Characterization of an Alkaline GH49 Dextranase from Marine Bacterium Arthrobacter oxydans KQ11 and Its Application in the Preparation of Isomalto-Oligosaccharide
Abstract
1. Introduction
2. Results
2.1. Gene Cloning
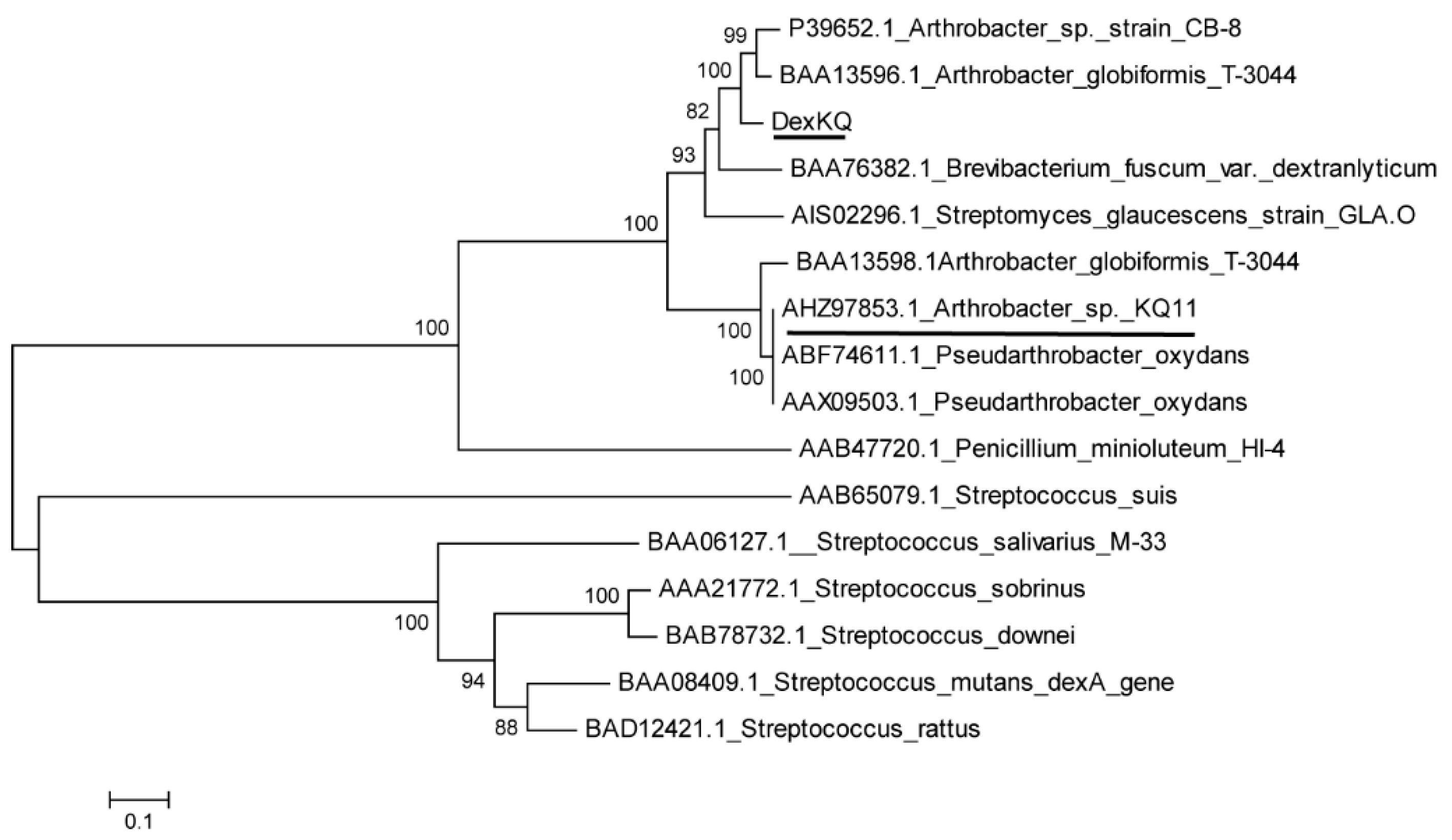
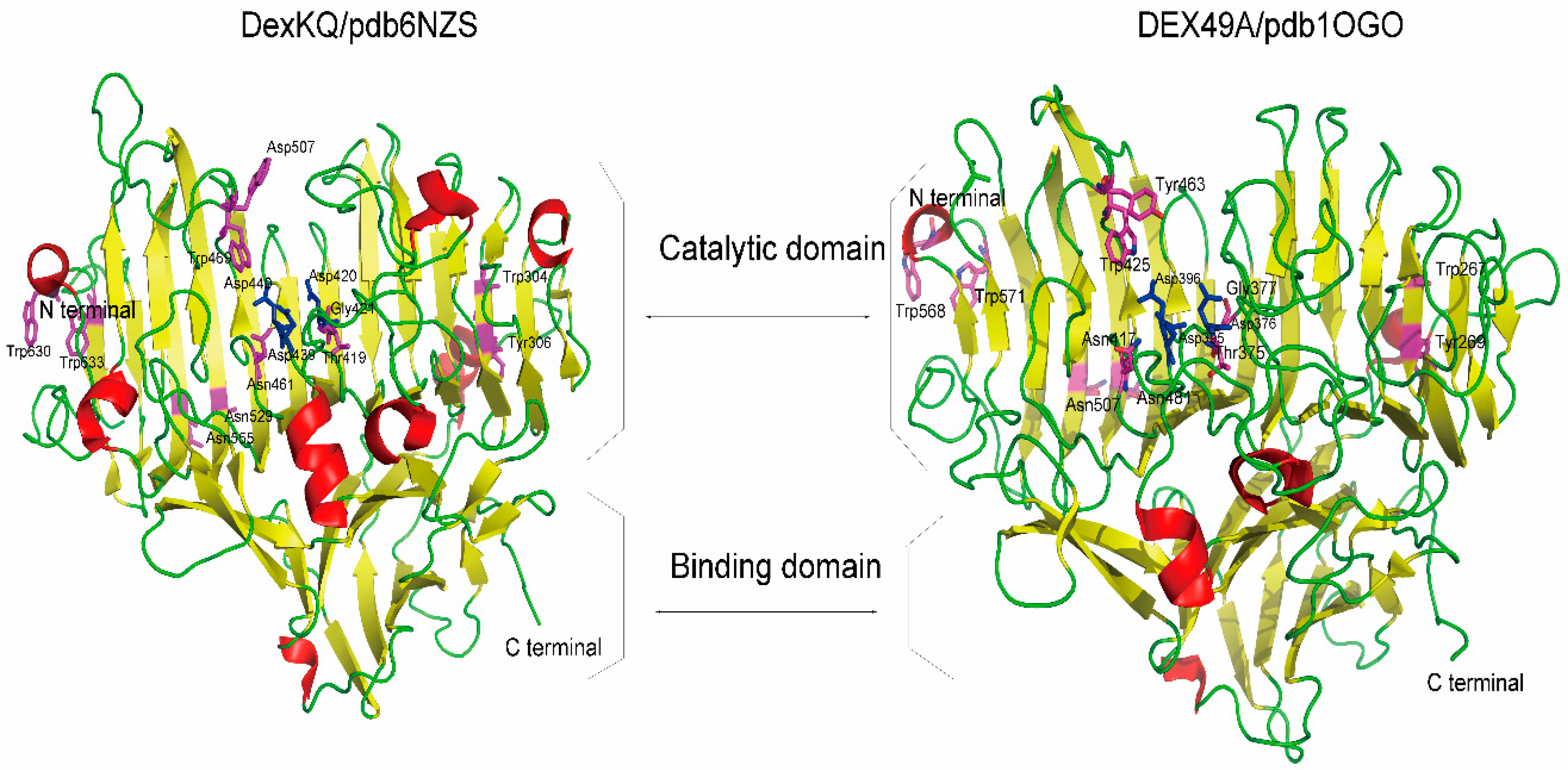
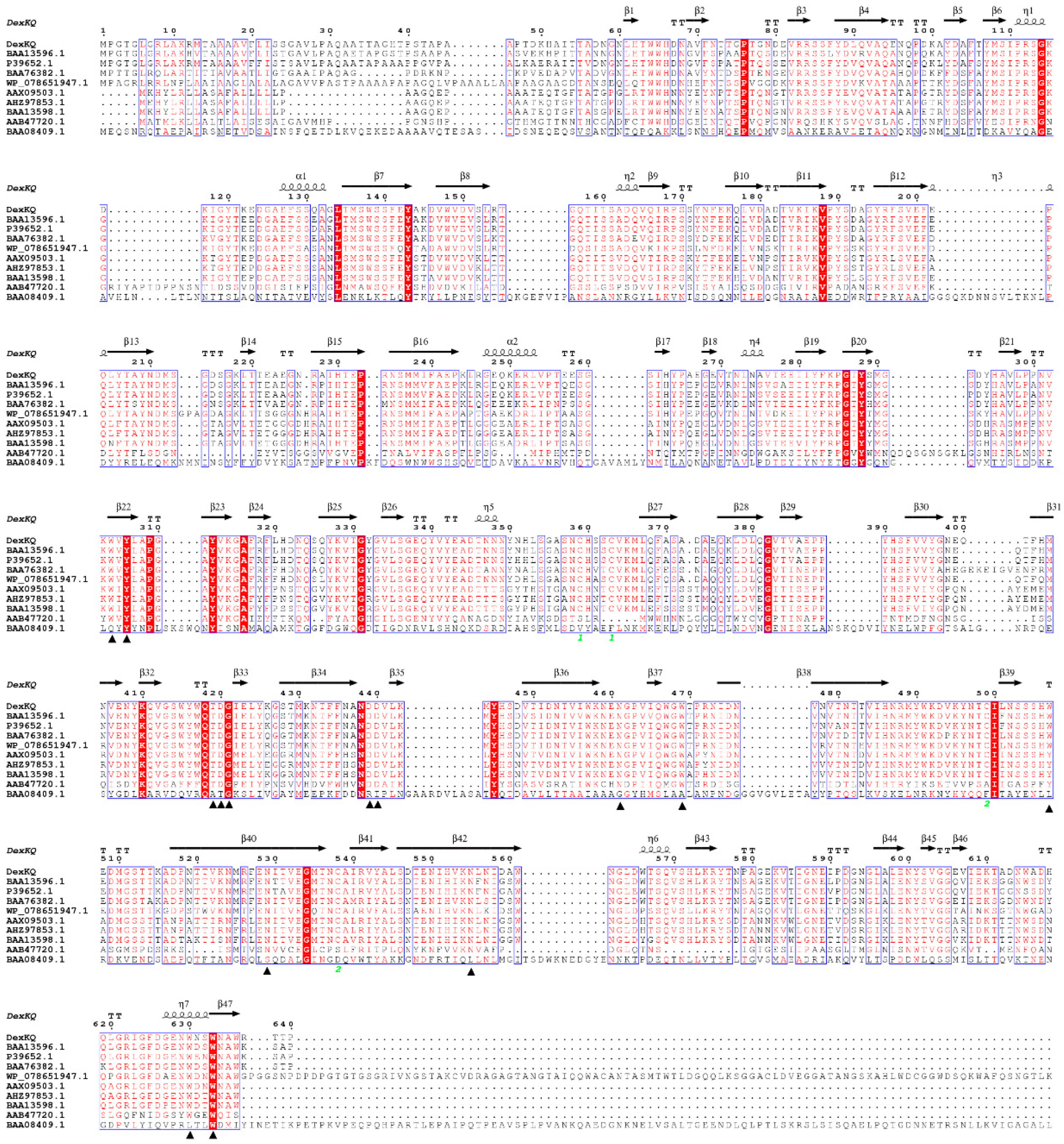
2.2. Bioinformatic Analysis
2.3. Recombinant Enzyme Expression and Property Characterization
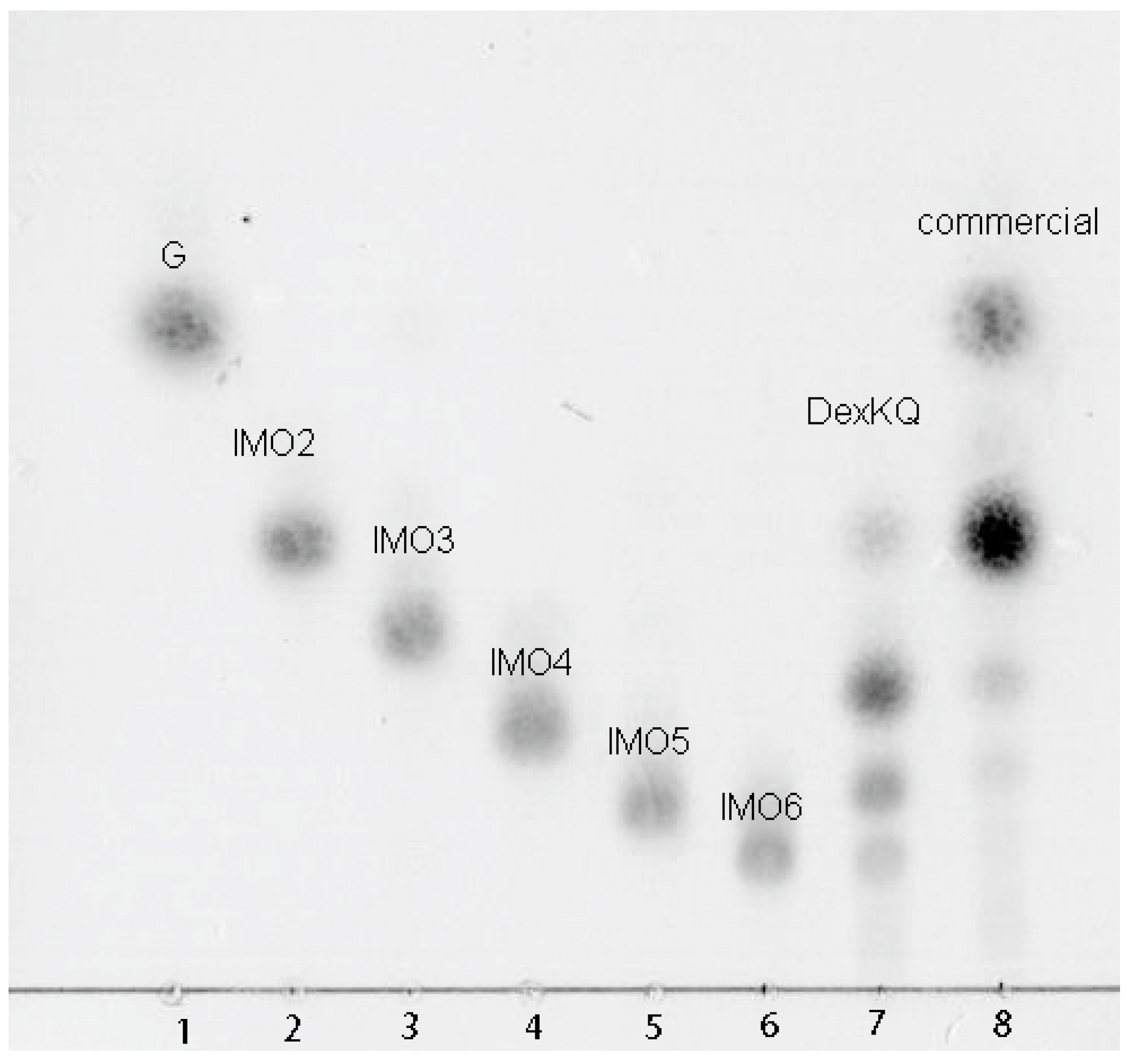
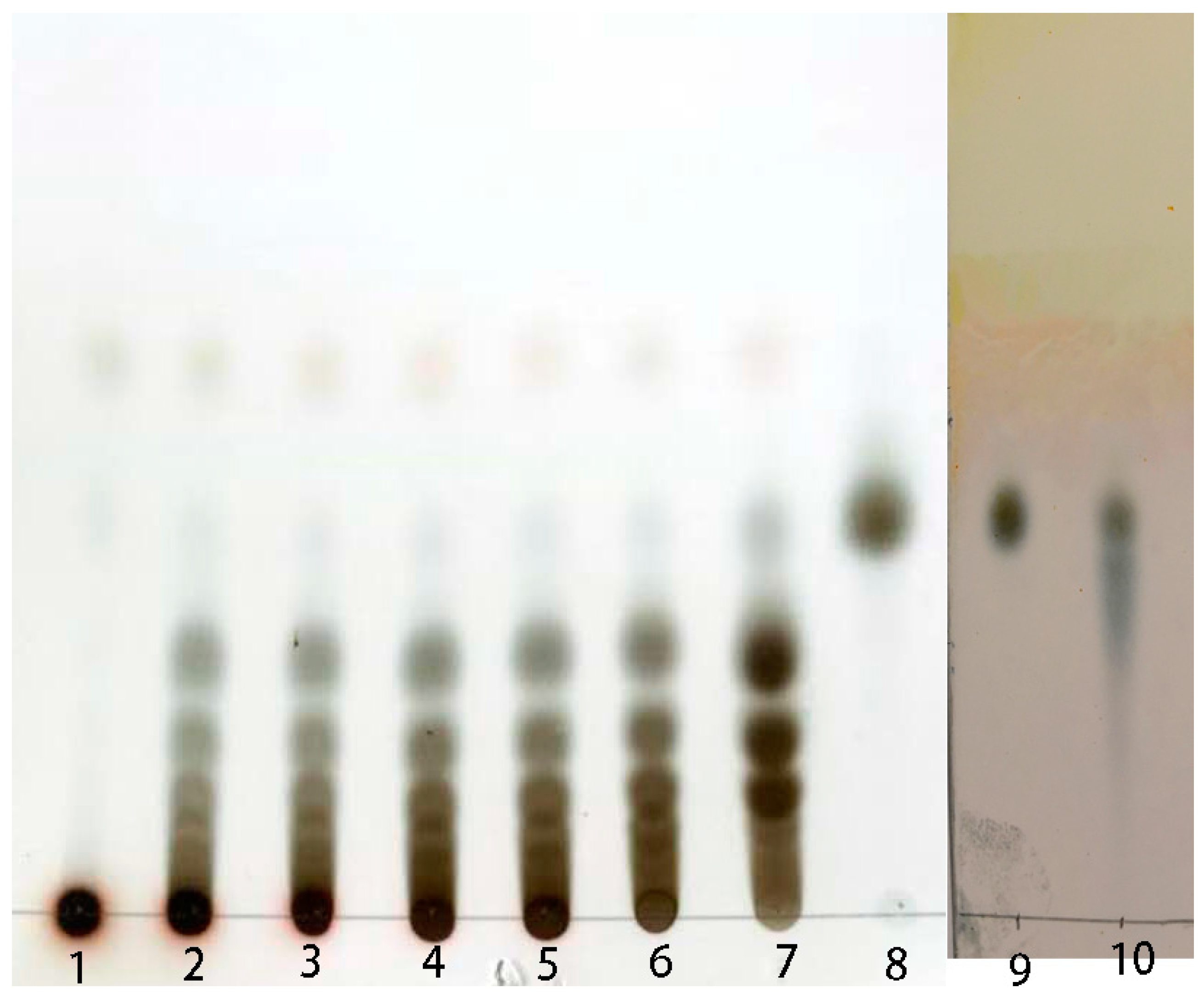
2.4. Preparation of Isomalto-Oligosaccharide and Application
3. Discussion and Conclusions
4. Materials and Methods
4.1. Molecular Cloning
4.2. Bioinformatic Analysis of the Dextranase Gene and Protein Sequence
4.3. Recombinant Expression of Dextranase
4.4. Dextranase Activity Assay
4.5. Effect of pH on Dextranase Activity and Stability
4.6. Effect of Temperature on Dextranase Activity and Stability
4.7. End Products Analysis
4.8. Enzyme Kinetics Assay
4.9. Preparation of Isomalto-Oligosaccharide (IMO)
4.10. Application of IMO in Culturing Microorganisms
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Khalikova, E.; Susi, P.; Korpela, T. Microbial dextran-hydrolyzing enzymes: Fundamentals and Applications. Microbiol. Mol. Biol. Rev. 2005, 69, 306–325. [Google Scholar] [CrossRef]
- Purushe, S.; Prakash, D.; Nawani, N.N.; Dhakephalkar, P.; Kapadnis, B. Biocatalytic potential of an alkalophilic and thermophilic dextranase as a remedial measure for dextran removal during sugar manufacture. Bioresour. Technol. 2012, 115, 2–7. [Google Scholar] [CrossRef]
- Gurpilhares, D.B.; Cinelli, L.P.; Simas, N.K.; Pessoa, A., Jr.; Sette, L.D. Marine prebiotics: Polysaccharides and oligosaccharides obtained by using microbial enzymes. Food Chem. 2019, 15, 280–336. [Google Scholar] [CrossRef]
- Wang, D.L.; Lv, M.S.; Wang, S.J.; Jiao, Y.L.; Li, W.J.; Zhu, Q.; Liu, Z.P. Purification and characterization of a novel marine Arthrobacter oxydans KQ11 dextranase. Carbohydr. Polym. 2014, 106, 71–76. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, 233–238. [Google Scholar] [CrossRef]
