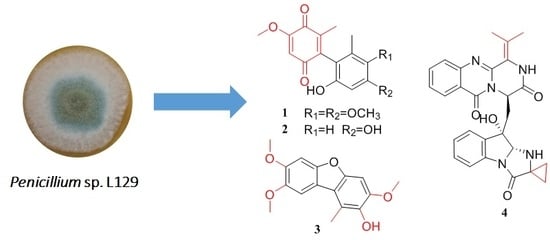
Dimeric 1,4-Benzoquinone Derivatives with Cytotoxic Activities from the Marine-Derived Fungus Penicillium sp. L129
Abstract
1. Introduction
2. Results and Discussion
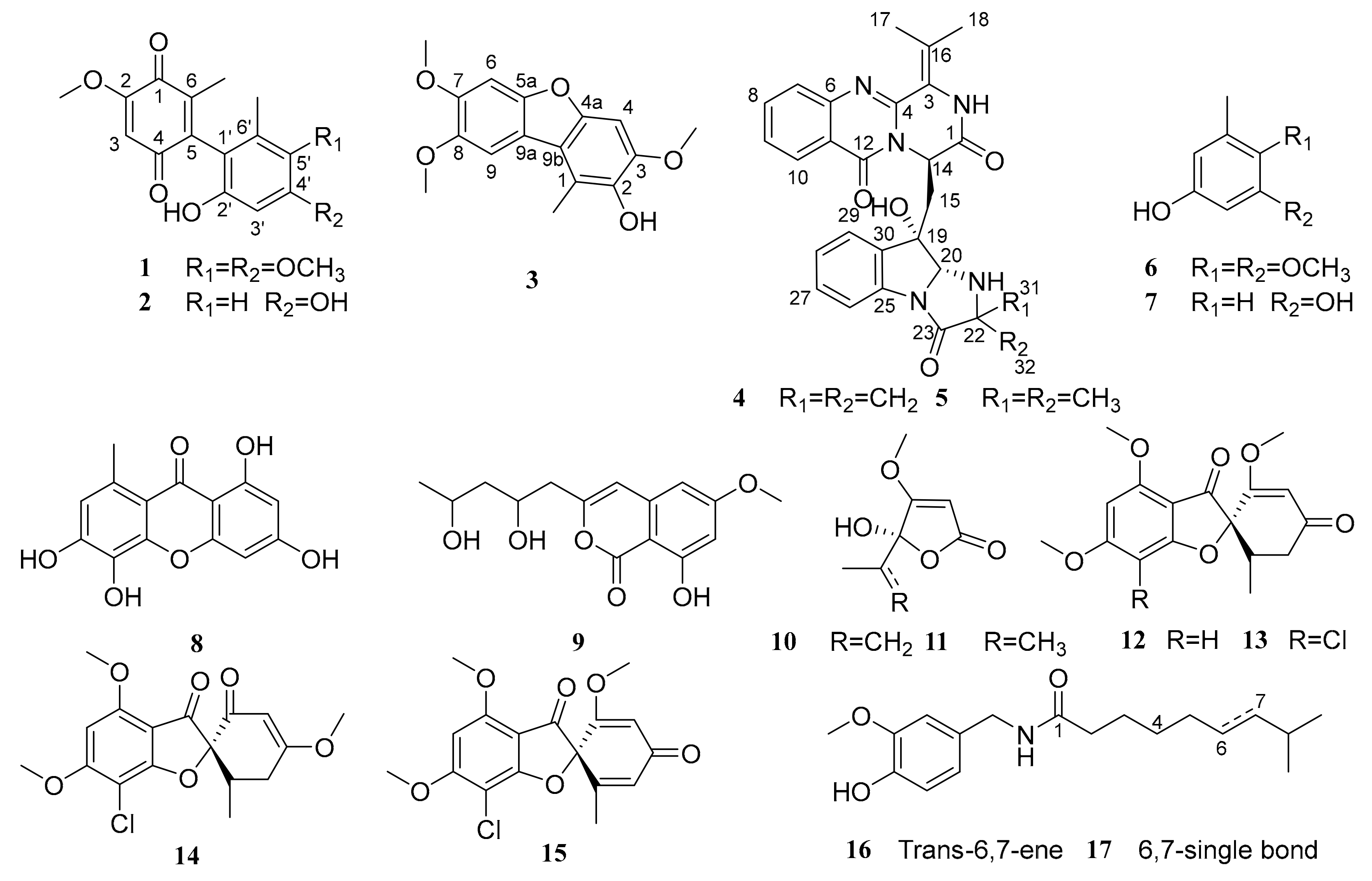
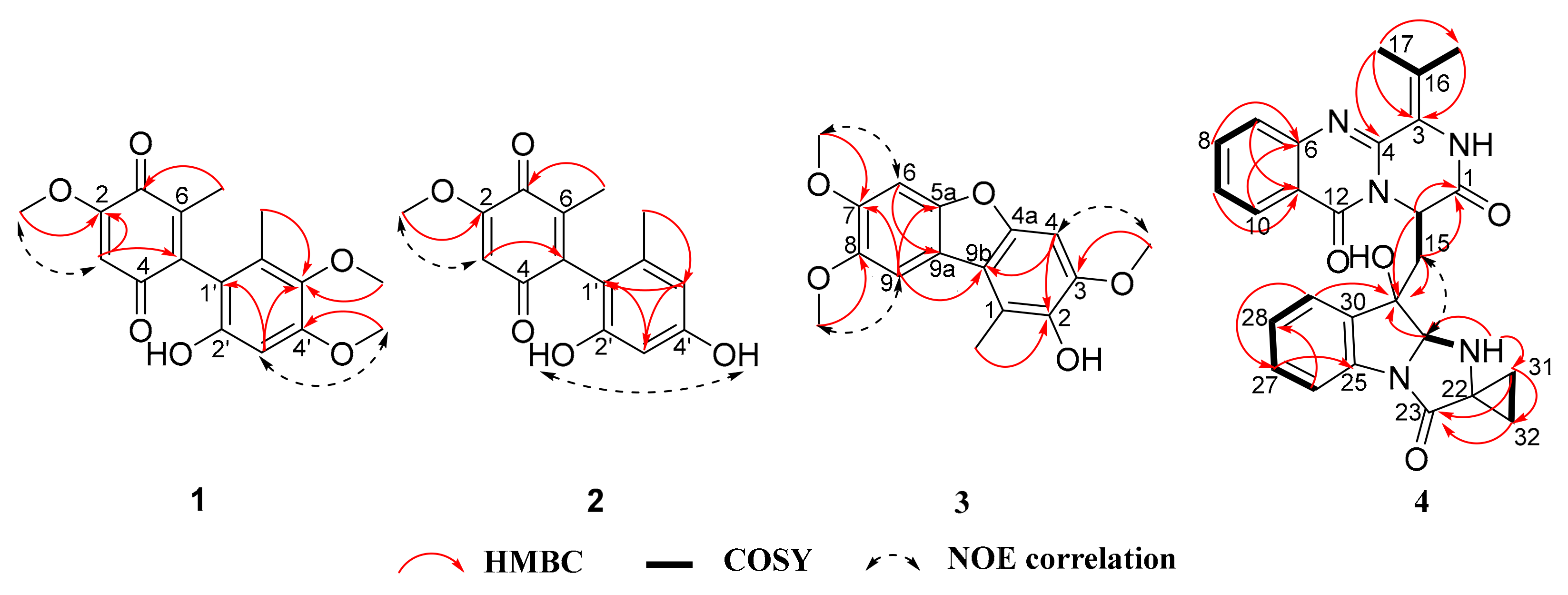
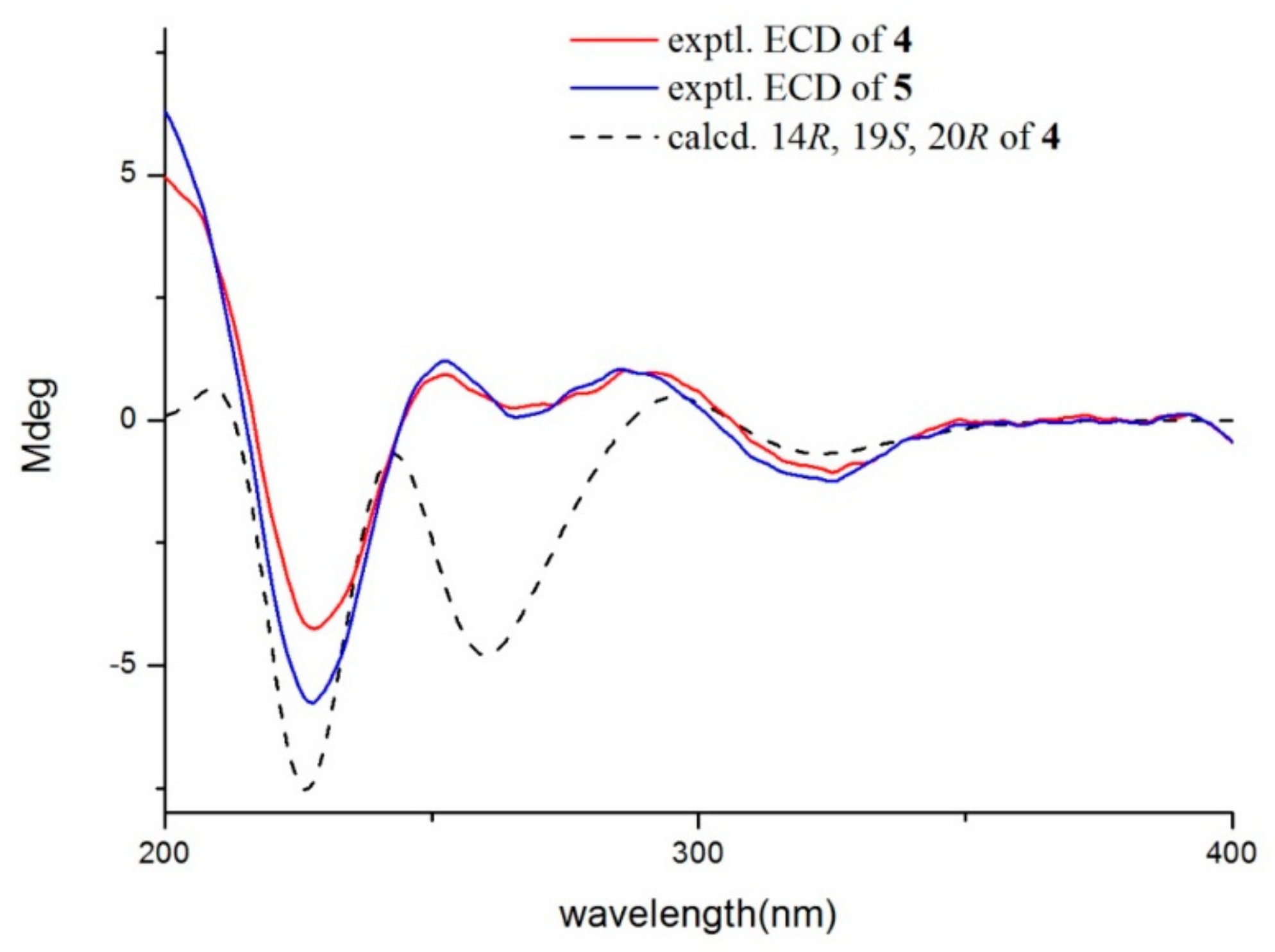
2.1. Structural Elucidation
2.2. The Bioactivities of Compounds
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal and Plant Material
3.3. Fermentation and Extraction
3.4. Purification and Identification
3.5. QS inhibitory Activity Essay
3.6. Cytotoxic Activity Essay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2019, 36, 122–173. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Su, M.; Song, S.J.; Jung, J.H. Marine-derived Penicillium species as producers of cytotoxic metabolites. Mar. Drugs 2017, 15, 329. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.H.; Wang, C.Y.; Mandi, A.; Li, X.M.; Hu, X.Y.; Kassack, M.U.; Kurtan, T.; Wang, B.G. Three diketopiperazine alkaloids with spirocyclic skeletons and one bisthiodiketopiperazine derivative from the mangrove-derived endophytic fungus Penicillium brocae MA-231. Org. Lett. 2016, 18, 5304–5307. [Google Scholar] [CrossRef]
- Liao, L.; Bae, S.Y.; Won, T.H.; You, M.; Kim, S.H.; Oh, D.C.; Lee, S.K.; Oh, K.B.; Shin, J. Asperphenins A and B, lipopeptidyl benzophenones from a marine-derived Aspergillus sp. Fungus. Org. Lett. 2017, 19, 2066–2069. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, J.; Lin, X.; Zhao, B.; Wei, X.; Li, G.; Kaliaperumal, K.; Liao, S.; Yang, B.; Zhou, X.; et al. Chrysamides A-C, three dimeric nitrophenyl trans-Epoxyamides produced by the deep-sea-derived fungus Penicillium chrysogenum SCSIO41001. Org. Lett. 2016, 18, 3650–3653. