [Google Scholar] [CrossRef]
- Ramnani, P.; Gupta, R. Keratinases vis-à-vis conventional proteases and feather degradation. World J. Microbiol. Biotechnol. 2007, 23, 1537–1540. [Google Scholar] [CrossRef]
- Claverías, F.P.; Undabarrena, A.; Gonzalez, M.; Seeger, M.; Camara, B. Culturable diversity and antimicrobial activity of Actinobacteria from marine sediments in Valparaíso bay, Chile. Front. Microbiol. 2015, 6, 737. [Google Scholar] [CrossRef]
- Undabarrena, A.; Beltrametti, F.; Claverías, F.P.; González, M. Exploring the Diversity and Antimicrobial Potential of Marine Actinobacteria from the Comau Fjord in Northern Patagonia, Chile. Front. Microbiol. 2016, 7, 1135. [Google Scholar] [CrossRef]
- Cumsille, A.; Undabarrena, A.; González, V.; Claverías, F.; Rojas, C.; Cámara, B. Biodiversity of actinobacteria from the South Pacific and the assessment of Streptomyces chemical diversity with metabolic profiling. Mar. Drugs 2017, 15, 286. [Google Scholar] [CrossRef]
- Valencia, R.; González, V.; Undabarrena, A.; Zamora-Leiva, L.; Ugalde, J.A.; Camara, B. An Integrative Bioinformatic Analysis for Keratinase Detection in Marine-derived Streptomyces. BMC Genom. 2020; under revision. [Google Scholar]
- Peng, Z.; Zhang, J.; Du, G.; Chen, J. Keratin Waste Recycling Based on Microbial Degradation: Mechanisms and Prospects. ACS Sustain. Chem. Eng. 2019, 7, 9727–9736. [Google Scholar] [CrossRef]
- Hassan, M.A.; Abol-Fotouh, D.; Omer, A.M.; Tamer, T.M.; Abbas, E. Comprehensive insights into microbial keratinases and their implication in various biotechnological and industrial sectors: A review. Int. J. Biol. Macromol. 2020, 154, 567–583. [Google Scholar] [CrossRef] [PubMed]
- Léchenne, B.; Reichard, U.; Zaugg, C.; Fratti, M.; Kunert, J.; Boulat, O.; Monod, M. Sulphite efflux pumps in Aspergillus fumigatus and dermatophytes. Microbiology 2007, 153, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Monod, M. Secreted proteases from dermatophytes. Mycopathologia 2008, 166, 285–294. [Google Scholar] [CrossRef]
- Grumbt, M.; Monod, M.; Yamada, T.; Hertweck, C.; Kunert, J.; Staib, P. Keratin degradation by dermatophytes relies on cysteine dioxygenase and a sulfite efflux pump. J. Investig. Dermatol. 2013, 133, 1550–1555. [Google Scholar] [CrossRef]
- Cowie, G.L.; Hedges, J.I. Carbohydrate sources in a coastal marine environment. Geochim. Cosmochim. Acta 1984, 48, 2075–2087. [Google Scholar] [CrossRef]
- Ruocco, N.; Costantini, S.; Guariniello, S.; Costantini, M. Polysaccharides from the marine environment with pharmacological, cosmeceutical and nutraceutical potential. Molecules 2016, 21, 551. [Google Scholar] [CrossRef] [PubMed]
