Abstract
Leptolyngbya, a well-known genus of cyanobacteria, is found in various ecological habitats including marine, fresh water, swamps, and rice fields. Species of this genus are associated with many ecological phenomena such as nitrogen fixation, primary productivity through photosynthesis and algal blooms. As a result, there have been a number of investigations of the ecology, natural product chemistry, and biological characteristics of members of this genus. In general, the secondary metabolites of cyanobacteria are considered to be rich sources for drug discovery and development. In this review, the secondary metabolites reported in marine Leptolyngbya with their associated biological activities or interesting biosynthetic pathways are reviewed, and new insights and perspectives on their metabolic capacities are gained.
1. Introduction
Cyanobacteria, also known as ‘blue-green algae’, rank among the oldest prokaryotes found on Earth and evolved to possess remarkable capabilities of oxygenic photosynthesis and nitrogen fixation, allowing them to both adapt to and affect the planet’s environmental conditions for over two billion years [1,2,3,4]. In traditional classification schema, filamentous cyanobacteria without akinetes, heterocysts or true-branching were generally assigned to the order Oscillatoriales (Section III) [5]. However, one specific genus of filamentous, nonheterocystous cyanobacteria with granular surface ornamentation was ultimately named Leptolyngbya in 1988, and placed in the order Pseudanabaenales [6,7,8]. The identification of this genus was supported by phylogenetic analysis, including 16S rRNA gene sequences, which at the time showed no clear relationship with any other defined genus of cyanobacteria [9].
Leptolyngbya originate from a diverse range of ecological habitats, including marine, fresh water, swamps, forests, rice fields, alkaline lakes, and even the polar desert or hot desert environments [10,11,12,13]. Members of this genus possess a variety of thin (0.5–3.5 μm), long filaments, and can grow solitarily or coiled into clusters and fine mats [14,15,16]. Heterocysts and akinetes are both absent in these organisms [17]. In the taxonomic databases Algaebase [18] and CyanoDB [19] there are 138 taxonomically accepted species of Leptolyngbya listed.
Marine collections of Leptolyngbya have been reported in Japan, Panama, Gulf of Thailand, the Red Sea, Hawaii, America Samoa, and others (Table 1), which illustrates that this genus is distributed widely in the world. A breadth of scientific studies have been conducted on these samples in recent decades, including morphological and phylogenetic characterization, photosynthesis and nitrogen fixation research, examination of the associated microbial community, growth modulation and natural product chemistry research [4,9,17,20,21,22]. It is well known that cyanobacteria produce many secondary metabolites, especially when grown in different environments and conditions. Variations such as temperature, pH, dissolved phosphorous levels, nitrogen content or associated symbionts can influence the expression of different natural products [22,23]. The metabolites of cyanobacteria are known to possess potent biological activity in areas such as cytotoxicity, anti-inflammation, neuromodulatory, antibacterial, and brine shrimp toxicity [24,25]. It is possible that these observed biological activities match or mimic some of the natural ecological roles of these secondary metabolites. A limited number of Leptolyngbya field collections have been propagated in laboratory cultures [26,27,28]. However, they grow slowly in this environment, making for additional challenges in secondary metabolite discovery from this resource [29]. This review summarizes recent research on the secondary metabolites found in the genus Leptolyngbya, and covers chemical, pharmacological, and biosynthetic aspects.

Table 1.
Secondary metabolites from Leptolyngbya and the reported bioactivities from each.
2. Chemical Diversity of the Secondary Metabolites Isolated from Leptolyngbya
2.1. Polypeptides
In 2008, McPhail and co-workers reported the discovery of coibamide A (1), a cyclic depsipeptide with a high degree of N- and O-methylation from a Leptolyngbya species collected in Panama (Figure 1) [32]. The absolute configuration was determined by a series of chiral HPLC analyses of the amino acids resulting from the hydrolyzed peptide as well as some computational modeling. Interest in coibamide A (1) mounted due to its exquisitely potent low-nanomolar in vitro inhibition activity against multiple cancer cell lines, including human NCI-H460, MDA-MB-231, H292, PC-3, SF-295, mouse neuro2a, LOX IMVI, HL-60(TB), and SNB-75 cell lines [32]. The number of cell lines was expanded to include human U87-MG, SF-295 glioblastoma cells and mouse embryonic fibroblasts (MEFs), and it was further revealed that the cyclized structure was crucial for potent biological activity [52]. Several studies subsequently reported the total synthesis of coibamide A (1). Yao and Lim et al. completed the total synthesis of the structure originally reported for coibamide A (2), as well as a synthetic O-desmethyl analogue. However, both the 1H and 13C NMR data for the synthetic product 2 differed from those of the natural product, which indicated that the absolute configuration of this compound required revision [33,34]. Additionally, Oishi and Fujii synthesized the d-N-Me-Ala epimer of coibamide A (4) due to an epimerization of this residue during the macrocyclization process (Figure 1) [35]. In 2015, Fang and Su were able to assign the correct configuration of coibamide A (1) with the revision of the l-HVA and l-N-Me-Ala residues to the d-HVA and d-N-Me-Ala after total synthesis of this alternative along with its diastereomeric analogues (Figure 1) [30]. The absolute configuration of coibamide A (1) proposed by Fang and Su was further confirmed by McPhail and Cheong using computational methods to calculate NMR data for the conformational space occupied by several possible diastereomers and comparison with experimental values [53]. This considerable effort to resolve the structure of coibamide A (1) was largely motivated by the potent cytotoxicity of this molecule, and as a byproduct, provided a number of new analogues for mechanistic and pharmacological studies. More recently, Su and Fang went on to synthesize 18 new analogues (2, 4, 5–20) including the originally proposed structure of coibamide A (2) as well as the revised correct structure (1) to perform a structure–activity relationship study (Figure 2) [31]. However, none of the coibamide A analogues were more potent cytotoxins than the natural product, indicating the strong correlation between the observed activity, the core molecular structure and optimization of this structure through natural evolutionary processes. The only analogue that exhibited similar inhibition as natural coibamide A was the [MeAla3-MeAla6]-coibamide (8), which significantly suppressed tumor growth in vivo [31,33,35].
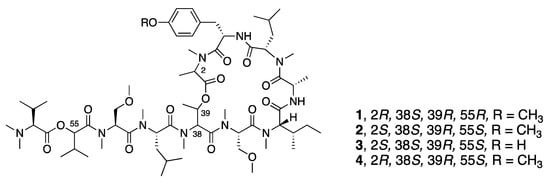
Figure 1.
Structure of coibamide A (2) from Leptolyngbya sp. and some synthetic analogues generated to validate its absolute configuration.
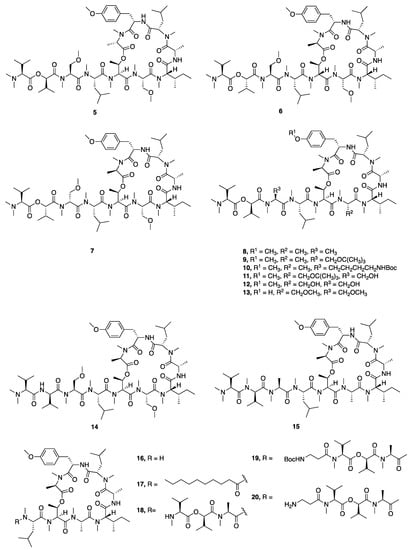
Figure 2.
Synthetic analogues of coibamide A produced to explore structure–activity relationships (SAR) in this molecular class.
The dolastatins are an expansive and well-known series of peptidic compounds. These were named after first being discovered from the sea hare Dolabella auricularia, but it was later found that the mollusk accumulates these natural products from cyanobacteria in their diets [37,39,54,55]. Dolastatin 12 (21) is one such compound, and has interesting 4-amino-2,2-dimethyl-3-oxopentanoic acid (Ibu) and (2S,3R)-3-amino-2-methylpentanoic acid (MAP) residues in its cyclic structure (Figure 3). The macrocycle also includes an ester linkage across a 2-hydroxy-3-methylpentanoate (Hmp) residue, and is thus a depsipeptide. While this compound was originally isolated from D. auricularia, it was noted to resemble the Lyngbya majuscula metabolite majusculamide C. Dolastatin 12 (21) was later re-discovered from a mixed cyanobacterial assemblage of L. majuscula and Schizothrix calcicola from Guam [36,39,56,57] as well as from a Leptolyngbya sp. RS03 collected in the Red Sea [36]. Dolastatin 12 (21) may be present as a mixture of diastereomers arising from epimerization of the acid-sensitive Ibu unit during the isolation process [57]. The configuration of this Ibu residue was determined by CD and 1H NMR analysis after hydrolysis and purification [39]. The correct configuration was determined to be R by comparing the free Ibu unit to Adhpa synthetic standards [57]. In the early studies, dolastatin 12 (21) was shown to inhibit actin polymerization [37,58]. Dolastatin 12 (21) has in vitro MICs <0.05 µg/mL against human nasopharyngeal carcinoma cells and 0.08 µg/mL against human colon adenocarcinoma cells [37]. Further cytotoxicity characterization showed that dolastatin 12 has an in vitro IC50 > 1 µM against HeLa cells [36,56] and a ED50 of 7.5 × 10−2 μg/mL against murine P388 lymphocytic leukemia [38,39].