- Oguma, T.; Kurokawa, T.; Tobe, K.; Kitao, S.; Kobayashi, M. Cloning and sequence analysis of the gene for glucodextranase from Arthrobacter globiformis T-3044 and expression in Escherichia coli cells. Biosci. Biotechnol. Biochem. 1999, 63, 2174–2182. [Google Scholar] [CrossRef][Green Version]
- Okushima, M.; Sugino, D.; Kouno, Y.; Nakano, S.; Miyahara, J.; Toda, H.; Kubo, S.; Matsushiro, A. Molecular cloning and nucleotide sequencing of the Arthrobacter dextranase gene and its expression in Escherichia coli and Streptococcus sanguis. Jpn. J. Genet. 1991, 66, 173–187. [Google Scholar] [CrossRef][Green Version]
- Kim, Y.M.; Ko, E.A.; Kang, H.K.; Kim, D. Construction, expression and characterization of fusion enzyme from Arthrobacter oxydans dextranase and Klebsiella pneumoniae amylase. Biotechnol. Lett. 2009, 31, 1019–1024. [Google Scholar] [CrossRef]
- Jin, H.L.; Nam, S.H.; Park, H.J.; Kim, Y.M.; Kim, N.; Kim, G.; Seo, E.S.; Kang, S.S.; Kim, D. Biochemical characterization of dextranase from Arthrobacter oxydans and its cloning and expression in Escherichia coli. Food Sci. Biotechnol. 2010, 19, 757–762. [Google Scholar]
- Ren, W.; Liu, L.; Gu, L.; Yan, W.; Feng, Y.L.; Dong, D.; Wang, S.; Lyu, M.; Wang, C. Crystal Structure of GH49 Dextranase from Arthrobacter oxidans KQ11: Identification of catalytic base and improvement of thermostability using semirational design based on b-factors. J. Agric. Food Chem. 2019, 67, 4355–4366. [Google Scholar] [CrossRef]
- Larsson, A.M.; Andersson, R.; Stahlberg, J.; Kenne, L.; Alwyn, J.T. Dextranase from Penicillium minioluteum: Reaction course, crystal structure, and product complex. Structure 2003, 11, 1111–1121. [Google Scholar] [CrossRef]
- Morisaki, H.; Igarashi, T.; Yamamoto, A.; Goto, N. Analysis of a dextran-binding domain of the dextranase of Streptococcus mutans. Lett. Appl. Microbiol. 2002, 35, 223–227. [Google Scholar] [CrossRef]
- Okazawa, Y.; Miyazaki, T.; Yokoi, G.; Ishizaki, Y.; Nishikawa, A.; Tonozuka, T. Crystal Structure and Mutational Analysis of Isomalto-dextranase, a Member of Glycoside Hydrolase Family 27. J. Biol. Chem. 2015, 290, 26339–26349. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, w320–w324. [Google Scholar] [CrossRef]