- Santhi, V.S.; Bhagat, A.K.; Saranya, S.; Govindarajan, G.; Jebakumar, S.R.D. Seaweed (Eucheuma cottonii) associated microorganisms, a versatile enzyme source for the lignocellulosic biomass processing. Int. Biodeterior. Biodegrad. 2014, 96, 144–151. [Google Scholar] [CrossRef]
- Santhi, V.S.; Gupta, A.; Saranya, S.; Jebakumar, S.R.D. A novel marine bacterium Isoptericola sp. JS-C42 with the ability to saccharifying the plant biomasses for the aid in cellulosic ethanol production. Biotechnol. Rep. 2014, 1, 8–14. [Google Scholar] [CrossRef][Green Version]
- Zhao, X.Q.; Xu, X.N.; Chen, L.Y. Production of Enzymes from Marine Actinobacteria. Adv. Food Nutr. Res. 2016, 78, 137–151. [Google Scholar] [CrossRef]
- Rajagopal, G.; Kannan, S. Systematic characterization of potential cellulolytic marine actinobacteria Actinoalloteichus sp. MHA15. Biotechnol. Rep. 2017, 13, 30–36. [Google Scholar] [CrossRef]
- Morales-Jiménez, J.; Zúñiga, G.; Ramírez-Saad, H.C.; Hernández-Rodríguez, C. Gut-Associated Bacteria Throughout the Life Cycle of the Bark Beetle Dendroctonus rhizophagus Thomas and Bright (Curculionidae: Scolytinae) and Their Cellulolytic Activities. Microb. Ecol. 2012, 64, 268–278. [Google Scholar] [CrossRef]
- Book, A.J.; Lewin, G.R.; McDonald, B.R.; Takasuka, T.E.; Doering, D.T.; Adams, A.S.; Blodgett, J.A.V.; Clardy, J.; Raffa, K.F.; Fox, B.G.; et al. Cellulolytic Streptomyces strains associated with herbivorous insects share a phylogenetically linked capacity to degrade lignocellulose. Appl. Environ. Microbiol. 2014, 80, 4692–4701. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Deng, G.; Yang, Y.; Wu, Z.; Wu, L. Optimization of endoglucanase production from a novel bacterial isolate, Arthrobacter sp. HPG166 and characterization of its properties. Braz. Arch. Biol. Technol. 2015, 58, 692–701. [Google Scholar] [CrossRef]
- Lopez-Ramirez, M.P.; Sanchez-Lopez, K.B.; Sarria-Guzman, Y.; Bello-Lopez, J.M.; Cano-Garcia, V.L.; Ruiz-Valdiviezo, V.M.; Dendooven, L. Haloalkalophilic cellulose-degrading bacteria isolated from an alkaline saline soil. J. Pure Appl. Microbiol. 2015, 9, 2879–2886. [Google Scholar]
- Acharyabhatta, A.; Kandula, S.K.; Terli, R. Taxonomy and polyphasic characterization of alkaline amylase producing marine actinomycete Streptomyces rochei BTSS 1001. Int. J. Microbiol. 2013, 2013, 276921. [Google Scholar] [CrossRef] [PubMed]
- Ilori, M.O.; Amund, O.O.; Omidiji, O. Purification and Properties of an α-Amylase Produced by a Cassava-Fermenting Strain of Micrococcus luteus. Folia Microbiol. (Praha) 1997, 42, 445–449. [Google Scholar] [CrossRef]
- Kim, S.-M.; Park, H.; Choi, J.-I. Cloning and Characterization of Cold-Adapted α-Amylase from Antarctic Arthrobacter agilis. Appl. Biochem. Biotechnol. 2016, 181, 1048–1059. [Google Scholar] [CrossRef]