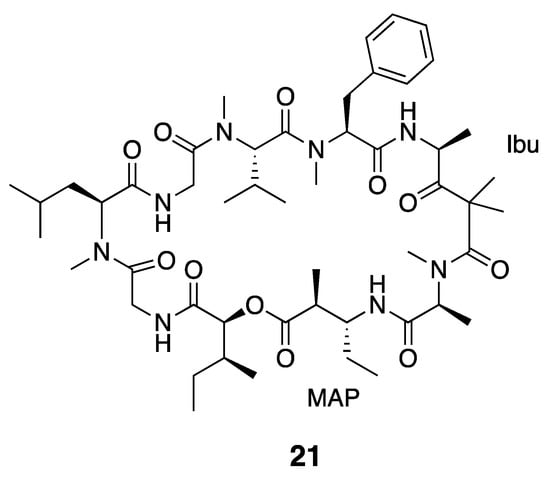
Figure 3.
Dolastatin 12, previously isolated from Leptolyngbya sp. as well as several other sources. See text for discussion of the unusual MAP and Ibu moieties in dolastatin 12.
Ibu-epidemethoxylyngbyastatin 3 (22) has a similar structural backbone to dolastatin 12 (21) and possesses the same Ibu residue (Figure 4). However, rather than containing the MAP as in dolastatin 12, Ibu-epidemethoxylyngbyastatin 3 has a molecular mass comparatively increased by 14 Da, which results from the presence of a 3-amino-2-methylhexanoic acid (Amha) moiety instead of MAP. In vitro cytotoxicity testing showed that Ibu-epidemethoxylyngbyastatin 3 (22) is at least 10-fold less toxic than dolastatin 12 (IC50 > 1 µM) against HeLa cells [36,56].
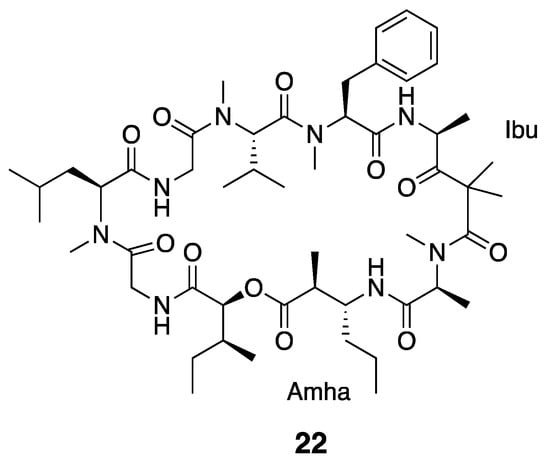
Figure 4.
Ibu-epidemethoxylyngbyastatin 3 from Leptolyngbya sp.
Grassypeptolides are a series of cyclic depsipeptides first isolated from the marine cyanobacterium Lyngbya confervoides. These peptides contain d-amino acids, thiazoline rings, and a β-amino acid (Figure 5) [59,60]. Further research led to the isolation and description of grassypeptolides D (23) and E (24) from a Red Sea Leptolyngbya sp. RS03, and grassypeptolides F and G from Lyngbya majuscula [61,62]. Grassypeptolides D (23) and E (24) showed cytotoxic activity to both HeLa cells (IC50 335 and 192 nM, respectively) and neuro2a cells (IC50 599 and 407 nM, respectively) [36,56,63]. By comparing with other grassypeptolide structures and their biological activities, an initial structure–activity relationship was deduced that indicated the N-Me-Phe-thn-ca-Aba-thn-ca tetrapeptide motif could be the key pharmacophore of the grassypeptolides [36,56].
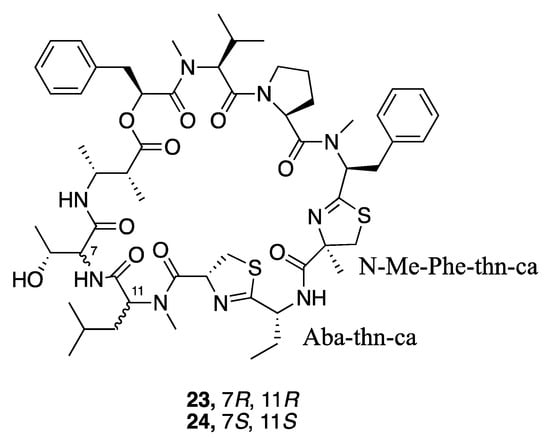
Figure 5.
Grassypeptolides from Leptolyngbya sp. RS03.
Loggerpeptins A–C (25–27) are cyclic depsipeptides with 3-amino-6-hydroxy-2-piperidone (Ahp) residues (Figure 6) that were isolated from a Florida cyanobacterial collection identified morphologically as Leptolyngbya sp. [40]. These compounds were screened for serine protease inhibitory activities to assess their antimetastatic effect against breast cancer cells. Loggerpeptin A (25) and B (26) were more potent than loggerpeptin C (27) with IC50s of 0.24 and 0.22 µM against bovine pancreatic chymotrypsin and 0.24 and 0.28 µM against porcine pancreatic elastase [40]. Loggerpeptin A (25) was 2- and 3-fold more potent than loggerpeptin B (26) and C (27) against human neutrophil elastase (HNE) [40]. All three compounds exhibited antiproteolytic activities, with IC50 values under 1 µM. As the major component in the collection, loggerpeptin C (27) was subject to a detailed molecular study [40]. Molecular docking showed that the Leu and N-terminal Thr-1, Abu and Ala residues of loggerpeptin C (27) were binding to the subsites S1-S4 of HNE and porcine pancreatic elastase [40].
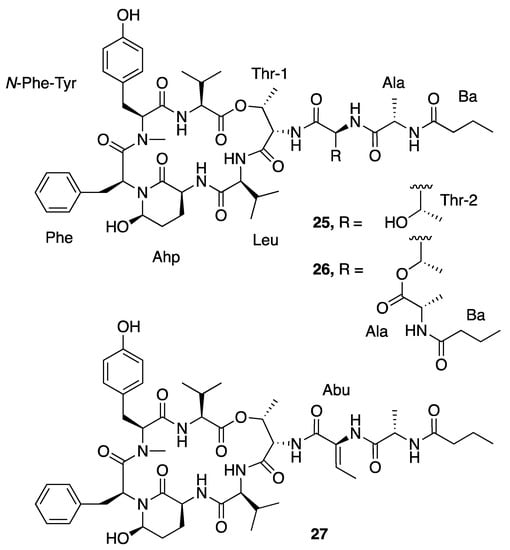
Figure 6.
Loggerpeptins from Leptolyngbya sp.
Molassamide (28) is another cyclic depsipeptide (Figure 7) that was isolated along with loggerpeptins A–C (25–27) [40]. Compared to loggerpeptins (25–27), molassamide (28) exhibited much more potent inhibition activity against porcine pancreatic elastase, with an IC50 value of 50 nM, indicating the Abu residue between Ahp and Thr-1 is important to the antiproteolytic selectivity [40]. Molassamide presents similar binding patterns in molecular docking as loggerpeptin C (27), with the Abu and N-terminal Thr-1, Thr-2 and Ala binding in subsites S1–S4 of HNE and porcine pancreatic elastase [40]. Molassamide also inhibited elastase from cleaving the substrate CD40 in both biochemical and cellular assays, and it also inhibited ICAM-1 cleavage and downregulated elastase-induced ICAM-1 gene expression. Overall, this profile of activity is indicative of molassamide being a promising candidate for potential treatment of breast cancer [40].
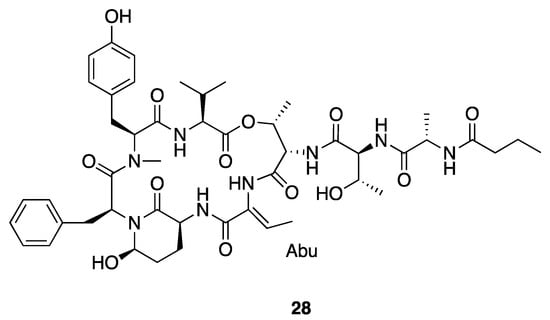
Figure 7.
Molassamide from Leptolyngbya sp.
2.2. Simple Esters
Lumyong et al. isolated an antibacterial compound, 2-hydroxyethyl-11-hydroxyhexadec-9-enoate (29) (Figure 8), from Leptolyngbya sp. LT19 [41], as a result of screening for antibacterial activities. They showed compound 25 to be active against Vibrio harveyi and V. parahaemolyticus, with MIC values of 250–1000 and 350–1000 µg/mL. This antibacterial activity could potentially be useful to the shrimp aquaculture industry that is often burdened by the highly damaging Vibrio spp. pathogens [41]. However, the absolute configuration of the single stereocenter in compound 25 has not yet been determined.
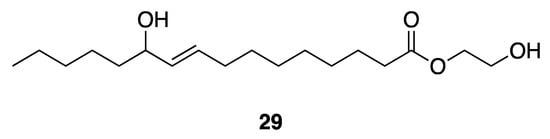
Figure 8.
2-Hydroxyethyl-11-hydroxyhexadec-9-enoate from Leptolyngbya sp. LT19.