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liao, Y.; Zhang, B.; Gao, M.; Ke, W.; Li, F.; Shao, Z. Citrinin monomer and dimer derivatives with antibacterial and cytotoxic activities isolated from the deep sea-derived fungus Penicillium citrinum NLG-S01-P1. Mar. Drugs 2019, 17, 46. [Google Scholar] [CrossRef]
- Lu, Z.Y.; Lin, Z.J.; Wang, W.L.; Du, L.; Zhu, T.J.; Fang, Y.C.; Gu, Q.Q.; Zhu, W.M. Citrinin dimers from the halotolerant fungus Penicillium citrinum B-57. J. Nat. Prod. 2008, 71, 543–546. [Google Scholar] [CrossRef]
- Leong, S.L.; Schnurer, J.; Broberg, A.; Verrucine, F. A quinazoline from Penicillium verrucosum. J. Nat. Prod. 2008, 71, 1455–1457. [Google Scholar] [CrossRef]
- Michael, J.P. Quinoline, quinazoline and acridone alkaloids. Nat. Prod. Rep. 2008, 25, 166–187. [Google Scholar] [CrossRef]
- Hu, X.Y.; Meng, L.H.; Li, X.; Yang, S.Q.; Li, X.M.; Wang, B.G. Three new indole diterpenoids from the sea-anemone-derived fungus Penicillium sp. AS-79. Mar. Drugs 2017, 15, 137. [Google Scholar] [CrossRef]
- Huang, L.H.; Xu, M.Y.; Li, H.J.; Li, J.Q.; Chen, Y.X.; Ma, W.Z.; Li, Y.P.; Xu, J.; Yang, D.P.; Lan, W.J. Amino acid-directed strategy for inducing the marine-derived fungus Scedosporium apiospermum F41-1 to maximize alkaloid diversity. Org. Lett. 2017, 19, 4888–4891. [Google Scholar] [CrossRef] [PubMed]
- Tadross, P.M.; Gilmore, C.D.; Bugga, P.; Virgil, S.C.; Stoltz, B.M. Regioselective reactions of highly substituted arynes. Org. Lett. 2010, 12, 1224–1227. [Google Scholar] [CrossRef] [PubMed]