- Chakraborty, S.; Jana, S.; Gandhi, A.; Sen, K.K.; Zhiang, W.; Kokare, C. Gellan gum microspheres containing a novel α-amylase from marine Nocardiopsis sp. strain B2 for immobilization. Int. J. Biol. Macromol. 2014, 70, 292–299. [Google Scholar] [CrossRef]
- Soto-Padilla, M.Y.; Gortáres-Moroyoqui, P.; Cira-Chávez, L.A.; Levasseur, A.; Dendooven, L.; Estrada-Alvarado, M.I. Characterization of extracellular amylase produced by haloalkalophilic strain Kocuria sp. HJ014. Int. J. Environ. Health Res. 2016, 26, 396–404. [Google Scholar] [CrossRef]
- Capella, J.; Vernazzani, B.G.; Gibbons, J.; Cabrera, E. Coastal migratory connections of Humpback Whales, Megaptera Novaeangliae Borowski, 1781, in southern Chile. An. Inst. Patagon. 2008, 36, 13–18. [Google Scholar] [CrossRef]
- Gaete, R.H.; Lobo, A.A.; Pakarati, S.Y.; Flores, M. Marine mammals of Easter Island (Rapa Nui) and Salas y Gomez Island (Motu Motiro Hiva), Chile: A review and new records. Lat. Am. J. Aquat. Res. 2014, 42, 743–751. [Google Scholar] [CrossRef]
- Sepúlveda, M.; Pérez-Álvarez, M.J.; Santos-Carvallo, M.; Pavez, G.; Olavarría, C.; Moraga, R.; Zerbini, A.N. From whaling to whale watching: Identifying fin whale critical foraging habitats off the Chilean coast. Aquat. Conserv. Mar. Freshw. Ecosyst. 2018, 28, 821–829. [Google Scholar] [CrossRef]
- Goldbogen, J.A.; Cade, D.E.; Calambokidis, J.; Friedlaender, A.S.; Potvin, J.; Segre, P.S.; Werth, A.J. How Baleen Whales Feed: The Biomechanics of Engulfment and Filtration. Ann. Rev. Mar. Sci. 2017, 9, 367–386. [Google Scholar] [CrossRef] [PubMed]
- Loch, C.; Viegas, S.V.; Waddell, J.N.; Kemper, C.; Cook, R.B.; Werth, A.J. Structure and properties of baleen in the Southern right (Eubalaena australis) and Pygmy right whales (Caperea marginata). J. Mech. Behav. Biomed. Mater. 2020, 110, 103939. [Google Scholar] [CrossRef]
- Herzog, B.; Overy, D.P.; Haltli, B.; Kerr, R.G. Discovery of keratinases using bacteria isolated from marine environments. Syst. Appl. Microbiol. 2016, 39, 49–57. [Google Scholar] [CrossRef]
- Cheng, S.W.; Hu, H.M.; Shen, S.W.; Takagi, H.; Asano, M.; Tsai, Y.C. Production and characterization of keratinase of a feather-degrading Bacillus licheniformis PWD-1. Biosci. Biotechnol. Biochem. 1995, 59, 2239–2243. [Google Scholar] [CrossRef]
- Alnahdi, H.S. Isolation and screening of extracellular proteases produced by new isolated bacillus sp. J. Appl. Pharm. Sci. 2012, 2, 071–074. [Google Scholar] [CrossRef]
- Awad, W.M.; Soto, A.R.; Siegel, S.; Skiba, W.E.; Bernstrom, G.G. The Proteolytic Enzymes of the K-1 Strain of Streptomyces griseus obtained from a Commercial Preparation (Pronase). J. Biol. Chem. 1972, 247, 4144–4154. [Google Scholar]