Choi et al. isolated honaucins A–C (30–32) from a Hawaiian collection of Leptolyngbya crossbyana which possess potent anti-inflammatory and quorum-sensing (QS) inhibitory activity (Figure 9) [42]. Chemical synthesis of honaucin and a number of analogs (30–32) revealed that the each of the functional groups is critical for both of these biological activities. Further, synthetic honaucin analogues 4-bromo-honaucin A (33) and 4′-iodohonaucin A (35) were discovered to have slightly more potent activity in cellular TRAP activity than honaucin A itself (30), with IC50 values of 0.54 and 0.61 μg/mL compared to 0.63 μg/mL for 30 (Figure 9), [43]. Mechanistic pharmacological investigations of honaucin A (30) indicated that the molecular target(s) involves the Nrf2-ARE (Antioxidant Response Element) pathway, and specifically involves interaction with Cys residues on Keap1 when it is complexed with Nrf2. This allows Nrf2 to be transported to the nucleus, where it activates cytoprotective genes and generates an anti-inflammatory response [64]. Additionally, further investigation of bromo-honaucin A (33) revealed that it provides a protective effect against bone loss in RANKL-treated murine monocyte/macrophage RAW264.7 cells [43].
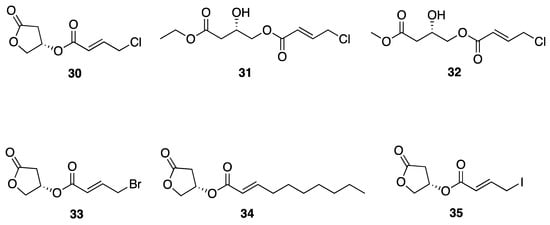
Figure 9.
Honaucins A–C from Leptolyngbya crossbyana and some synthetic derivatives.
2.3. Macrolides
Leptolyngbyolides A–D (36–39), a series of 22-membered macrolides, were isolated from Leptolyngbya sp. collected in Okinawa, Japan. The absolute configuration of each was assigned following an asymmetric total synthesis of leptolyngbyolide C (38) (Figure 10) [44]. Leptolyngbyolides A–D (36–39) were screened for cytotoxic activity against HeLa S3 cells in vitro and were found to be moderately active, with IC50 values of 0.99, 0.16, 0.64 and 0.15 µM, respectively [44]. Furthermore, these metabolites possessed actin-depolymerizing activity with an EC50 of 12.6, 11.6, 26.9 and 21.5 µM, and this may represent the mechanism for the observed cellular apoptosis caused by these compounds [44].
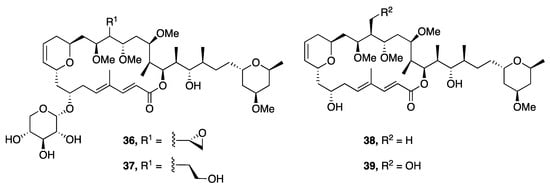
Figure 10.
Leptolyngbyolides from Leptolyngbya sp.
A new macrolide, palmyrolide A (40), was isolated from an environmental assemblage of a Leptolyngbya cf. and Oscillatoria spp. collected from Palmyra Atoll in the Central Pacific Ocean (Figure 11). Palmyrolide A (40) comprises a rare N-methyl enamide functionality, as well as an intriguing t-butyl branch that likely results biosynthetically from the incorporation of malonate and three methyl groups from S-adenosyl-l-methionine (SAM), as shown for apratoxin [45,65]. The absolute configuration of this molecule was assigned by total synthesis of both the natural (−)-palmyrolide A (40) and its enantiomer, (+)-ent-palmyrolide A. This was necessitated because the t-butyl substituent apparently provides the lactone ester bond with resistance to hydrolysis, precluding chemical degradation studies [66,67,68]. It was speculated that cyanobacterial secondary metabolites possessing this motif, a t-butyl adjacent to an ester, might be stable under a wide variety of environmental conditions, similar to what was observed in the laboratory environment. Palmyrolide A (40) exhibited potent inhibition of calcium oscillations in murine cerebrocortical neurons and sodium channel-blocking activity in neuroblastoma (neuro2a) cells [45].
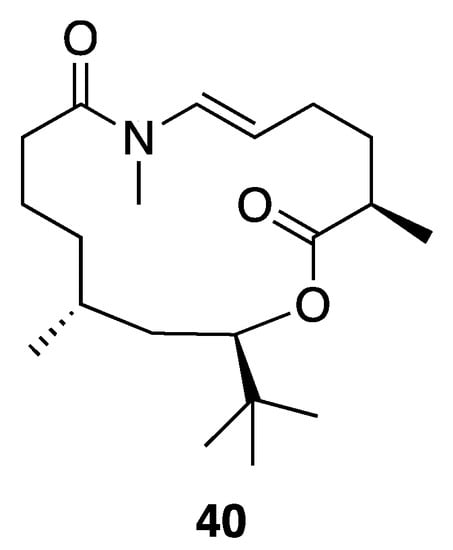
Figure 11.
Palmyrolide A (40) from Leptolyngbya cf. sp.
Phormidolide (41) is a 16-membered macrocyclic lactone polyketide-derived metabolite that was discovered from an Indonesian Leptolyngbya sp. (ISB3NOV94-8A) (Figure 12). This molecule has several unique structural features including a large number of hydroxy and methyl groups on the carbon backbone, several points of both cis and trans unsaturation, and a vinyl bromide at one terminus. Phormidolide (41) showed in vitro brine shrimp toxicity, with an LC50 of 1.5 µM. Multiple NMR experiments, including GHMBC, 2D INADEQUATE, and ACCORD-ADEQUATE, G-BIRDR, X-HSQMBC NMR experiments, enabled a J-based configuration analysis and deduction of relative configuration of stereocenters; a variable temperature Mosher ester analysis was used to assign the absolute configuration [46]. An investigation of the biosynthetic pathway for phormidolide (41) used a genome sequencing approach, and identified the phormidolide biosynthetic gene cluster (phm). The phormidolide (41) gene cluster was found to be of the trans-AT PKS type, which has been relatively rarely reported in cyanobacteria. This was based on finding two discrete trans-AT open reading frames along with KS-AT adaptor regions (ATd) within the PKS megasynthase. The megasynthase possesses ketosynthases, ketoreductases, KS-AT adaptor regions, dehydratases, methyltransferases, O-methyltransferase, enoyl-CoA hydratases, an FkbH-like domain, a pyran synthase, an NRPS-like condensation domain and an acyl carrier protein that were consistent with structure of phormidolide (41). The biosynthetic pathway also provided further supporting evidence for the absolute configuration of phormidolide (41), due to the stereospecificity of the ketoreductases observed in phm compared with known homologues [26]. However, subsequent chemical synthesis of key fragments of phormidolide revealed the need for a revision in configuration in at least one stereocenter [69]. Simultaneously, a reanalysis of the biosynthetic gene cluster suggested additional revisions in configuration were possibly required [70]. These findings stimulated a concerted computational and NMR-based re-investigation of phormidolide’s complex 3-dimensional structure, leading to a revision in several stereocenters (42) [71].
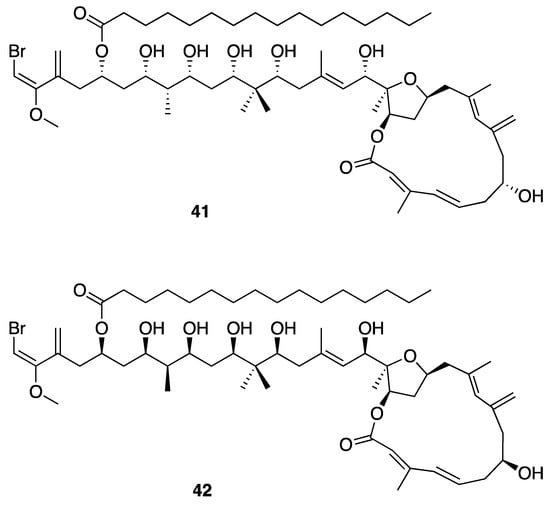
Figure 12.
Phormidolide from Leptolyngbya sp. with the original proposed stereostructure (41) and revised stereostructure (42).
2.4. Pyrones
Kalkipyrone A (43) was first reported in a mixed assemblage of Lyngbya majuscula and Tolypothrix sp. (Figure 13), and was found to have potent brine shrimp toxicity (LD50 = 1 µg/mL) and ichthyotoxicity against Carassius auratus goldfish (LD50 = 2 µg/mL) [48]. Kalkipyrone A (43) and its analogue kalkipyrone B (44) were later found as metabolites of a Leptolyngbya sp. (ASG15JUL14-6) collected from America Samoa (Figure 12) [47]. Kalkipyrone A (43) and B (44) were moderately toxic to a Saccharomyces cerevisiae strain lacking 16 ATP-binding cassette transporter pump genes (ABC16-monster strain; IC50 = 14.6 and 13.4 µM, respectively), while the in vitro cytotoxicity of these two natural products against H460 human lung cancer cells was somewhat more potent (EC50 = 0.9 and 9.0 µM, respectively) [47].
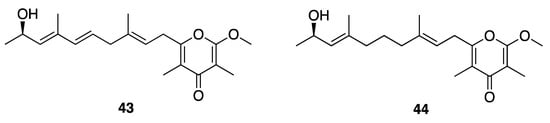
Figure 13.
Kalkpyrones from Leptolyngbya sp.
