Slow-Binding Inhibition of Tyrosinase by Ecklonia cava Phlorotannins
Abstract
1. Introduction
2. Results and Discussion
2.1. Isolation and Identification
2.2. Inhibition of Phlorotannins on Tyrosinase
2.3. Slow-Binding Inhibition
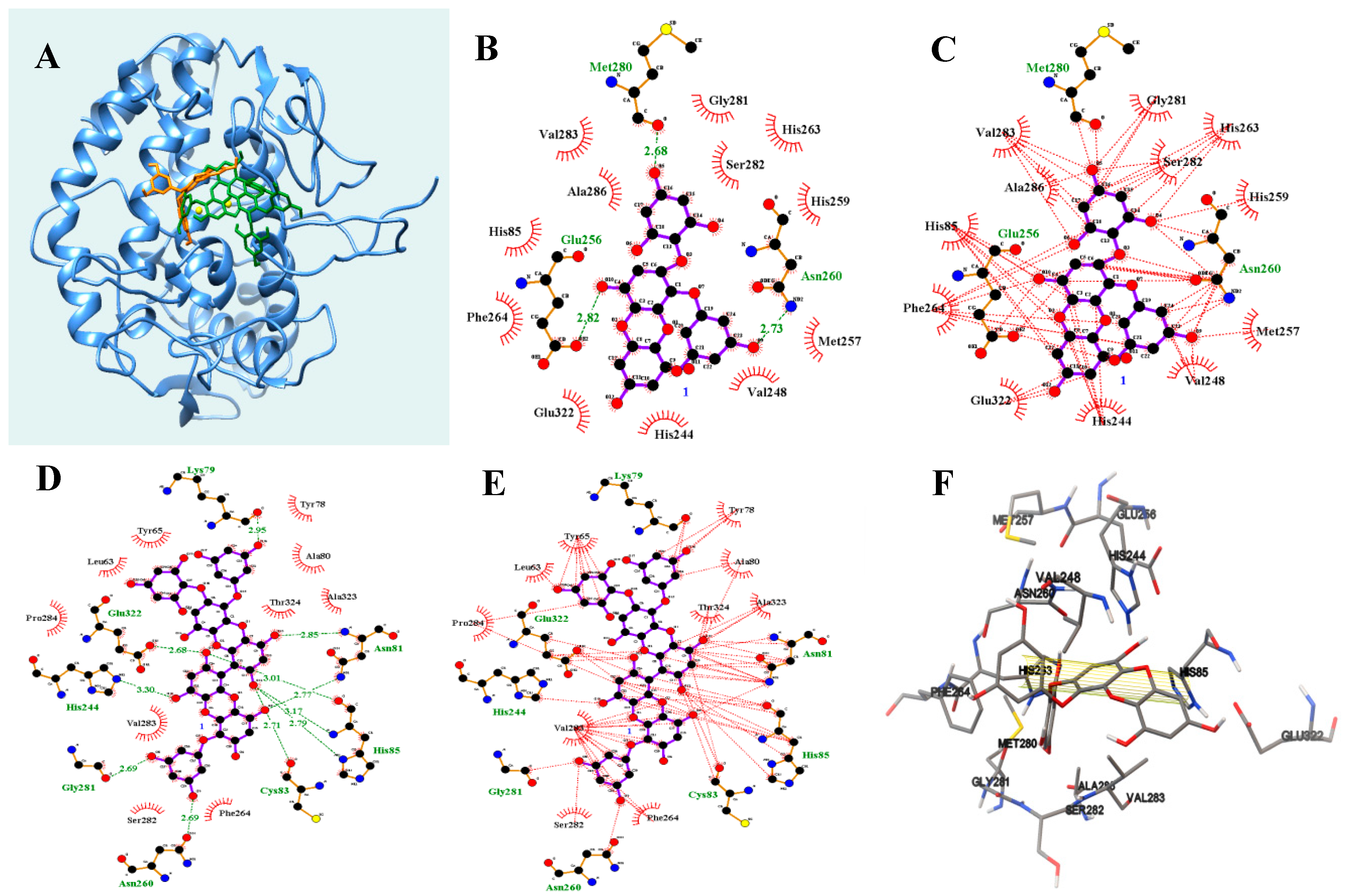
2.4. Molecular Docking
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Plant Material
3.3. Extraction and Isolation
Compound 1
Compound 2
Compound 3
Compound 4
Compound 5
Compound 6
Compound 7
3.4. Tyrosinase Assay
3.5. Slow-Binding Inhibition Analysis
3.6. Molecular Docking
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Şöhretoğlu, D.; Sari, S.; Barut, B.; Özel, A. Tyrosinase inhibition by some flavonoids: Inhibitory activity, mechanism by in vitro and in silico studies. Bioorg. Chem. 2018, 81, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Bukhari, S.N.A.; Jantan, I.; Tan, O.U.; Sher, M.; Naeem-ul-Hassan, M.; Qin, H.-L. Biological activity and molecular docking studies of curcumin-related α,β-unsaturated carbonyl-based synthetic compounds as anticancer agents and mushroom tyrosinase inhibitors. J. Agric. Food Chem. 2014, 62, 5538–5547. [Google Scholar] [CrossRef] [PubMed]
- Ha, Y.M.; Park, Y.J.; Lee, J.Y.; Park, D.; Choi, Y.J.; Lee, E.K.; Kim, J.M.; Kim, J.-A.; Park, J.Y.; Lee, H.J.; et al. Design, synthesis and biological evaluation of 2-(substituted phenyl)thiazolidine-4-carboxylic acid derivatives as novel tyrosinase inhibitors. Biochimie 2012, 94, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, G.; Yan, J.; Gong, D. Inhibitory effect of morin on tyrosinase: Insights from spectroscopic and molecular docking studies. Food Chem. 2014, 163, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Lan, W.-C.; Tzeng, C.-W.; Lin, C.-C.; Yen, F.-L.; Ko, H.-H. Prenylated flavonoids from Artocarpus altilis: Antioxidant activities and inhibitory effects on melanin production. Phytochemistry 2013, 89, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-M.; Chen, C.-Y.; Chen, C.-Y.; Ho, M.-L.; Chou, Y.-T.; Chang, H.-C.; Lee, C.-H.; Wang, C.-Z.; Chu, I.-M. (-)-N-Formylanonaine from Michelia alba as a human tyrosinase inhibitor and antioxidant. Bioorg. Med. Chem. 2010, 18, 5241–5247. [Google Scholar] [CrossRef] [PubMed]
- Zolghadri, S.; Bahrami, A.; Khan, M.T.H.; Munoz-Munoz, J.; Garcia-Molina, F.; Garcia-Canovas, F.; Saboury, A.A. A comprehensive review on tyrosinase inhibitors. J. Enzyme Inhib. Med. Chem. 2019, 34, 279–309. [Google Scholar] [CrossRef]
- Lee, J.; Jun, M. Dual BACE1 and cholinesterase inhibitory effects of phlorotannins from Ecklonia cava-An in vitro and in silico study. Mar. Drugs 2019, 17, 91. [Google Scholar] [CrossRef]