- Michotey, V.; Blanco, C. Characterization of an endoserine protease secreted by Arthrobacter aureus. Appl. Environ. Microbiol. 1994, 60, 341–343. [Google Scholar] [CrossRef]
- Kuddus, M.; Ramteke, P.W. A cold-active extracellular metalloprotease from Curtobacterium luteum (MTCC 7529): Enzyme production and characterization. J. Gen. Appl. Microbiol. 2008, 54, 385–392. [Google Scholar] [CrossRef]
- Gesheva, V.; Vasileva-Tonkova, E. Production of enzymes and antimicrobial compounds by halophilic Antarctic Nocardioides sp. grown on different carbon sources. World J. Microbiol. Biotechnol. 2012, 28, 2069–2076. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Chen, X.; Li, J.-J.; Ren, D. An alkaline protease from Kocuria kristinae F7: Properties and characterization of its hydrolysates from soy protein. Eur. Food Res. Technol. 2013, 236, 293–301. [Google Scholar] [CrossRef]
- Adamitsch, B.F.; Hampel, W.A. Formation of lipolytic enzymes by Brevibacterium linens. Biotechnol. Lett. 2000, 22, 1643–1646. [Google Scholar] [CrossRef]
- Hantsis-Zacharov, E.; Halpern, M. Culturable psychrotrophic bacterial communities in raw milk and their proteolytic and lipolytic traits. Appl. Environ. Microbiol. 2007, 73, 7162–7168. [Google Scholar] [CrossRef] [PubMed]
- Cadirci, B.H.; Yasa, I.; Kocyigit, A. Streptomyces sp. TEM 33 possesses high lipolytic activity in solid-state fermentation in comparison with submerged fermentation. Prep. Biochem. Biotechnol. 2016, 46, 23–29. [Google Scholar] [CrossRef]
- Yuan, D.; Lan, D.; Xin, R.; Yang, B.; Wang, Y. Biochemical properties of a new cold-active mono- and diacylglycerol lipase from marine member Janibacter sp. strain HTCC2649. Int. J. Mol. Sci. 2014, 15, 10554–10566. [Google Scholar] [CrossRef]
- Lan, D.; Qu, M.; Yang, B.; Wang, Y. Enhancing production of lipase MAS1 from marine Streptomyces sp. strain in Pichia pastoris by chaperones co-expression. Electron. J. Biotechnol. 2016, 22, 62–67. [Google Scholar] [CrossRef]
- Cervantes-González, E.; Rojas-Avelizapa, N.G.; Cruz-Camarillo, R.; García-Mena, J.; Rojas-Avelizapa, L.I. Oil-removal enhancement in media with keratinous or chitinous wastes by hydrocarbon-degrading bacteria isolated from oil-polluted soils. Environ. Technol. 2008, 29, 171–182. [Google Scholar] [CrossRef]
- Laba, W.; Choinska, A.; Rodziewicz, A.; Piegza, M. Keratinolytic abilities of Micrococcus luteus from poultry waste. Braz. J. Microbiol. 2015, 46, 691–700. [Google Scholar] [CrossRef]
- Scott, J.A.; Untereiner, W.A. Determination of keratin degradation by fungi using keratin azure. Med. Mycol. 2004, 42, 239–246. [Google Scholar] [CrossRef]
- Daroit, D.J.; Brandelli, A. A current assessment on the production of bacterial keratinases. Crit. Rev. Biotechnol. 2014, 34, 372–384. [Google Scholar] [CrossRef] [PubMed]