Three related γ-pyrone-containing polyketides described as yoshinones A, B1, and B2 (45–47), were isolated from Leptolyngbya sp. collected from Ishigaki island Okinawa, Japan (Figure 14). The absolute configuration of each compound was attempted by a modified Mosher’s method; however, these failed possibly as a result of the low amounts of samples available [49]. Thus, to assign the absolute configuration and provide sufficient amounts for further research, the absolute stereochemistry was achieved through total synthesis and comparison of NMR and chiroptical properties between the natural product and synthetic standards [72]. Yoshinone A (45) was found to inhibit adipogenic differentiation against 3T3-L1 cells, with an EC50 of 420 nM and with little cytotoxicity (IC50 = 63.8 µM to S. cerevisiae ABC16-monster cell) [47,49]. The adipogenic differentiation against 3T3-L1 cells of yoshinone B1 (46) and B2 (47) was considerably less potent, with less than 50% activity observed at tested concentrations up to 5 µM [48]. Further examination of structure–activity relationships in this drug class indicated that the position of the pyrone ring and side chain olefin are important for the inhibition of adipogenic differentiation. Further in vitro and in vivo experiments showed that yoshinone A (45) stimulates lactate accumulation deriving from the glycolytic system, and increases fat utilization to compensate for an insufficient energy supply [73]. These properties could possibly support the utility of yoshinone A in anti-obesity indications.
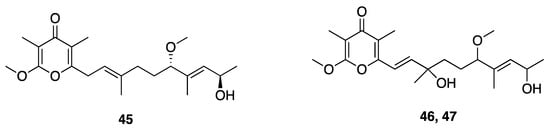
Figure 14.
Yoshinones from Leptolyngbya sp.
2.5. Polyaromatics
A series of brominated polyphenolics, crossbyanols A–D (48–51), were isolated from an extensive benthic Leptolyngbya crossbyana bloom in Hawaii (Figure 15) [50]. In addition to the high level of bromination, crossbyanol B–D (49–51) also have sulfated phenolic functionalities. These metabolites were suggested to play a role in the observed coral toxicity caused by the overgrowing cyanobacteria. Crossbyanol A (48) was found to activate sodium influx in mouse neuroblastoma (neuro2a) cells, with an EC50 20 µg/mL, whereas crossbyanol B (49) possessed antibiotic activity, with an MIC of 2.0-3.9 µg/mL against methicillin-resistant Staphylococcus aureus (MRSA). The latter metabolite also showed moderately potent brine shrimp toxicity (IC50 2.8 µg/mL). Crossbyanols C (50) and D (51) were not observed to have biological activity in these tests, and all four compounds were inactive as cytotoxins to H460 human lung cancer cells [50].
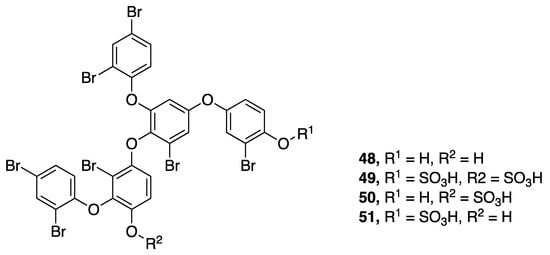
Figure 15.
Crossbyanols from Leptolyngbya crossbyana.
2.6. Oxazolines
A series of polar oxazolines, named leptazolines A–D (52–55), were isolated from the culture media of a Leptolyngbya sp. (Figure 16) [51]. Their planar structures were characterized by MS and NMR along with formation of acetate derivatives. Relative configuration was determined by comparison of carbon shifts with those calculated by density functional theory (DFT). Interestingly, the calculations were found to vary as a function of the computer operating system (Ubuntu 16, Windows 10, MAC Mavericks, MAC Mojave). Biological assay showed that leptazoline B (53) modestly inhibited the growth of PANC-1 cells, with a GI50 of 10 µM [51], whereas leptazoline A (52) with its aromatic chlorine atom did not show any significant activity to this cell line.
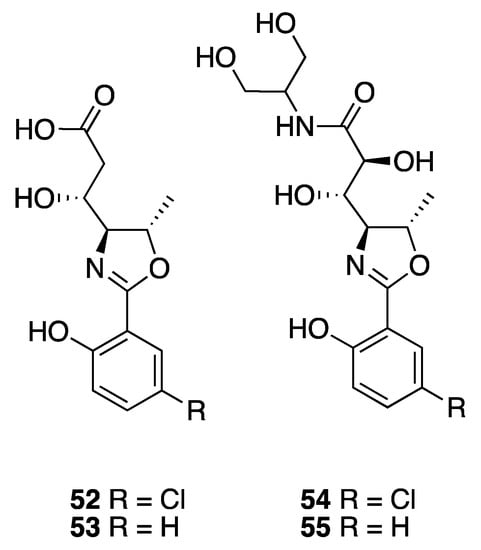
Figure 16.
Leptazolines from Leptolyngbya sp.
2.7. Other
2.7.1. Toxins
Cyanobacteria are known to produce a variety of toxins, including those that are hepatotoxic, neurotoxic, or cardiotoxic, and which generally increase economic burdens and impact public health [23]. Cyanobacterial populations are known to sporadically grow excessively to form blooms, and in some cases these are harmful. A total of 34 species from 15 genera and five families were screened for known toxins including the neurotoxic saxitoxin (56) and the hepatotoxic microcystins (e.g., microcystin-LR, 57) (Figure 17) [17,23,74]. Leptolyngbya collections from the Red Sea had on average 58.9 μg/g dry wt. and 438–489 μg/g dry wt. of these two toxin classes, respectively. Toxin production by marine Leptolyngbya poses toxicological risks to marine organisms that may feed on them, or that may be exposed to the cyanotoxins present in seawater [74].
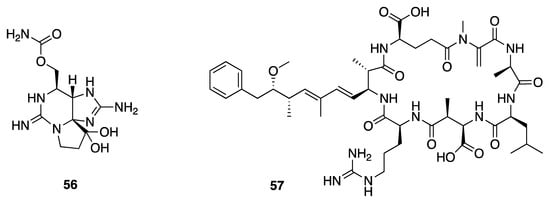
Figure 17.
Two toxins, saxitoxin (56) and microcystin-LR (57), isolated from Leptolyngbya sp.
2.7.2. Non-Toxic Metabolites
Non-toxic secondary metabolites from cyanobacteria include various chemical classes such as phytohormones, siderophores, and UV-absorbing compounds such as mycosporine amino acids (MAAs) and scytonemin (58); all of these have been reported in Leptolyngbya sp. (Figure 18) [23,75,76]. These latter two series of compounds have been shown to protect photosynthetic cyanobacteria from solar UV damage [23]. An investigation of UV-B photoprotective compounds in marine Leptolyngbya discovered shinorine (59) (Figure 18), which is now realized to be one of the most dominant MAAs present in several species of cyanobacteria [75]. Scytonemin has a broader absorption profile than the MAAs, protecting against the solar irradiance damage across the UV (UV-A, -B and -C; 250–425 nm). Interestingly, scytonemin also shows interesting anti-inflammatory activity through inhibition of polo-like kinase stimulated cell proliferation pathways [77]. The biosynthesis of scytonemin, deriving from the assembly of tyrosine and tryptophan derived components, has been studied at the genomic and mechanistic level in several studies; however, all of these studies have been conducted in other genera of cyanobacteria such as Nostoc punctiforme ATCC 29133 and Lyngbya aestuarii [78,79,80,81].
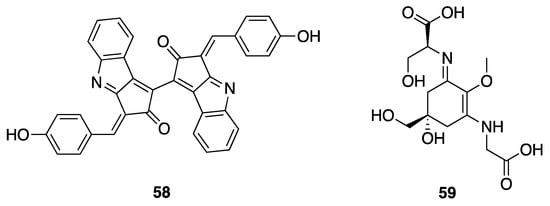
Figure 18.
Scytonemin (58) and the MAA shinorine (59) from Leptolyngbya sp.
2.7.3. Phenolic Compounds
Phenolic compounds, including flavonoids and lignans, are typically natural antioxidants as well as an important group of bioactive compounds [82]. The extract from a thermophilic cyanobacterium Leptolyngbya sp. collected from northern Tunisia was screened by HPLC and showed the presence of 25 phenolic compounds—gallic acid (60), hydroxytyrosol (61), protocatechuic acid (62), vanillic acid (63), isovanillic acid (64), 3-hydroxybenzoic acid (3-HBA) (65), 4-hydroxybenzoic acid (4-HBA) (66), resorcinol (67), naphtoresorcinol (68), syringic acid (69), catechol (70), and oleuropein (71) (Figure 19); chlorogenic acid (72), dihyrdro-p-coumaric acid (73), dihyrdro-m-coumaric acid (74), ferulic acid (75), and rosamerinic acids (76) (Figure 20); catechin (77), luteolin-7-glucoside (78), apigenin-7-glucoside (79), flavone (80), naringenin (81), luteolin (82), and apigenin (83) (Figure 21); resveratrol (84) and pinoresinol (85) (Figure 22)—demonstrating that Leptolyngbya may constitute a rich source of antioxidant natural products [83,84]. These compounds also have inherent UV-absorbing properties, albeit at more restricted wavelengths than scytonemin (58).