- Rojas, I.S.; Lotina-Hennsen, B.; Mata, R. Effect of lichen metabolites on thylakoid electron transport and photophosphorylation in isolated spinach chloroplasts. J. Nat. Prod. 2000, 63, 1396–1399. [Google Scholar] [CrossRef] [PubMed]
- Belofsky, G.N.; Gloer, K.B.; Gloer, J.B.; Wicklow, D.T.; Dowd, P.F. New p-terphenyl and polyketide metabolites from the sclerotia of Penicillium raistrickii. J. Nat. Prod. 1998, 61, 1115–1119. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.C.; Chen, G.D.; Zhao, Y.Q.; Xin, S.C.; Li, S.; Tang, J.S.; Li, X.X.; Hu, D.; Liu, X.Z.; Gao, H. New isocoumarins from a cold-adapted fungal strain Mucor sp. and their developmental toxicity to zebrafish embryos. Chem. Biodivers. 2014, 11, 1099–1108. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Nakahara, S.; Fujioka, S. Aspyrone, a nematicidal compound isolated from the fungus, Aspergillus melleus. Biosci. Biotechnol. Biochem. 1996, 60, 1375–1376. [Google Scholar] [CrossRef]
- Shang, Z.; Li, X.M.; Li, C.S.; Wang, B.G. Diverse secondary metabolites produced by marine-derived fungus Nigrospora sp. MA75 on various culture media. Chem. Biodivers. 2012, 9, 1338–1348. [Google Scholar] [CrossRef]
- Zhao, J.H.; Zhang, Y.L.; Wang, L.W.; Wang, J.Y.; Zhang, C.L. Bioactive secondary metabolites from Nigrospora sp. LLGLM003, an endophytic fungus of the medicinal plant Moringa oleifera lam. World J. Microb. Biot. 2012, 28, 2107–2112. [Google Scholar] [CrossRef]
- Levine, S.G.; Hicks, R.E.; Gottlieb, H.E.; Wenkert, E. Carbon-13 nuclear magnetic resonance spectroscopy of naturally occurring substances. XXX. Griseofulvin. J. Org. Chem. 1975, 40, 2540–2542. [Google Scholar] [CrossRef]