- Valencia, P.; Pinto, M.; Almonacid, S. Identification of the key mechanisms involved in the hydrolysis of fish protein by Alcalase. Process Biochem. 2014, 49, 258–264. [Google Scholar] [CrossRef]
- Bressollier, P.; Letourneau, F.; Urdaci, M.; Verneuil, B. Purification and characterization of a keratinolytic serine proteinase from Streptomyces albidoflavus. Appl. Environ. Microbiol. 1999, 65, 2570–2576. [Google Scholar] [CrossRef] [PubMed]
- Tatineni, R.; Doddapaneni, K.K.; Potumarthi, R.C.; Vellanki, R.N.; Kandathil, M.T.; Kolli, N.; Mangamoori, L.N. Purification and characterization of an alkaline keratinase from Streptomyces sp. Bioresour. Technol. 2008, 99, 1596–1602. [Google Scholar] [CrossRef]
- Jain, R.; Jain, A.; Rawat, N.; Nair, M.; Gumashta, R. Feather hydrolysate from Streptomyces sampsonii GS 1322: A potential low cost soil amendment. J. Biosci. Bioeng. 2016, 121, 672–677. [Google Scholar] [CrossRef]
- Monod, M.; Capoccia, S.; Léchenne, B.; Zaugg, C.; Holdom, M.; Jousson, O. Secreted proteases from pathogenic fungi. Int. J. Med. Microbiol. 2002, 292, 405–419. [Google Scholar] [CrossRef]
- Huang, Y.; Busk, P.K.; Herbst, F.A.; Lange, L. Genome and secretome analyses provide insights into keratin decomposition by novel proteases from the non-pathogenic fungus Onygena corvina. Appl. Microbiol. Biotechnol. 2015, 99, 9635–9649. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, T.; Song, N.; Li, Q.; Wang, Z.; Zhang, X.; Lu, X.; Fang, J.; Chen, J. Purification and characterization of four key enzymes from a feather-degrading Bacillus subtilis from the gut of tarantula Chilobrachys guangxiensis. Int. Biodeterior. Biodegrad. 2014, 96, 26–32. [Google Scholar] [CrossRef]
- Huang, Y.; Łężyk, M.; Herbst, F.A.; Busk, P.K.; Lange, L. Novel keratinolytic enzymes, discovered from a talented and efficient bacterial keratin degrader. Sci. Rep. 2020, 10, 10033. [Google Scholar] [CrossRef]
- Li, Z.W.; Liang, S.; Ke, Y.; Deng, J.J.; Zhang, M.S.; Lu, D.L.; Li, J.Z.; Luo, X.C. The feather degradation mechanisms of a new Streptomyces sp. isolate SCUT-3. Commun. Biol. 2020, 3, 191. [Google Scholar] [CrossRef]
- Menasria, T.; Aguilera, M.; Hocine, H.; Benammar, L.; Ayachi, A.; Bachir, A.S.; Dekak, A.; Monteoliva-Sánchez, M. Diversity and bioprospecting of extremely halophilic archaea isolated from Algerian arid and semi-arid wetland ecosystems for halophilic-active hydrolytic enzymes. Microbiol. Res. 2018, 207, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Kasana, R.C.; Salwan, R.; Dhar, H.; Dutt, S.; Gulati, A. A rapid and easy method for the detection of microbial cellulases on agar plates using Gram’s iodine. Curr. Microbiol. 2008, 57, 503–507. [Google Scholar] [CrossRef]
- dela Cruz, T.E.E.; Torres, J.M. Gelatin Hydrolysis Test Protocol. Am. Soc. Microbiol. 2012, 1–10. [Google Scholar]