- Yang, H.; Lee, S.Y.; Lee, S.R.; Pyun, B.-J.; Kim, H.J.; Lee, Y.H.; Kwon, S.W.; Suh, D.H.; Lee, C.H.; Hong, E.-J.; et al. Therapeutic effect of Ecklonia cava extract in letrozole-induced polycystic ovary syndrome rats. Front. Pharmacol. 2018, 9, 1325. [Google Scholar] [CrossRef]
- Oh, S.; Son, M.; Lee, H.S.; Kim, H.-S.; Jeon, Y.-J.; Byun, K. Protective effect of pyrogallol-phloroglucinol-6,6-bieckol from Ecklonia cava on monocyte-associated vascular dysfunction. Mar. Drugs 2018, 16, 441. [Google Scholar] [CrossRef]
- Yang, Y.-I.; Woo, J.-H.; Seo, Y.-J.; Lee, K.-T.; Lim, Y.; Choi, J.-H. Protective effect of brown alga not italic phlorotannins against hyperinflammatory responses in lipopolysaccharide-induced sepsis models. J. Agric. Food Chem. 2016, 64, 570–578. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.S.; Kim, H.R.; Byun, D.S.; Son, B.W.; Nam, T.J.; Choi, J.S. Tyrosinase inhibitors isolated from the edible brown alga Eckloina stolonifera. Arch. Pharmcal. Res. 2004, 27, 1226–1232. [Google Scholar] [CrossRef]
- Yoon, N.Y.; Eom, T.-K.; Kim, M.-M.; Kim, S.-K. Inhibitory effect of phlorotannins isolated from Ecklonia cava on mushroom tyrosinase activity and melanin formation in mouse B16F10 melanoma cells. J. Agric. Food Chem. 2009, 57, 4124–4129. [Google Scholar] [CrossRef] [PubMed]
- Wijesinghe, W.A.J.P.; Ko, S.-C.; Jeon, Y.-J. Effect of phlorotannins isolated from Ecklonia cava on angiotensin enzyme (ACE) inhibitory activity. Nutr. Res. Pract. 2011, 5, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Ko, J.Y.; Oh, J.Y.; Kim, C.Y.; Lee, H.J.; Kim, J.; Jeon, Y.J. Preparative isolation and purification of phlorotannins from Ecklonia cava using centrifugal partition chromatography by one-step. Food Chem. 2014, 158, 433–437. [Google Scholar] [CrossRef]
- Park, J.Y.; Kim, J.H.; Kwon, J.M.; Kwon, H.J.; Jeong, H.J.; Kim, Y.M.; Kim, D.; Lee, W.S.; Ryu, Y.B. Dieckol, a SARS-CoV 3CL pro inhibitor, isolated from the edible brown algae Ecklonia cava. Bioorg. Med. Chem. 2013, 21, 3730–3737. [Google Scholar] [CrossRef] [PubMed]
- Fukuyama, Y.; Kodama, M.; Miura, I.; Kinzyo, Z.; Mori, H.; Nakayama, Y.; Takahashi, M. Anti-plasmin Inhibitor. V.: Structures of novel dimeric eckols isolated from the brown alga Ecklonia kurome OKAMURA. Chem. Pharm. Bull. 1989, 37, 2438–2440. [Google Scholar] [CrossRef]
- Sugiura, Y.; Matsuda, K.; Yamada, Y.; Nishikawa, M.; Shioya, K.; Katsuzaki, H.; Imai, K.; Amano, H. Anti-allergic phlorotannins from the edible brown alga, Eisenia arborea. Food Sci. Technol. Res. 2007, 13, 54–60. [Google Scholar] [CrossRef]
- Kang, S.-M.; Heo, S.-J.; Kim, K.-N.; Yang, S.-H.; Kim, A.-D.; Jeon, Y.-J. Molecular docking studies of a phlorotannin, dieckol isolated from Eckloina cava with tyrosinase inhibitory activity. Bioorg. Med. Chem. 2012, 20, 311–316. [Google Scholar] [CrossRef]