- Kim, Y.M.; Kim, D. Characterization of novel thermostable dextranase from Thermotoga lettingae TMO. Appl. Microbiol. Biotechnol. 2010, 85, 581–587. [Google Scholar] [CrossRef]
- Finnegan, P.M.; Brumbley, S.M.; OShea, M.G.; Nevalainen, H.; Bergquist, P.L. Isolation and characterization of genes encoding thermoactive and thermostable dextranases from two thermotolerant soil bacteria. Curr. Microbiol. 2004, 49, 327–333. [Google Scholar] [CrossRef]
- Ko, J.A.; Nam, S.H.; Kim, D.; Lee, J.H.; Kim, Y.M. Identification of catalytic amino acid residues by chemical modification in dextranase. J. Microbiol. Biotechnol. 2016, 26, 837–845. [Google Scholar] [CrossRef]
- Igarashi, T.; Yamamoto, A.; Goto, N. Characterization of the dextranase gene (dex) of Streptococcus mutans and its recombinant product in an Escherichia coli host. Microbiol. Immunol. 1995, 39, 387–391. [Google Scholar] [CrossRef]
- Park, T.S.; Jeong, H.J.; Ko, J.A.; Ryu, Y.B.; Park, S.J.; Kim, D.; Kim, Y.M.; Lee, W.S. Biochemical characterization of thermophilic dextranase from a thermophilic bacterium, Thermoanaerobacter pseudethanolicus. J. Microbiol. Biotechnol. 2012, 22, 637–641. [Google Scholar] [CrossRef]
- Iwai, A.; Ito, H.; Mizuno, T.; Mori, H.; Matsui, H.; Honma, M.; Okada, G.; Chiba, S. Molecular cloning and expression of an isomalto-dextranase gene from Arthrobacter globiformis T6. J. Bacteriol. 1994, 176, 7730–7734. [Google Scholar] [CrossRef][Green Version]
- Morimoto, N.; Yasukawa, Y.; Watanabe, K.; Unno, T.; Ito, H.; Matsui, H. Cloning and heterologous expression of a glucodextranase gene from Arthrobacter globiformis I42, and experimental evidence for the catalytic diad of the recombinant enzyme. J. Biosci. Bioeng. 2004, 97, 127–130. [Google Scholar] [CrossRef]
- Yamamoto, T.; Terasawa, K.; Kim, Y.M.; Kimura, A.; Kitamura, Y.; Kobayashi, M.; Funane, K. Identification of catalytic amino acids of cyclodextran glucanotransferase from Bacillus circulans T-3040. Biosci. Biotechnol. Biochem. 2006, 70, 1947–1953. [Google Scholar] [CrossRef][Green Version]