- Letourneau, F.; Soussotte, V.; Bressollier, P.; Branland, P.; Verneuil, B. Keratinolytic activity of Streptomyces sp. S.K1-02: A new isolated strain. Lett. Appl. Microbiol. 1998, 26, 77–80. [Google Scholar] [CrossRef]
- Thys, R.C.S.; Brandelli, A. Purification and properties of a keratinolytic metalloprotease from Microbacterium sp. J. Appl. Microbiol. 2006, 101, 1259–1268. [Google Scholar] [CrossRef] [PubMed]
- Ellman, G.L. Tissue Sulfhydryl Groups. Arch. Biochem. Biophys. 1959, 82, 70–77. [Google Scholar] [CrossRef]
- Bradford, M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Heussen, C.; Dowdle, E.B. Electrophoretic analysis of plasminogen activators in polyacrylamide gels containing sodium dodecyl sulfate and copolymerized substrates. Anal. Biochem. 1980, 102, 196–202. [Google Scholar] [CrossRef]
- Pan, D.; Hill, A.P.; Kashou, A.; Wilson, K.A.; Tan-Wilson, A. Electrophoretic transfer protein zymography. Anal. Biochem. 2011, 411, 277–283. [Google Scholar] [CrossRef]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Mitteer, D.R.; Greer, B.D.; Fisher, W.W.; Cohrs, V.L. Teaching behavior technicians to create publication-quality, single-case design graphs in Graphpad prism 7. J. Appl. Behav. Anal. 2018, 51, 998–1010. [Google Scholar] [CrossRef] [PubMed]

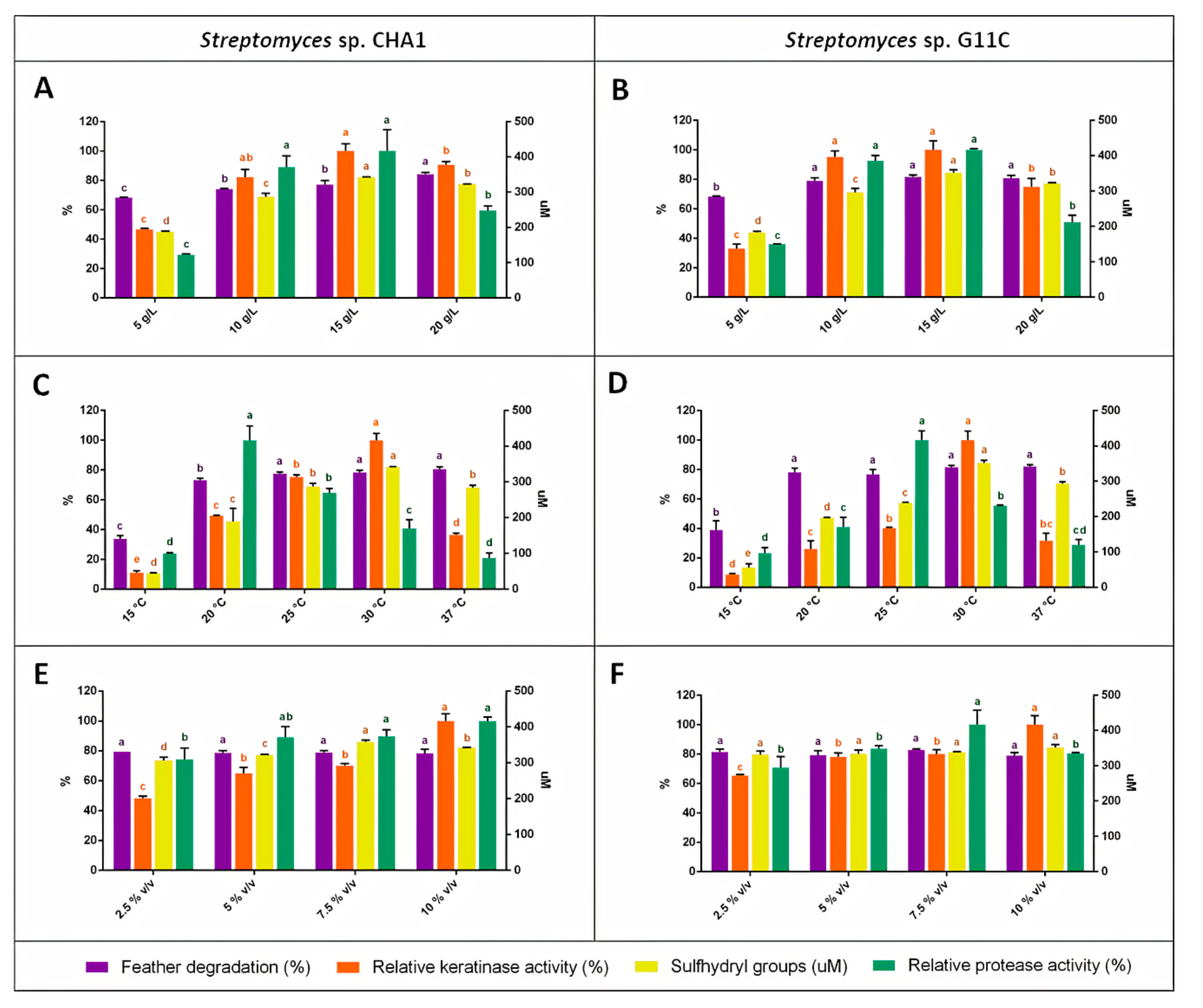
| Strain | Activity | Substrate | Temperature | Inoculum | |||
|---|---|---|---|---|---|---|---|
| Max. Value | Parameter g/L | Max. Value | Parameter °C | Max. Value | Parameter % v/v | ||
| CHA1 | Feather deg. | 84.3 ± 1.0 * | 20 | 79.1 ± 1.8 | 37 | 78.9 ± 1.3 | 7.5 |
| Keratinase act. | 62.3 ± 3.0 | 15 | 63.5 ± 3.0 * | 30 | 63.5 ± 3.0 * | 10 | |
| R-SH groups | 342.4 ± 3.3 * | 15 | 342.6 ± 4.2 * | 30 | 358.4 ± 5.0 * | 7.5 | |
| Protease act. | 11.85 ± 1.7 | 15 | 29.1 ± 2.8 * | 20 | 13.9 ± 0.4 | 10 | |
| G11C | Feather deg. | 81.7 ± 1.1 | 15 | 81.7 ± 1.1 | 30 | 82.8 ± 0.6 | 7.5 |
| Keratinase act. | 78.3 ± 4.7 | 15 | 78.3 ± 4.7 * | 30 | 78.3 ± 4.7 * | 10 | |
| R-SH groups | 352.0 ± 7.5 | 15 | 352.0 ± 7.5 * | 30 | 352.0 ± 7.5 | 10 | |
| Protease act. | 15.2 ± 0.1 | 15 | 27.4 ± 1.7 * | 25 | 18.9 ± 1.8 * | 7.5 | |
| Seq-Illumina Locus | Description | MEROPS Classification | Molecular Function | Length | MolWt (MH) | Spectral Counts |
|---|---|---|---|---|---|---|
| G11C_03013 | S8 family peptidase | s08 | Serine endopeptidase | 528 | 53,486.9 | 79 |
| G11C_00267 | Serine protease | s01 | Serine endopeptidase | 1427 | 152,855.9 | 60 |
| G11C_05333 | S1 family peptidase | s01 | Serine endopeptidase | 384 | 38,911.3 | 27 |
| G11C_03247 | M56 family metallopeptidase | m56 | Metalloendopeptidase | 311 | 32,205.8 | 18 |