- Wubshet, S.G.; Nyberg, N.T.; Tejesvi, M.V.; Pirttila, A.M.; Kajula, M.; Mattila, S.; Staerk, D. Targeting high-performance liquid chromatography-high-resolution mass spectrometry-solid-phase extraction-nuclear magnetic resonance analysis with high-resolution radical scavenging profiles-bioactive secondary metabolites from the endophytic fungus Penicillium namyslowskii. J. Chromatogr. A. 2013, 1302, 34–39. [Google Scholar]
- Lin, L.Z.; West, D.P.; Cordell, G.A. NMR assignments of cis- and trans-capsaicin. Nat. Prod. Lett. 2006, 3, 5–8. [Google Scholar] [CrossRef]
- Gomez-Calvario, V.; Garduno-Ramirez, M.L.; Leon-Rivera, I.; Rios, M.Y. 1H and 13C NMR data on natural and synthetic capsaicinoids. Magn. Reson. Chem. 2016, 54, 268–290. [Google Scholar] [PubMed]
- Liu, H.; Zhao, F.; Yang, R.; Wang, M.; Zheng, M.; Zhao, Y.; Zhang, X.; Qiu, F.; Wang, H. Dimeric 1,4-benzoquinone derivatives and a resorcinol derivative from Ardisia gigantifolia. Phytochemistry 2009, 70, 773–778. [Google Scholar] [CrossRef] [PubMed]
- Fukuyama, Y.; Kiriyama, Y.; Okino, J.; Kodama, M. Belamcandaquinones A and B, novel dimeric 1,4-benzoquinone derivatives possessing cyclooxygenase inhibitory activity. Tetrahedron Lett. 1993, 34, 7633–7636. [Google Scholar] [CrossRef]
- Ko, R.K.; Lee, S.; Hyun, C.G.; Lee, N.H. New dibenzofurans from the branches of Distylium racemosum Sieb. et Zucc. Bull. Korean Chem. Soc. 2009, 30, 1376–1378. [Google Scholar]
- Xu, K.; Gao, Y.; Li, Y.L.; Xie, F.; Zhao, Z.T.; Lou, H.X. Cytotoxic p-terphenyls from the endolichenic fungus Floricola striata. J. Nat. Prod. 2018, 81, 2041–2049. [Google Scholar] [CrossRef]