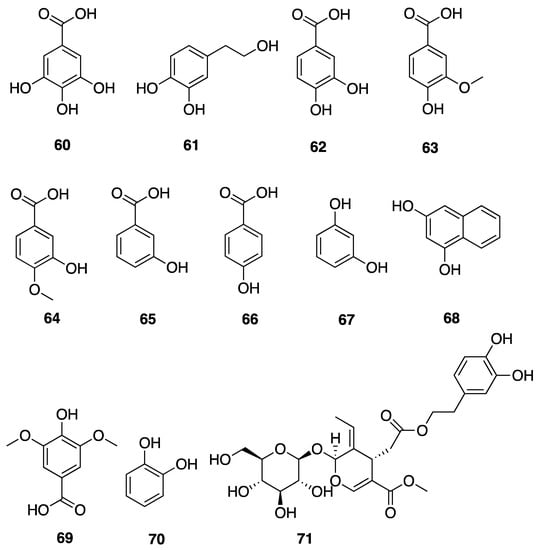
Figure 19.
Hydroxybenzoic acids (HBAs) from Leptolyngbya sp.
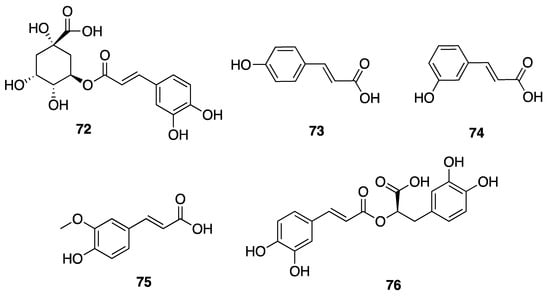
Figure 20.
Hydroxycinnamic acids (HCAs) from Leptolyngbya sp.
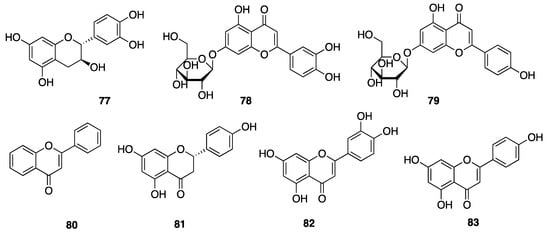
Figure 21.
Flavonoids from Leptolyngbya sp.
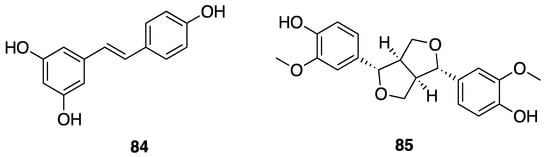
Figure 22.
Stilbene and a lignan from Leptolyngbya sp.
2.7.4. Odorous Metabolites
Geosmin (86) and 2-methylisoborneol (87) are two earthy-musty odorous terpenoid secondary metabolites that were first isolated from actinomycetes (Figure 23). Later, these same molecules were found to have a significant presence in many cyanobacteria species. These simple terpenoids have each been reported numerous times as being among the main causes for off-flavors in water and other products [85]. Wang et al. isolated 86 and 87 from Leptolyngbya bijugata strains, and quantified each at 13.6–22.4 and 12.3–57.5 μg/L, respectively, demonstrating their production by Leptolyngbya [86].
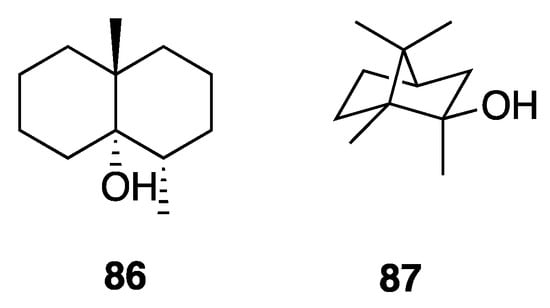
Figure 23.
Geosmin and 2-methylisoborneol (MIB) from Leptolyngbya bijugata.
2.7.5. Pigments
Phycocyanin is a pigment–protein complex found in cyanobacteria and eukaryotic algae that functions as a light-harvesting pigment. It is widely used in biotechnological, food and pharmaceutical industries [87]. Schipper et al. analyzed the phycocyanin content in Leptolyngbya sp. QUCCCM 56 from a desert environment to reveal that it possesses higher and purer phycocyanin compared to the current commercial source, Arthrospira platensis [87]. Other than absorbing light, phycocyanin also shows potential anti-aging and proteostasis-suppressive activities. With phycocyanin treatment, the life span of wild-type (N2) C. elegans was extended from 14.8 to 19.1 days [88].
3. Laboratory Cultivation
Despite many attempts, only a few environmental collections of Leptolyngbya have been propagated under laboratory culture conditions and, from these, there has been a relatively low rate of natural product isolation. Rather, most of the compounds reported in this genus have been discovered from environmental samples (Table 1). Martins et al. screened five Leptolyngbya strains among 28 cyanobacteria samples collected from the Portuguese Coast and cultured in Z8 medium, and showed cytotoxic activities of the extracts against multiple human tumor cell lines [15,16,89]. The most commonly used media for cyanobacteria culture is BG11 [53], originally formulated in 1988 to possess synthetic sea salt, microelements mixture, deionized water and vitamin mixtures [28]. In most cases, even with suitable media and abiotic factors, filamentous cyanobacteria are slow-growing life forms, normally with growth rates much slower than algae and other bacteria, thereby presenting a challenge for secondary metabolite discovery due to the small quantities of biomass produced in cultures [28,90]. The 2-hydroxyethyl-11-hydroxyhexadec-9-enoate, by contrast, was able to be isolated from the laboratory culture of Leptolyngbya with the relatively small biomass of 383.6 g because of its reduced structural complexity and higher production yield [41].
4. Conclusions
Leptolyngbya is a widely distributed genus of cyanobacteria that has emerged to be a very rich source of structurally novel and biologically active natural products. However, to date, this genus appears to be underexplored for its chemical, biological and biosynthetic potential when compared to some other genera of filamentous cyanobacteria, such as Moorena and Symploca [90]. Working with Leptolyngbya has been challenging due to the difficulty in bringing it into culture in the laboratory environment. This has impeded not only new compound discovery, but also exploration of its genomic characteristics, including those that are responsible for natural product biosynthesis. It is possible that developments with the heterologous expression of cyanobacterial natural product pathways will enable a more extensive exploration of the rich secondary metabolome of this genus in the future [91,92].
Author Contributions
Conceptualization, Y.L. and W.H.G.; formal analysis, Y.L. and W.H.G.; investigation, Y.L. and W.H.G.; resources, Y.L. and W.H.G.; data curation, Y.L. and W.H.G.; writing—original draft preparation, Y.L.; writing—review and editing, Y.L., C.B.N., K.L.A., H.G., W.H.G; visualization, Y.L. and W.H.G.; supervision, H.G. and W.H.G.; project administration, W.H.G; funding acquisition, W.H.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by NIH grant GM107550 (to W.H.G).
Acknowledgments
Y.L. thanks financial support provided by the fellowship from the Ocean University of China. We thank E. Glukhov for providing the photomicrographs of Leptolyngbya in culture.
Conflicts of Interest
William H. Gerwick declares a competing financial interest as a cofounder of NMR Finder LLC.