- Zhang, Y.Q.; Li, R.H.; Zhang, H.B.; Wu, M.; Hu, X.Q. Purification, characterization, and application of a thermostable dextranase from Talaromyces pinophilus. J. Ind. Microbiol. Biotechnol. 2017, 44, 317–327. [Google Scholar] [CrossRef]
- Frank, A.E.; Hans, J.J. Immobilization of dextranase from Chaetomium erraticum. J. Biotechnol. 2007, 131, 440–447. [Google Scholar]
- Roca, H.; Garcia, B.M.; Rodriguez, E.; Mateu, D.; Coroas, L.; Cremata, J.A.; Garcia, R.; Pons, T.; Delgado, J. Cloning of the Penicillium minioluteum gene encoding dextranase and its expression in Pichia pastoris. Yeast 1996, 12, 1187–1200. [Google Scholar] [CrossRef]
- Betancourta, L.H.; García, R.; Gonzalez, J.; Montesinoa, R.; Quinteroa, O.; Takaob, T.; Shimonishib, Y.; Cremataa, J.A. Dextranase (alpha-1,6 glucanglucanohydrolase) from Penicillium minioluteum expressed in Pichia pastoris: Two host cells with minor differencesin N-glycosylation. FEMS Yeast Res. 2001, 1, 151–160. [Google Scholar]
- Volkov, P.V.; Gusakov, A.V.; Rubtsova, E.A.; Rozhkova, A.M.; Matys, V.Y.; Nemashkalov, V.A.; Sinitsyn, A.P. Properties of a recombinant GH49 family dextranase heterologously expressed in two recipient strains of Penicillium species. Biochimie 2019, 157, 123–130. [Google Scholar] [CrossRef]
- Kang, H.K.; Kim, S.H.; Park, J.Y.; Jin, X.J.; Oh, D.K.; Kang, S.S.; Kim, D. Cloning and characterization of a dextranase gene from Lipomyces starkeyi and its expression in Saccharomyces cerevisiae. Yeast 2005, 22, 1239–1248. [Google Scholar] [CrossRef][Green Version]
- Shimizu, E.; Unno, T.; Ohba, M.; Okada, G. Purification and characterization of an isomaltotriose-producing endo dextranase from a Fusarium sp. Biosci. Biotechnol. Biochem. 1998, 62, 117–122. [Google Scholar] [CrossRef]
- Cai, R.; Lv, M.S.; Fang, Y.W.; Jiao, Y.L.; Zhu, Q.; Liu, Z.P.; Wang, S.J. Screening, production, and characterization of dextranase from Catenovulum sp. Ann. Microbiol. 2014, 64, 147–155. [Google Scholar] [CrossRef]
- Tecker, S.R.; Adney, W.S. Alkaline Tolerant Dextranase from Streptomyces anulatus. U.S. Patent 6509184 B1, 12 January 2003. [Google Scholar]