- Michael, A.; Pfaller, M.D.; Ronald, N.; Jones, M.D. Performance accuracy of antibacterial and antifungal susceptibility test methods: Report from the College of American Pathologists (CAP) microbiology surveys program (2001-2003). Arch. Pathol. Lab. Med. 2006, 130, 767–778. [Google Scholar]
- Li, G.; Kusari, S.; Lamshoft, M.; Schuffler, A.; Laatsch, H.; Spiteller, M. Antibacterial secondary metabolites from an endophytic fungus, Eupenicillium sp. LG41. J. Nat. Prod. 2014, 77, 2335–2341. [Google Scholar] [CrossRef]
- Zheng, H.; Dong, Y.; Li, L.; Sun, B.; Liu, L.; Yuan, H.; Lou, H. Novel benzo[a]quinolizidine analogs induce cancer cell death through paraptosis and apoptosis. J. Med. Chem. 2016, 59, 5063–5076. [Google Scholar] [CrossRef]
- Sun, B.; Liu, J.; Gao, Y.; Zheng, H.B.; Li, L.; Hu, Q.W.; Yuan, H.Q.; Lou, H.X. Design, synthesis and biological evaluation of nitrogen-containing macrocyclic bisbibenzyl derivatives as potent anticancer agents by targeting the lysosome. Eur. J. Med. Chem. 2017, 136, 603–618. [Google Scholar] [CrossRef]
- Zhu, F.H.; Liu, Y.Q.; Lou, H.X. Bisbibenzyl-induced autophagy delays cell death in PC-3 cells. J. Shandong Univ. Health Sci. 2013, 51, 11–17. [Google Scholar]
| Position | 1 a | 2 b | ||
|---|---|---|---|---|
| δC, Type | δH (J in Hz) | δC, Type | δH (J in Hz) | |
| 1 | 182.6, C | 182.3, C | ||
| 2 | 158.8, C | 158.3, C | ||
| 3 | 107.7, CH | 6.01, s | 107.6, CH | 6.11, s |
| 4 | 186.5, C | 186.1, C | ||
| 5 | 140.9, C | 141.7, C | ||
| 6 | 143.0, C | 140.7, C | ||
| 1′ | 112.7, C | 111.7, C | ||
| 2′ | 148.7, C | 155.1, C | ||
| 3′ | 98.8, CH | 6.36, s | 99.9, CH | 6.16, d (1.9) |
| 4′ | 153.7, C | 157.9, C | ||
| 5′ | 141.6, C | 107.8, CH | 6.13, d (1.9) | |