References
- Gademann, K.; Portmann, C. Secondary metabolites from cyanobacteria: Complex structures and powerful bioactivities. Curr. Org. Chem. 2008, 12, 326–341. [Google Scholar] [CrossRef]
- Summons, R.E.; Jahnke, L.L.; Hope, J.M.; Logan, G.A. 2-Methylhopanoids as biomarkers forcyanobacterial oxygenic photosynthesis. Nature 1999, 400, 554–557. [Google Scholar] [CrossRef] [PubMed]
- Blankenship, R.E.; Hartman, H. The origin and evolution of oxygenic photosynthesis. Trends Biochem. Sci. 1998, 23, 94–97. [Google Scholar] [CrossRef]
- Berman-Frank, I.; Lundgren, P.; Falkowski, P. Nitrogen fixation and photosynthetic oxygen evolution in cyanobacteria. Res. Microbiol. 2003, 154, 157–164. [Google Scholar] [CrossRef]
- Kim, J.H.; Choi, W.; Jeon, S.M.; Kim, T.; Park, A.; Kim, J.; Heo, S.J.; Oh, C.; Shim, W.B.; Kang, D.H. Isolation and characterization of Leptolyngbya sp. KIOST-1, a basophilic and euryhaline filamentous cyanobacterium from an open paddle-wheel raceway Arthrospira culture pond in Korea. J. Appl. Microbiol. 2015, 119, 1597–1612. [Google Scholar] [CrossRef]
- Mai, T.; Johansen, J.R.; Pietrasiak, N.; Bohunická, M.; Martin, M.P. Revision of the Synechococcales (Cyanobacteria) through recognition of four families including Oculatellaceae fam. nov. and Trichocoleaceae fam. nov. and six new genera containing 14 species. Phytotaxa 2018, 365, 1–59. [Google Scholar] [CrossRef]
- Bruno, L.; Billi, D.; Bellezza, S.; Albertano, P. Cytomorphological and genetic characterization of troglobitic Leptolyngbya strains isolated from roman hypogea. Appl. Environ. Microbiol. 2009, 75, 608–617. [Google Scholar] [CrossRef]
- Anagnostidis, K.; Komárek, J. Modern approach to the classification system of cyanophytes. 3—Oscillatoriales. Algol. Stud. /Arch. Hydrobiol. Suppl. Vol. 1988, 50–53, 327–472. [Google Scholar]
- Komárek, J.; Kaštovský, J.; Mareš, J.; Johansen, J.R. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia 2014, 86, 295–335. [Google Scholar]
- Cirés, S.; Casero, M.C.; Quesada, A. Toxicity at the edge of life: A review on cyanobacterial toxins from extreme environments. Mar. Drugs 2017, 15, 233. [Google Scholar] [CrossRef]
- Ahmed, M.; Stal, L.J.; Hasnain, S. The morphology and bioactivity of the rice field cyanobacterium Leptolyngbya. Rev. Biol. Trop. 2014, 62, 1251–1260. [Google Scholar] [PubMed]
- Veerabadhran, M.; Chakraborty, S.; Mitra, S.; Karmakar, S.; Mukherjee, J. Effects of flask configuration on biofilm growth and metabolites of intertidal cyanobacteria isolated from a mangrove forest. J. Appl. Microbiol. 2018, 125, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, Z.A.; Al-Shehri, A.M. Biodiversity and toxin production of cyanobacteria in mangrove swamps in the Red Sea off the southern coast of Saudi Arabia. Bot. Mar. 2015, 58, 23–34. [Google Scholar] [CrossRef]
- Yuu, H.; Fujisawa, T.; Ohtsubo, Y.; Katayama, M.; Misawa, N.; Wakazuki, S.; Shimura, Y.; Nakamura, Y.; Kawachi, M.; Yoshikawa, H.; et al. Complete genome sequence of cyanobacterium Leptolyngbya sp. NIES-3755. Am. Soc. Microbiol. 2016, 4, e0009-16. [Google Scholar]
- Brito, Â.; Ramos, V.; Seabra, R.; Santos, A.; Santos, C.L.; Lopo, M.; Ferreira, S.; Martins, A.; Mota, R.; Frazão, B.; et al. Culture-dependent characterization of cyanobacterial diversity in the intertidal zones of the Portuguese coast: A polyphasic study. Syst. Appl. Microbiol. 2012, 35, 110–119. [Google Scholar] [CrossRef]
- Costa, M.; Garcia, M.; Costa-Rpdrigues, J.; Costa, M.S.; Ribeiro, M.J.; Fernandes, M.H.; Barros, P.; Barreiro, A.; Vasconcelos, V.; Martins, R. Exploring bioactive properties of marine cyanobacteria isolated from the Portuguese Coast: High potential as a source of anticancer compounds. Mar. Drugs 2014, 12, 98–114. [Google Scholar] [CrossRef]
- Semary, N.A. El Microscopic, molecular, and biochemical investigations to characterize a benthic cyanoprokaryote Leptolyngbya strain from Egypt. Microsc. Res. Tech. 2013, 76, 249–257. [Google Scholar] [CrossRef]
- Guiry, M.D.; Guiry, G.M. AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. Available online: https://www.algaebase.org (accessed on 3 October 2020).
- Komárek, J.; Hauer, T. The On-line Database of Cyanobacterial Genera. Available online: http://www.cyanodb.cz (accessed on 3 October 2020).
- Singh, D.P.; Khattar, J.I.S.; Gupta, M.; Kaur, G. Evaluation of toxicological impact of cartap hydrochloride on some physiological activities of a non-heterocystous cyanobacterium Leptolyngbya foveolarum. Pestic. Biochem. Physiol. 2014, 110, 63–70. [Google Scholar] [CrossRef]
- Pramanik, A.; Sundararaman, M.; Das, S.; Ghosh, U.; Mukherjee, J. Isolation and characterization of cyanobacteria possessing antimicrobial activity from the sundarbans, the world’s largest tidal mangrove forest. J. Phycol. 2011, 47, 731–743. [Google Scholar] [CrossRef]
- Xue, Y.; Zhao, P.; Quan, C.; Zhao, Z.; Gao, W.; Li, J.; Zu, X.; Fu, D.; Feng, S.; Bai, X.; et al. Cyanobacteria-derived peptide antibiotics discovered since 2000. Peptides 2018, 107, 17–24. [Google Scholar] [CrossRef]
- Haque, F.; Banayan, S.; Yee, J.; Chiang, Y.W. Extraction and applications of cyanotoxins and other cyanobacterial secondary metabolites. Chemosphere 2017, 183, 164–175. [Google Scholar] [CrossRef]
- Vijayakumar, S.; Menakha, M. Pharmaceutical applications of cyanobacteria-A review. J. Acute Med. 2015, 5, 15–23. [Google Scholar] [CrossRef]
- Engene, N.; Gunasekera, S.P.; Gerwick, W.H.; Paul, V.J. Phylogenetic inferences reveal a large extent of novel biodiversity in chemically rich tropical marine cyanobacteria. Appl. Environ. Microbiol. 2013, 79, 1882–1888. [Google Scholar] [CrossRef]
- Bertin, M.J.; Vulpanovici, A.; Monroe, E.A.; Korobeynikov, A.; Sherman, D.H.; Gerwick, L.; Gerwick, W.H. The Phormidolide biosynthetic gene cluster: A trans-AT PKS pathway encoding a toxic macrocyclic polyketide. ChemBioChem 2016, 17, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Castenholz, R.W. Culturing methods for cyanobacteria. Methods Enzymol. 1988, 167, 68–93. [Google Scholar]
- Moss, N.A.; Leao, T.; Glukhov, E.; Gerwick, L.; Gerwick, W.H. Collection, Culturing, and Genome Analyses of Tropical Marine Filamentous Benthic Cyanobacteria, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2018; Volume 604, ISBN 9780128139592. [Google Scholar]
- Leão, P.N.; Ramos, V.; Goncalves, P.B.; Viana, F.; Lage, O.M.; Gerwick, W.H.; Vasconcelos, V.M. Chemoecological screening reveals high bioactivity in diverse culturable portuguese marine cyanobacteria. Mar. Drugs 2013, 11, 1316–1335. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Pan, Z.; Wu, C.; Wang, W.; Fang, L.; Su, W. Efficient synthesis and stereochemical revision of Coibamide, A. J. Am. Chem. Soc. 2015, 137, 13488–13491. [Google Scholar] [CrossRef]
- Yao, G.; Wang, W.; Ao, L.; Cheng, Z.; Wu, C.; Pan, Z.; Liu, K.; Li, H.; Su, W.; Fang, L. Improved total synthesis and biological evaluation of coibamide A analogues. J. Med. Chem. 2018, 61, 8908–8916. [Google Scholar] [CrossRef]
- Medina, R.A.; Goeger, D.E.; Hills, P.; Mooberry, S.L.; Huang, N.; Romero, L.I.; Ortega-Barría, E.; Gerwick, W.H.; McPhail, K.L. Coibamide A, a potent antiproliferative cyclic depsipeptide from the panamanian marine cyanobacterium Leptolyngbya sp. J. Am. Chem. Soc. 2008, 130, 6324–6325. [Google Scholar] [CrossRef]
- Sable, G.A.; Park, J.; Kim, H.; Lim, S.J.; Jang, S.; Lim, D. Solid-Phase Total synthesis of the proposed structure of coibamide A and its derivative: Highly methylated cyclic depsipeptides. Eur. J. Org. Chem. 2015, 2015, 7043–7052. [Google Scholar] [CrossRef]