- Aslan, Y.; Tanriseven, A. Immobilization of Penicillium lilacinum dextranase to produce isomalto-oligosaccharides from dextran. Biochem. Eng. J. 2007, 34, 8–14. [Google Scholar] [CrossRef]
- Bertrand, E.; Pierre, G.; Delattre, C.; Gardarin, C.; Bridiau, N.; Maugard, T.; Strancar, A.; Michaud, P. Dextranase immobilization on epoxy CIM(®) disk for the production of isomaltooligosaccharides from dextran. Carbohydr. Polym. 2014, 13, 707–713. [Google Scholar] [CrossRef]
- Singh, D.P.; Singh, J.; Boparai, R.K.; Zhu, J.H.; Mantri, S.; Khare, P.; Khardori, R.; Kondepudi, K.K.; Chopra, K.; Bishnoi, M. Isomalto-oligosaccharides, a prebiotic, functionally augment green tea effects against high fat diet-induced metabolic alterations via preventing gut dysbacteriosis in mice. Pharmacol. Res. 2017, 123, 103–113. [Google Scholar] [CrossRef]
- Basu, A.; Mutturi, S.; Prapulla, S.G. Production of isomaltooligosaccharides (IMO) using simultaneous saccharification and transglucosylation from starch and sustainable sources. Process Biochem. 2016, 51, 1464–1471. [Google Scholar] [CrossRef]
- Niu, D.D.; Qiao, J.; Li, P.J.; Tian, K.M.; Liu, X.G.; Sing, S.R.; Lu, F.P. Highly efficient enzymatic preparation of isomalto-oligosaccharides from starch using an enzyme cocktail. Electron. J. Biotechnol. 2017, 26, 46–51. [Google Scholar] [CrossRef]
- Wang, W.D.; Xin, H.X.; Fang, X.C.; Dou, H.T.; Liu, F.G.; Huang, D.; Han, S.M.; Fei, G.J.; Zhu, L.M.; Zha, S.H.; et al. Isomalto-oligosaccharides ameliorate visceral hyperalgesia with repair damage of ileal epithelial ultrastructure in rats. PLoS ONE 2017, 12, e0175276. [Google Scholar] [CrossRef]
- Ketabi, A.; Dieleman, L.A.; Ganzle, M.G. Influence of isomalto-oligosaccharides on intestinal microbiota in rats. J. Appl. Microbiol. 2011, 110, 1297–1306. [Google Scholar] [CrossRef]
- Kaneko, T.; Kohmoto, T.; Kikuchi, H.; Shiota, M.; Iino, H.; Mitsuoka, T. Effects of isomaltooligosaccha-rides with different degrees of polymerization on human fecal bifidobacteria. Biosci. Biotechnol. Biochem. 1994, 58, 2288–2290. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manunal, 4th ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2012; Volume 2, pp. 166–205. [Google Scholar]