| 6′ | 131.0, C | 137.3, C | ||
| 6-CH3 | 13.6, CH3 | 1.88, s | 13.0, CH3 | 1.69, s |
| 6′-CH3 | 13.4, CH3 | 1.96, s | 19.5, CH3 | 1.82, s |
| 2-OCH3 | 56.4, OCH3 | 3.85, s | 56.3, OCH3 | 3.80, s |
| 4′-OCH3 | 55.8, OCH3 | 3.83, s | ||
| 5′-OCH3 | 60.6, OCH3 | 3.75, s | ||
| 2′-OH | 9.06, s | |||
| 4′-OH | 9.24, s | |||
| Position | 3a | |
|---|---|---|
| δC, Type | δH (J in Hz) | |
| 1 | 116.6, C | |
| 2 | 140.1, C | |
| 3 | 147.0, C | |
| 4 | 93.1, CH | 7.13, s |
| 4a | 149.1, C | |
| 5a | 150.3, C | |
| 6 | 96.2, CH | 7.30, s |
| 7 | 148.1, C | |
| 8 | 145.5, C | |
| 9 | 104.1, CH | 7.45, s |
| 9a | 116.3, C | |
| 9b | 115.5, C | |
| 1-CH3 | 12.3, CH3 | 2.56, s |
| 3-OCH3 | 56.2, OCH3 | 3.87, s |
| 7-OCH3 | 56.0, OCH3 | 3.84, s |
| 8-OCH3 | 56.3, OCH3 | 3.86, s |
| Position | 4a | |
|---|---|---|
| δC, Type | δH (J in Hz) | |
| 1 | 166.5, C | |
| 3 | 121.5, C | |
| 4 | 146.8, C | |
| 6 | 146.8, C | |
| 7 | 127.1, CH | 7.66, d (8.2) |
| 8 | 134.6, CH | 7.85, t (7.7) |
| 9 | 126.9, CH | 7.55, t (7.4) |
| 10 | 126.3, CH | 8.16, d (8.3) |
| 11 | 119.6, C | |
| 12 | 159.9, C | |
| 14 | 52.5, CH | 5.43, t (6.5) |
| 15 | 37.5, CH2 | 15a: 2.48, dd (8.2, 14.8) 15b: 2.57, dd (5.4, 14.8) |
| 16 | 130.8, C | |
| 17 | 21.5, CH3 | 2.31, s |
| 18 | 21.1, CH3 | 1.96, s |
| 19 | 75.1, C | |
| 20 | 80.5, CH | 5.25, d (9.5) |
| 22 | 46.6, C | |
| 23 | 173.5, C | |
| 25 | 138.3, C | |
| 26 | 124.3, CH | 7.10, m |
| 27 | 129.7, CH | 7.33, m |
| 28 | 114.4, CH | 7.33, m |
| 29 | 124.6, CH | 7.35, m |
| 30 | 137.1, C | |
| 31 | 14.3, CH2 | 0.84, m |
| 32 | 10.9, CH2 | 1.01, m; 0.93, m |
| 19-OH | 5.68, br s | |
| 2-NH | 10.04, br s | |