| G11C_03889 | X-Pro dipeptidyl peptidase | s15 | Aminopeptidase | 597 | 62,112.5 | 15 |
| G11C_02143 | PrsW family intramembrane metalloprotease | m82 | Metalloendopeptidase | 318 | 34,048.5 | 12 |
| G11C_05273 | S8 family peptidase | s08 | Serine endopeptidase | 404 | 41,273.7 | 10 |
| G11C_01742 | Lon protease 2 | s16 | ATP-dependent serine endopeptidase | 359 | 37,268.1 | 9 |
| G11C_01512 | S1 family peptidase | s01 | Serine endopeptidase | 360 | 36,229.1 | 9 |
| G11C_03649 | Hypothetical protein | nd | 242 | 25,244.8 | 8 | |
| G11C_03746 | PepSY domain-containing protein | nd | Peptidase propeptide | 229 | 23,855.7 | 8 |
| G11C_02264 | Extracellular serine proteinase | s08 | Serine endopeptidase | 404 | 40,851.8 | 7 |
| G11C_03951 | Calpain family cysteine protease | c02 | Cysteine endopeptidase | 498 | 54,800.4 | 7 |
| G11C_04177 | S8_S53 family peptidase | s53 | Acid-acting serine endopeptidase | 456 | 47,500.6 | 7 |
| G11C_02546 | Trypsin-like serine protease | s01 | Serine endopeptidase | 296 | 29,365.6 | 6 |
| G11C_03446 | Class F sortase | nd | Sortase cysteine transpeptidase | 140 | 13,909.3 | 5 |
| G11C_02426 | M50 family metallopeptidase | m50 | Metalloendopeptidase | 247 | 26,291.5 | 4 |
| G11C_03494 | Papain-like cysteine peptidase | nd | Cysteine endopeptidase/exopeptidase | 219 | 23,970 | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
González, V.; Vargas-Straube, M.J.; Beys-da-Silva, W.O.; Santi, L.; Valencia, P.; Beltrametti, F.; Cámara, B. Enzyme Bioprospection of Marine-Derived Actinobacteria from the Chilean Coast and New Insight in the Mechanism of Keratin Degradation in Streptomyces sp. G11C. Mar. Drugs 2020, 18, 537. https://doi.org/10.3390/md18110537
González V, Vargas-Straube MJ, Beys-da-Silva WO, Santi L, Valencia P, Beltrametti F, Cámara B. Enzyme Bioprospection of Marine-Derived Actinobacteria from the Chilean Coast and New Insight in the Mechanism of Keratin Degradation in Streptomyces sp. G11C. Marine Drugs. 2020; 18(11):537. https://doi.org/10.3390/md18110537
Chicago/Turabian StyleGonzález, Valentina, María José Vargas-Straube, Walter O. Beys-da-Silva, Lucélia Santi, Pedro Valencia, Fabrizio Beltrametti, and Beatriz Cámara. 2020. "Enzyme Bioprospection of Marine-Derived Actinobacteria from the Chilean Coast and New Insight in the Mechanism of Keratin Degradation in Streptomyces sp. G11C" Marine Drugs 18, no. 11: 537. https://doi.org/10.3390/md18110537
APA StyleGonzález, V., Vargas-Straube, M. J., Beys-da-Silva, W. O., Santi, L., Valencia, P., Beltrametti, F., & Cámara, B. (2020). Enzyme Bioprospection of Marine-Derived Actinobacteria from the Chilean Coast and New Insight in the Mechanism of Keratin Degradation in Streptomyces sp. G11C. Marine Drugs, 18(11), 537. https://doi.org/10.3390/md18110537