| Internal Peptide Sequences of Native Dextranase Enzyme | Partial AA Sequence of ORF 4194 |
|---|---|
| 1: AYDAFTYMSIPR | SFYDLQVAQENQPDKAYDAFTYMSIPRSGKDKIWVDVSLRTGQTITSADQVQIRPSSYNFEKQLVDADTVKIKVPYSDAGYR FSVEFEPQLYTAYNDMSGDSGKLTTEAEGNRAIHTEPRNSMMIFAEPKLRGEQKERLVPTEESGSIHQVGSWYWQTDGIELYKGST |
| 2: TGQTITSADQVQIRPSSYNFEK | |
| 3: IKVPYSDAGYR | |
| 4: FSVEFEPQLYTAYNDMSGDSGK | |
| 5: NSMMIFAEPK | |
| 6: QVGSWYWQTDGIELYK |
| DEX49A | DexKQ | DEX49A | DexKQ |
|---|---|---|---|
| Asp395 | Asp439 | Tyr463 | Trp507 |
| Asp376 | Asp420 | Trp267 | Trp304 |
| Asp396 | Asp440 | Tyr269 | Tyr306 |
| Asn481 | Asn529 | Trp568 | Trp630 |
| Asn507 | Asn555 | Trp571 | Trp633 |
| Asn417 | Asn461 | Thr375 | Thr419 |
| Trp425 | Trp469 | Gly377 | Gly421 |
| Origin | Temperature and pH Parameters | Classification | Main End Product | Reference |
|---|---|---|---|---|
| Arthrobacter globiformis T-3044 E. coli recombinant | unknown | Endodextranase GH49 | unknown | [6] |
| Arthrobacter globiformis CB-8 E. coli recombinant | unknown | Endodextranase GH49 | unknown | [7] |
| Arthrobacter oxydans E. coli recombinant | optimum activity at 37 °C, pH 7.5 | Endodextranase GH49 | IMO2,3 | [8] |
| Arthrobacter oxydans E. coli recombinant | stable below 40 °C and optimum activity at pH 6.0 | Endodextranase GH49 | unknown | [9] |
| Thermotoga lettingae TMO | optimal pH and temperature were 4.3 and 55–60 °C stable from pH 4.3–10.0 | Endodextranase GH66 | IMO2,3 | [15] |
| Paenibacillus sp. E. coli recombinant | optimal pH and temperature were 5.5 °C and 60 °C | Endodextranase GH66 | IMO2,3 | [16] |
| Paenibacillus sp. native | optimal pH was 5.5 | Endodextranase GH66 | IMO4 | [17] |
| Streptococcus mutans E. coli recombinant | optimal pH and temperature were 5.0 °C and 40 °C | Endodextranase GH66 | IMO2,3,4 | [18] |
| Thermoanaerobacter pseudethanolicus E. coli recombinant | optimal pH was 5.2 and a half-life of 7.4 h at 70 °C between pH 3.1 and 8.5 | Endodextranase GH66 | IMO2,3 | [19] |