- He, W.; Qiu, H.B.; Chen, Y.J.; Xi, J.; Yao, Z.J. Total synthesis of proposed structure of coibamide A, a highly N- and O-methylated cytotoxic marine cyclodepsipeptide. Tetrahedron Lett. 2014, 55, 6109–6112. [Google Scholar] [CrossRef]
- Nabika, R.; Suyama, T.L.; Hau, A.M.; Misu, R.; Ohno, H.; Ishmael, J.E.; McPhail, K.L.; Oishi, S.; Fujii, N. Synthesis and biological evaluation of the [d-MeAla11]-epimer of coibamide A. Bioorg. Med. Chem. Lett. 2015, 25, 302–306. [Google Scholar] [CrossRef] [PubMed]
- Thornburg, C.C.; Thimmaiah, M.; Shaala, L.A.; Hau, A.M.; Malmo, J.M.; Ishmael, J.E.; Youssef, D.T.A.; McPhail, K.L. Cyclic depsipeptides, grassypeptolides D and E and Ibu- epidemethoxylyngbyastatin 3, from a Red Sea Leptolyngbya cyanobacterium. J. Nat. Prod. 2011, 74, 1677–1685. [Google Scholar] [CrossRef]
- Harrigan, G.G.; Yoshida, W.Y.; Moore, R.E.; Nagle, D.G.; Park, P.U.; Biggs, J.; Paul, V.J.; Mooberry, S.L.; Corbett, T.H.; Valeriote, F.A. Isolation, structure determination, and biological activity of dolastatin 12 and lyngbyastatin 1 from Lyngbya majuscula/Schizothrix calcicola cyanobacterial assemblages. J. Nat. Prod. 1998, 61, 1221–1225. [Google Scholar] [CrossRef] [PubMed]
- Pettit, G.R.; Kamano, Y.; Kizu, H.; Dufresne, C.; Herald, C.L.; Bontems, R.J.; Schmidt, J.M.; Boettner, F.E.; Nieman, R.A. Isolation and structure of the cell growth inhibitory depsipeptides dolastatins 11 and 12. Heterocycles 1989, 28, 3–8. [Google Scholar] [CrossRef]
- Bates, R.B.; Brusoe, K.G.; Burns, J.J.; Caldera, S.; Cui, W.; Gangwar, S.; Gramme, M.R.; McClure, K.J.; Rouen, G.P.; Schadow, H.; et al. Dolastatins. 26. synthesis and stereochemistry of dolastatin 11. J. Am. Chem. Soc. 1997, 119, 2111–2113. [Google Scholar] [CrossRef]
- Al-Awadhi, F.H.; Paul, V.J.; Luesch, H. Structural diversity and anticancer activity of marine-derived elastase inhibitors: Key features and mechanisms mediating the antimetastatic effects in invasive breast cancer. ChemBioChem 2018, 19, 815–825. [Google Scholar] [CrossRef]
- Maneechote, N.; Yingyongnarongkul, B.E.; Suksamran, A.; Lumyong, S. Inhibition of Vibrio spp. by 2-Hydroxyethyl-11-hydroxyhexadec-9-enoate of marine cyanobacterium Leptolyngbya sp. LT19. Aquac. Res. 2017, 48, 2088–2095. [Google Scholar] [CrossRef]
- Choi, H.; Mascuch, S.J.; Villa, F.A.; Byrum, T.; Teasdale, M.E.; Smith, J.E.; Preskitt, L.B.; Rowley, D.C.; Gerwick, L.; Gerwick, W.H. Honaucins A-C, potent inhibitors of inflammation and bacterial quorum sensing: Synthetic derivatives and structure-activity relationships. Chem. Biol. 2012, 19, 589–598. [Google Scholar] [CrossRef]
- Sapkota, M.; Li, L.; Choi, H.; Gerwick, W.H.; Soh, Y. Bromo-honaucin A inhibits osteoclastogenic differentiation in RAW 264.7 cells via Akt and ERK signaling pathways. Eur. J. Pharmacol. 2015, 769, 100–109. [Google Scholar] [CrossRef]
- Cui, J.; Morita, M.; Ohno, O.; Kimura, T.; Teruya, T.; Watanabe, T.; Suenaga, K.; Shibasaki, M. Leptolyngbyolides, cytotoxic macrolides from the marine cyanobacterium Leptolyngbya sp.: Isolation, biological activity, and catalytic asymmetric total synthesis. Chem. A Eur. J. 2017, 23, 8500–8509. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.R.; Cao, Z.; Engene, N.; Soria-Mercado, I.E.; Murray, T.F.; Gerwick, W.H. Palmyrolide A, an unusually stabilized neuroactive macrolide from palmyra atoll cyanobacteria. Org. Lett. 2010, 12, 4490–4493. [Google Scholar] [CrossRef] [PubMed]
- Williamson, R.T.; Boulanger, A.; Vulpanovici, A.; Roberts, M.A.; Gerwick, W.H. Structure and absolute stereochemistry of phormidolide, a new toxic metabolite from the marine cyanobacterium Phormidium sp. J. Org. Chem. 2002, 67, 7927–7936. [Google Scholar] [CrossRef] [PubMed]
- Bertin, M.J.; Demirkiran, O.; Navarro, G.; Moss, N.A.; Lee, J.; Goldgof, G.M.; Vigil, E.; Winzeler, E.A.; Valeriote, F.A.; Gerwick, W.H. Kalkipyrone B, a marine cyanobacterial γ-pyrone possessing cytotoxic and anti-fungal activities. Phytochemistry 2016, 122, 113–118. [Google Scholar] [CrossRef]
- Graber, M.A.; Gerwick, W.H. Kalkipyrone, a toxic γ-pyrone from an assemblage of the marine cyanobacteria Lyngbya majuscula and Tolypothrix sp. J. Nat. Prod. 1998, 61, 677–680. [Google Scholar] [CrossRef] [PubMed]
- Inuzuka, T.; Yamamoto, K.; Iwasaki, A.; Ohno, O.; Suenaga, K.; Kawazoe, Y.; Uemura, D. An inhibitor of the adipogenic differentiation of 3T3-L1 cells, yoshinone A, and its analogs, isolated from the marine cyanobacterium Leptolyngbya sp. Tetrahedron Lett. 2014, 55, 6711–6714. [Google Scholar] [CrossRef]
- Choi, H.; Engene, N.; Smith, J.E.; Preskitt, L.B.; Gerwick, W.H. Crossbyanols A-D, toxic brominated polyphenyl ethers from the hawai’ian bloom-forming cyanobacterium Leptolyngbya crossbyana. J. Nat. Prod. 2010, 73, 517–522. [Google Scholar] [CrossRef]
- Bhandari Neupane, J.; Neupane, R.P.; Luo, Y.; Yoshida, W.Y.; Sun, R.; Williams, P.G. Characterization of leptazolines A-D, polar oxazolines from the cyanobacterium Leptolyngbya sp., reveals a glitch with the “willoughby-hoye” scripts for calculating NMR chemical shifts. Org. Lett. 2019, 21, 8449–8453. [Google Scholar] [CrossRef]
- Hau, A.M.; Greenwood, J.A.; Löhr, C.V.; Serrill, J.D.; Proteau, P.J.; Ganley, I.G.; McPhail, K.L.; Ishmael, J.E. Coibamide A induces mTOR-independent autophagy and cell death in human glioblastoma cells. PLoS ONE 2013, 8, e65250. [Google Scholar] [CrossRef]
- Snyder, K.M.; Sikorska, J.; Ye, T.; Fang, L.; Su, W.; Carter, R.G.; McPhail, K.L.; Cheong, P.H.Y. Towards theory driven structure elucidation of complex natural products: Mandelalides and coibamide A. Org. Biomol. Chem. 2016, 14, 5826–5831. [Google Scholar] [CrossRef]
- Pettit, G.R.; Kamano, Y.; Fujii, Y.; Herald, C.L.; Inoue, M.; Brown, P.; Gust, D.; Kitahara, K.; Schmidt, J.M.; Michael, C.; et al. Marine animal biosynthetic constituents for cancer chemotherapy. J. Nat. Prod. 1981, 44, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.; Pörzgen, P.; Pittman, E.; Yoshida, W.Y.; Westenburg, H.E.; Horgen, F.D. Isolation and structure determination of malevamide E, a dolastatin 14 analogue, from the marine cyanobacterium Symploca laete-viridis. J. Nat. Prod. 2008, 71, 750–754. [Google Scholar] [CrossRef] [PubMed]
- Thornburg, C.C.; Mcphail, K.L. Investigation of Unique Marine Environments for Microbial Natural Products; Oregon State University: Corvallis, OR, USA, 2013. [Google Scholar]
- Williams, P.G.; Moore, R.E.; Paul, V.J. Isolation and structure determination of lyngbyastatin 3, a lyngbyastatin 1 homologue from the marine cyanobacterium Lyngbya majuscula. Determination of the configuration of the 4-Amino-2,2-dimethyl-3-oxopentanoic acid unit in majusculamide C, dolastatin 12, lyngbyastatin 1, and lyngbyastatin 3 from cyanobacteria. J. Nat. Prod. 2003, 66, 1356–1363. [Google Scholar] [PubMed]
- Gerwick, W.H.; Tan, L.T.; Sitachitta, N. Nitrogen-Containing Metabolites Marine Cyanobacteria; Elsevier: Amsterdam, The Netherlands, 2001; Volume 57. [Google Scholar]
- Kwan, J.C.; Ratnayake, R.; Abboud, K.A.; Paul, V.J.; Luesch, H. Grassypeptolides A-C, cytotoxic bis-thiazoline containing marine cyclodepsipeptides. J. Org. Chem. 2010, 75, 8012–8023. [Google Scholar] [CrossRef] [PubMed]
- Kwan, J.C.; Rocca, J.R.; Abboud, K.A.; Paul, V.J.; Luesch, H. Total structure determination of grassypeptolide, a new marine cyanobacterial cytotoxin. Org. Lett. 2008, 10, 789–792. [Google Scholar] [CrossRef]
- Gogineni, V.; Hamann, M.T. Marine natural product peptides with therapeutic potential: Chemistry, biosynthesis, and pharmacology. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 81–196. [Google Scholar] [CrossRef]