- Masson, P.; Lushchekina, S.V. Slow-binding inhibition of cholinesterase, pharmacological and toxicological relevance. Arch. Biochem. Biophys. 2016, 593, 60–68. [Google Scholar] [CrossRef]
- Menon, V.; Rao, M. Slow-tight binding inhibition of pepsin by an aspartic protease inhibitor from Streptomyces sp. MBR04. Inter. J. Biol. Macromol. 2012, 51, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.-L.; Lim, G.T.; Yin, S.-J.; Lee, J.; Si, Y.-X.; Yang, J.-M.; Park, Y.-D.; Qian, G.-Y. The inhibitory effect of pyrogallol on tyrosinase activity and structure: Integration study of inhibition kinetics with molecular dynamics simulation. Int. J. Biol. Macromol. 2019, 121, 463–471. [Google Scholar] [CrossRef] [PubMed]




| Inhibitory Activity of Compounds on Tyrosinase a | |||
|---|---|---|---|
| 100 μM (%) | IC50 (μM) | Binding Mode (Ki, μM) | |
| 1 | 26.7 ± 1.0 | N.T. c | N.T. c |
| 2 | 86.0 ± 4.7 | 13.5 ± 0.1 | Reported as non-competitive d |
| 3 | 94.7 ± 1.4 | 7.0 ± 0.2 | Competitive (8.2 ± 1.1) |
| 4 | 57.1 ± 3.3 | 66.4 ± 0.1 | N.T. c |
| 5 | 86.9 ± 0.9 | 8.8 ± 0.1 | Competitive (5.8 ± 0.8) |
| 6 | 41.2 ± 4.3 | N.T. c | N.T. c |
| 7 | 43.3 ± 1.5 | N.T. c | N.T. c |
| Kojic acid b | 25.0 ± 0.4 | ||
| Compound | K3 (mMs−1) | k4 (s−1) | K5 (s−1) | K6 (s−1) | kapp i (μM) |
|---|---|---|---|---|---|
| 3 | 0.0002 | 0.0013 | - | - | 6.5 μM |
| 5 | - | - | 0.0047 | 0.0003 | 4.4 μM |
| Compound | Hydrogen Bonds (Å) | Binding Energy (kcal/mol) |
|---|---|---|
| 3 | Glu256(2.82), Asn260(2.73), Met280(2.68) | −6.59 |
| 5 | Lys79(2.95), Asn81(2.85), 2.77), Cys83(2.71), His85(2.77, 3.17, 2.79), His244(3.30), Asn260(2.69), Gly281(2.69), Glu322(2.68) | −6.68 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.H.; Lee, S.; Park, S.; Park, J.S.; Kim, Y.H.; Yang, S.Y. Slow-Binding Inhibition of Tyrosinase by Ecklonia cava Phlorotannins. Mar. Drugs 2019, 17, 359. https://doi.org/10.3390/md17060359
Kim JH, Lee S, Park S, Park JS, Kim YH, Yang SY. Slow-Binding Inhibition of Tyrosinase by Ecklonia cava Phlorotannins. Marine Drugs. 2019; 17(6):359. https://doi.org/10.3390/md17060359
Chicago/Turabian StyleKim, Jang Hoon, Sunggun Lee, Saerom Park, Ji Soo Park, Young Ho Kim, and Seo Young Yang. 2019. "Slow-Binding Inhibition of Tyrosinase by Ecklonia cava Phlorotannins" Marine Drugs 17, no. 6: 359. https://doi.org/10.3390/md17060359
APA StyleKim, J. H., Lee, S., Park, S., Park, J. S., Kim, Y. H., & Yang, S. Y. (2019). Slow-Binding Inhibition of Tyrosinase by Ecklonia cava Phlorotannins. Marine Drugs, 17(6), 359. https://doi.org/10.3390/md17060359