| Arthrobacter globiformis T6 E. coli recombinant | optimal pH and temperature were 3.5 °C and 60 °C | Isomaltose exo-dextranase GH27 | IMO2 | [20] |
| Arthrobacter globiformis I42 E. coli recombinant | optimum pH and temperature were 6.0 and 45 °C | Glucodextranase GH15 | unknown | [21] |
| Bacillus circulans T-3040 E. coli recombinant | optimal pH and temperature were 5.2 °C and 60 °C | Cyclodextran glucanotransferase GH66 | cyclodextran | [22] |
| Talaromyces pinophilus native | optimum temperature of 45 °C and an optimum pH of 6.0, stable over pH range 3.0 to 10.0 | Endodextranase GH49 | IMO2,3 | [23] |
| Chaetomium erraticum native | optimal pH and temperature were 5.2 °C and 60 °C | Endodextranase GH49 | IMO2,3 | [24] |
| Penicillium minioluteum Pichia pastoris recombinant | optimum activity at pH 5.0 and 60 °C | Endodextranase GH49 | IMO2,3 | [25,26] |
| Penicillium funiculosum recombiant in other Pencillium strain | activity optimal at pH 4.5–5.0 and 55–60 °C | Endodextranase GH49 | Glucose and IMO2 | [27] |
| Lipomyces starkeyi S. cerevisiae recombinant | maximum activity at pH 6.0 and 37 °C | Endodextranase GH49 | IMO2,3 | [28] |
| Fusarium sp. native | maximum activity at pH 6.5 and 35 °C, stable under 4 °C and at pH ranging from 4.5 to 11.8 | Endodextranase | IMO3 | [29] |
| Catenovulum sp. native | maximum activity at pH 8.0 and 40 °C, stable under 30 °C and pH ranging from 5.0 to 11.0 | Endodextranase GH49 | IMO2 | [30] |
| Streptomyces sp. NK458 native | maximum activity at pH 9.0 and 60 °C stable at pH 5.0–10.0 | unknown | unknown | [2] |
| Streptomyces anulatus native | retained 50% of initial activity at pH 5.1–10.1 under 30 °C for 1.5 h | Endodextranase GH49 | unknown | [31] |
| Arthrobacter oxydans KQ11 E. coli recombinant | maximum activity at pH 9.0 and 55 °C, stable under 40 °C and at pH ranging from 6.0 to 13.0 | Endodextranase GH49 | IMO4,5,6 | This study |
| Substrate | Main Linkage | Relative Activity (%) | |
|---|---|---|---|
| DexKQ Dextranase | Sunshine Dextranase | ||
| 3%Dextran T5 | α-1,6 | 70.5 ± 2.1 | 102.5 ± 0.8 |
| 3% Dextran T11 | α-1,6 | 44.4 ± 0.3 | 126.6 ± 1.5 |