- Popplewell, W.L.; Ratnayake, R.; Wilson, J.A.; Beutler, J.A.; Colburn, N.H.; Henrich, C.J.; McMahon, J.B.; McKee, T.C. Grassypeptolides F and G, cyanobacterial peptides from Lyngbya majuscula. J. Nat. Prod. 2011, 74, 1686–1691. [Google Scholar] [CrossRef]
- Kwan, J.C.; Liu, Y.; Ratnayake, R.; Hatano, R.; Kuribara, A.; Morimoto, C.; Ohnuma, K.; Paul, V.J.; Ye, T.; Luesch, H. Grassypeptolides as natural inhibitors of dipeptidyl peptidase 8 and T-cell activation. ChemBioChem 2014, 15, 799–804. [Google Scholar] [CrossRef]
- Mascuch, S.J.; Boudreau, P.D.; Carland, T.M.; Pierce, N.T.; Olson, J.; Hensler, M.E.; Choi, H.; Campanale, J.; Hamdoun, A.; Nizet, V.; et al. Marine natural product honaucin A attenuates inflammation by activating the Nrf2-ARE pathway. J. Nat. Prod. 2018, 81, 506–514. [Google Scholar] [CrossRef]
- Skiba, M.A.; Sikkema, A.P.; Moss, N.A.; Lowell, A.N.; Su, M.; Sturgis, R.M.; Gerwick, L.; Gerwick, W.H.; Sherman, D.H.; Smith, J.L. Biosynthesis of t-butyl in apratoxin A: Functional analysis and architecture of a PKS loading module. ACS Chem. Biol. 2018, 13, 1640–1650. [Google Scholar] [CrossRef]
- Wadsworth, A.D.; Furkert, D.P.; Brimble, M.A. Total synthesis of the macrocyclic N-Methyl enamides palmyrolide A and 2S-sanctolide A. J. Org. Chem. 2014, 79, 11179–11193. [Google Scholar] [CrossRef]
- Tello-Aburto, R.; Johnson, E.M.; Valdez, C.K.; Maio, W.A. Asymmetric total synthesis and absolute stereochemistry of the neuroactive marine macrolide palmyrolide A. Org. Lett. 2012, 14, 2150–2153. [Google Scholar] [CrossRef]
- Tello-Aburto, R.; Newar, T.D.; Maio, W.A. Evolution of a protecting-group-free total synthesis: Studies en route to the neuroactive marine macrolide (-)-palmyrolide A. J. Org. Chem. 2012, 77, 6271–6289. [Google Scholar] [CrossRef]
- Lam, N.Y.S.; Muir, G.; Challa, V.R.; Britton, R.; Paterson, I. A counterintuitive stereochemical outcome from a chelation-controlled vinylmetal aldehyde addition leads to the configurational reassignment of phormidolide A. Chem. Commun. 2019, 55, 9717–9720. [Google Scholar] [CrossRef]
- Helfrich, E.J.N.; Ueoka, R.; Dolev, A.; Rust, M.; Meoded, R.A.; Bhushan, A.; Califano, G.; Costa, R.; Gugger, M.; Steinbeck, C.; et al. Automated structure prediction of trans-acyltransferase polyketide synthase products. Nat. Chem. Biol. 2019, 15, 813–821. [Google Scholar] [CrossRef]
- Ndukwe, I.E.; Wang, X.; Lam, N.Y.S.; Ermanis, K.; Alexander, K.L.; Bertin, M.J.; Martin, G.E.; Muir, G.; Paterson, I.; Britton, R.; et al. Synergism of anisotropic and computational NMR methods reveals the likely configuration of phormidolide A. Chem. Commun. 2020, 56, 7565–7568. [Google Scholar] [CrossRef]
- Shinomiya, S.; Iwasaki, A.; Ohno, O.; Suenaga, K. Total synthesis and stereochemical determination of yoshinone A. Phytochemistry 2016, 132, 109–114. [Google Scholar] [CrossRef]
- Koyama, T.; Kawazoe, Y.; Iwasaki, A.; Ohno, O.; Suenaga, K.; Uemura, D. Anti-obesity activities of the yoshinone A and the related marine γ-pyrone compounds. J. Antibiot. 2016, 69, 348–351. [Google Scholar] [CrossRef]
- Mohamed, Z. Harmful cyanobacteria and their cyanotoxins in Egyptian fresh waters–state of knowledge and research needs. Afr. J. Aquat. Sci. 2016, 41, 361–368. [Google Scholar] [CrossRef]
- Joshi, D.; Mohandass, C.; Dhale, M. Effect of UV-B radiation and desiccation stress on photoprotective compounds accumulation in marine Leptolyngbya sp. Appl. Biochem. Biotechnol. 2018, 184, 35–47. [Google Scholar] [CrossRef]
- Babu, S.V.; Ashokkumar, B.; Sivakumar, N.; Sudhakarsamy, P.; Varalakshmi, P. Indole-3-acetic acid from filamentous cyanobacteria: Screening, strain identification and production. J. Sci. Ind. Res. 2013, 72, 581–584. [Google Scholar]
- Stevenson, C.S.; Capper, E.A.; Roshak, A.K.; Marquez, B.; Eichman, C.; Jackson, J.R.; Mattern, M.; Gerwick, W.H.; Jacobs, R.S.; Marshall, L.A. The identification and characterization of the marine natural product scytonemin as a novel antiproliferative pharmacophore. J. Pharmacol. Exp. Ther. 2002, 303, 858–866. [Google Scholar] [CrossRef] [PubMed]
- Soule, T.; Stout, V.; Swingley, W.D.; Meeks, J.C.; Garcia-Pichel, F. Molecular genetics and genomic analysis of scytonemin biosynthesis in Nostoc punctiforme ATCC 29133. J. Bacteriol. 2007, 189, 4465–4472. [Google Scholar] [CrossRef] [PubMed]
- Balskus, E.P.; Walsh, C.T. Investigating the initial steps in the biosynthesis of cyanobacterial sunscreen scytonemin. J. Am. Chem. Soc. 2008, 130, 15260–15261. [Google Scholar] [CrossRef]
- Sorrels, C.M.; Proteau, P.J.; Gerwick, W.H. Organization, evolution, and expression analysis of the biosynthetic gene cluster for scytonemin, a cyanobacterial UV-absorbing pigment. Appl. Environ. Microbiol. 2009, 75, 4861–4869. [Google Scholar] [CrossRef]
- Jones, C.S.; Esquenazi, E.; Dorrestein, P.C.; Gerwick, W.H. Probing the in vivo biosynthesis of scytonemin, a cyanobacterial ultraviolet radiation sunscreen, through small scale stable isotope incubation studies and MALDI-TOF mass spectrometry. Bioorg. Med. Chem. 2011, 19, 6620–6627. [Google Scholar] [CrossRef]
- Martins, S.; Mussatto, S.I.; Martínez-Avila, G.; Montañez-Saenz, J.; Aguilar, C.N.; Teixeira, J.A. Bioactive phenolic compounds: Production and extraction by solid-state fermentation. A review. Biotechnol. Adv. 2011, 29, 365–373. [Google Scholar] [CrossRef]
- Ljaz, S.; Hasnain, S. Antioxidant potential of indigenous cyanobacterial strains in relation with their phenolic and flavonoid contents. Nat. Prod. Res. 2016, 30, 1297–1300. [Google Scholar]
- Trabelsi, L.; Mnari, A.; Abdel-Daim, M.M.; Abid-Essafi, S.; Aleya, L. Therapeutic properties in Tunisian hot springs: First evidence of phenolic compounds in the cyanobacterium Leptolyngbya sp. biomass, capsular polysaccharides and releasing polysaccharides. BMC Complement. Altern. Med. 2016, 16, 515. [Google Scholar] [CrossRef]
- Smith, J.L.; Boyer, G.L.; Zimba, P.V. A review of cyanobacterial odorous and bioactive metabolites: Impacts and management alternatives in aquaculture. Aquaculture 2008, 280, 5–20. [Google Scholar] [CrossRef]
- Wang, Z.; Xiao, P.; Song, G.; Li, Y.; Li, R. Isolation and characterization of a new reported cyanobacterium Leptolyngbya bijugata coproducing odorous geosmin and 2-methylisoborneol. Environ. Sci. Pollut. Res. 2015, 22, 12133–12140. [Google Scholar] [CrossRef] [PubMed]
- Schipper, K.; Fortunati, F.; Oostlander, P.C.; Al Muraikhi, M.; Al Jabri, H.M.S.J.; Wijffels, R.H.; Barbosa, M.J. Production of phycocyanin by Leptolyngbya sp. in desert environments. Algal Res. 2020, 47, 101875. [Google Scholar] [CrossRef]
- Singh, N.K.; Sonani, R.R.; Awasthi, A.; Prasad, B.; Patel, A.R.; Kumar, J.; Madamwar, D. Phycocyanin moderates aging and proteotoxicity in caenorhabditis elegans. J. Appl. Phycol. 2016, 28, 2407–2417. [Google Scholar] [CrossRef]
- Kim, S.-K. Handbook of Anticancer Drugs from Marine Origin; Springer: Berlin/Heidelberg, Germany, 2015; ISBN 9783319071442. [Google Scholar]
- Dittmann, E.; Gugger, M.; Sivonen, K.; Fewer, D.P. Natural product biosynthetic diversity and comparative genomics of the cyanobacteria. Trends Microbiol. 2015, 23, 642–652. [Google Scholar] [CrossRef]
- Videau, P.; Wells, K.N.; Singh, A.J.; Eiting, J.; Proteau, P.J.; Philmus, B. Expanding the natural products heterologous expression repertoire in the model cyanobacterium Anabaena sp. strain PCC 7120: Production of pendolmycin and teleocidin B-4. ACS Synth. Biol. 2020, 9, 63–75. [Google Scholar] [CrossRef]
- Taton, A.; Ecker, A.; Diaz, B.; Moss, N.A.; Anderson, B.; Leão, T.F.; Simkovsky, R.; Dorrestein, P.C.; Gerwick, L.; William, H. Heterologous expression of cryptomaldamide in a cyanobacterial host. BioRxiv 2020. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).