| 21-NH | 3.57, d (9.4) | |
| Compounds | IC50 | |||
|---|---|---|---|---|
| MCF-7 | A549 | U87 | PC3 | |
| 1 | 12.39 ± 2.43 | >40 | 9.01 ± 2.36 | 14.59 ± 2.75 |
| 2 | 25.32 ± 3.22 | >40 | 13.45 ± 3.12 | 19.93 ± 3.48 |
| 3 | >40 | >40 | >40 | >40 |
| 4 | >40 | >40 | >40 | >40 |
| Adriamycin | 2.03 ± 0.42 | 1.53 ± 0.37 | 1.21 ± 0.50 | 0.98 ± 0.23 |
| Organism | 12 | 13 | 14 | 15 |
|---|---|---|---|---|
| Colletotrichum musae (ACCC 31244) | 1 | 0.1 | <10 | >1 |
| Colletotrichum coccodes (ACCC 36067) | NA | NA | <10 | NA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.-M.; Ju, C.-X.; Li, G.; Sun, Y.; Peng, Y.; Li, Y.-X.; Peng, X.-P.; Lou, H.-X. Dimeric 1,4-Benzoquinone Derivatives with Cytotoxic Activities from the Marine-Derived Fungus Penicillium sp. L129. Mar. Drugs 2019, 17, 383. https://doi.org/10.3390/md17070383
Zhang H-M, Ju C-X, Li G, Sun Y, Peng Y, Li Y-X, Peng X-P, Lou H-X. Dimeric 1,4-Benzoquinone Derivatives with Cytotoxic Activities from the Marine-Derived Fungus Penicillium sp. L129. Marine Drugs. 2019; 17(7):383. https://doi.org/10.3390/md17070383
Chicago/Turabian StyleZhang, Hui-Min, Chuan-Xia Ju, Gang Li, Yong Sun, Yu Peng, Ying-Xia Li, Xiao-Ping Peng, and Hong-Xiang Lou. 2019. "Dimeric 1,4-Benzoquinone Derivatives with Cytotoxic Activities from the Marine-Derived Fungus Penicillium sp. L129" Marine Drugs 17, no. 7: 383. https://doi.org/10.3390/md17070383
APA StyleZhang, H.-M., Ju, C.-X., Li, G., Sun, Y., Peng, Y., Li, Y.-X., Peng, X.-P., & Lou, H.-X. (2019). Dimeric 1,4-Benzoquinone Derivatives with Cytotoxic Activities from the Marine-Derived Fungus Penicillium sp. L129. Marine Drugs, 17(7), 383. https://doi.org/10.3390/md17070383