| 3% Dextran T20 | α-1,6 | 90.5 ± 0.5 | 101.2 ± 1.3 |
| 3% Dextran T40 | α-1,6 | 85.0 ± 2.3 | 152.5 ± 0.4 |
| 1% Dextran T70 | α-1,6 | 55.0 ± 1.2 | 33.1 ± 0.5 |
| 3% Dextran T70 | α-1,6 | 100.0 | 100.0 |
| 5% Dextran T70 | α-1,6 | 76.3 ± 0.4 | 116.4 ± 1.2 |
| 8% Dextran T70 | α-1,6 | 87.4 ± 0.5 | 94.5 ± 1.6 |
| 10% Dextran T70 | α-1,6 | 181.2 ± 1.4 | 303.0 ± 0.4 |
| 15% Dextran T70 | α-1,6 | 120.8 ± 0.4 | 163.5±1.5 |
| 20% Dextran T70 | α-1,6 | 147.8 ± 1.1 | 485.9 ± 2.3 |
| 3% Dextran T100 | α-1,6 | 71.9 ± 4.5 | 78.734 ± 3.1 |
| 3% Dextran T200 | α-1,6 | 76.3 ± 3.3 | 90.5 ± 0.6 |
| 3% Dextran T500 | α-1,6 | 85.9 ± 0.3 | 75.2 ± 2.2 |
| Blue dextran 2000 | α-1,6 | 75.6 ± 1.1 | 67.3 ± 0.9 |
| pullulan | α-1,6 | 0 | 0 |
| laminaran | β-1,3 | 0 | 0 |
| Amylose | α-1,4 | 0 | 0 |
| Amyloid | α-1,4 and α-1,6 | 0 | 0 |
| Sucrose | α-1,4 | 0 | 0 |
| Trehalose | α-1,2 | 0 | 0 |
| Carboxymethyl cellulose | β-1,4 | 0 | 0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Ren, W.; Ly, M.; Li, H.; Wang, S. Characterization of an Alkaline GH49 Dextranase from Marine Bacterium Arthrobacter oxydans KQ11 and Its Application in the Preparation of Isomalto-Oligosaccharide. Mar. Drugs 2019, 17, 479. https://doi.org/10.3390/md17080479
Liu H, Ren W, Ly M, Li H, Wang S. Characterization of an Alkaline GH49 Dextranase from Marine Bacterium Arthrobacter oxydans KQ11 and Its Application in the Preparation of Isomalto-Oligosaccharide. Marine Drugs. 2019; 17(8):479. https://doi.org/10.3390/md17080479
Chicago/Turabian StyleLiu, Hongfei, Wei Ren, Mingsheng Ly, Haifeng Li, and Shujun Wang. 2019. "Characterization of an Alkaline GH49 Dextranase from Marine Bacterium Arthrobacter oxydans KQ11 and Its Application in the Preparation of Isomalto-Oligosaccharide" Marine Drugs 17, no. 8: 479. https://doi.org/10.3390/md17080479
APA StyleLiu, H., Ren, W., Ly, M., Li, H., & Wang, S. (2019). Characterization of an Alkaline GH49 Dextranase from Marine Bacterium Arthrobacter oxydans KQ11 and Its Application in the Preparation of Isomalto-Oligosaccharide. Marine Drugs, 17(8), 479. https://doi.org/10.3390/md17080479




