Cloning, Expression, and Characterization of a New Glycosaminoglycan Lyase from Microbacterium sp. H14
Abstract
1. Introduction
2. Results
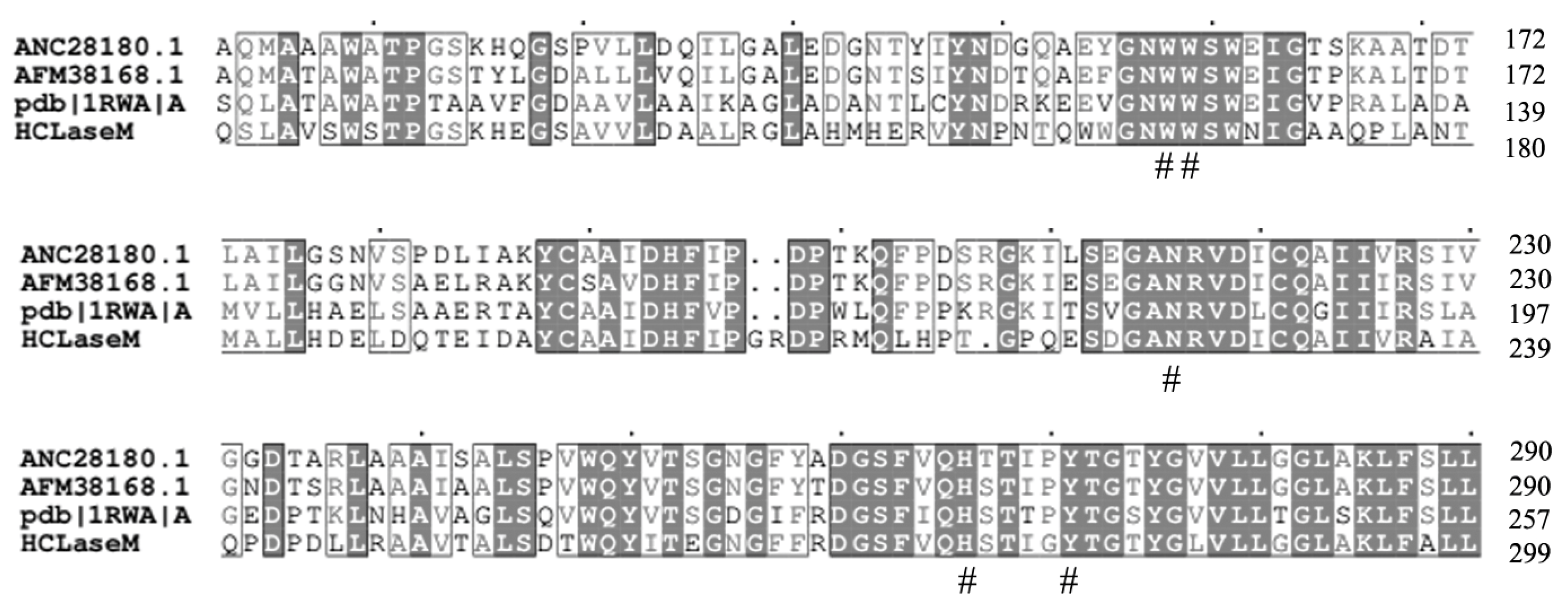
2.1. Cloning and Sequence Analysis of HCLaseM Gene
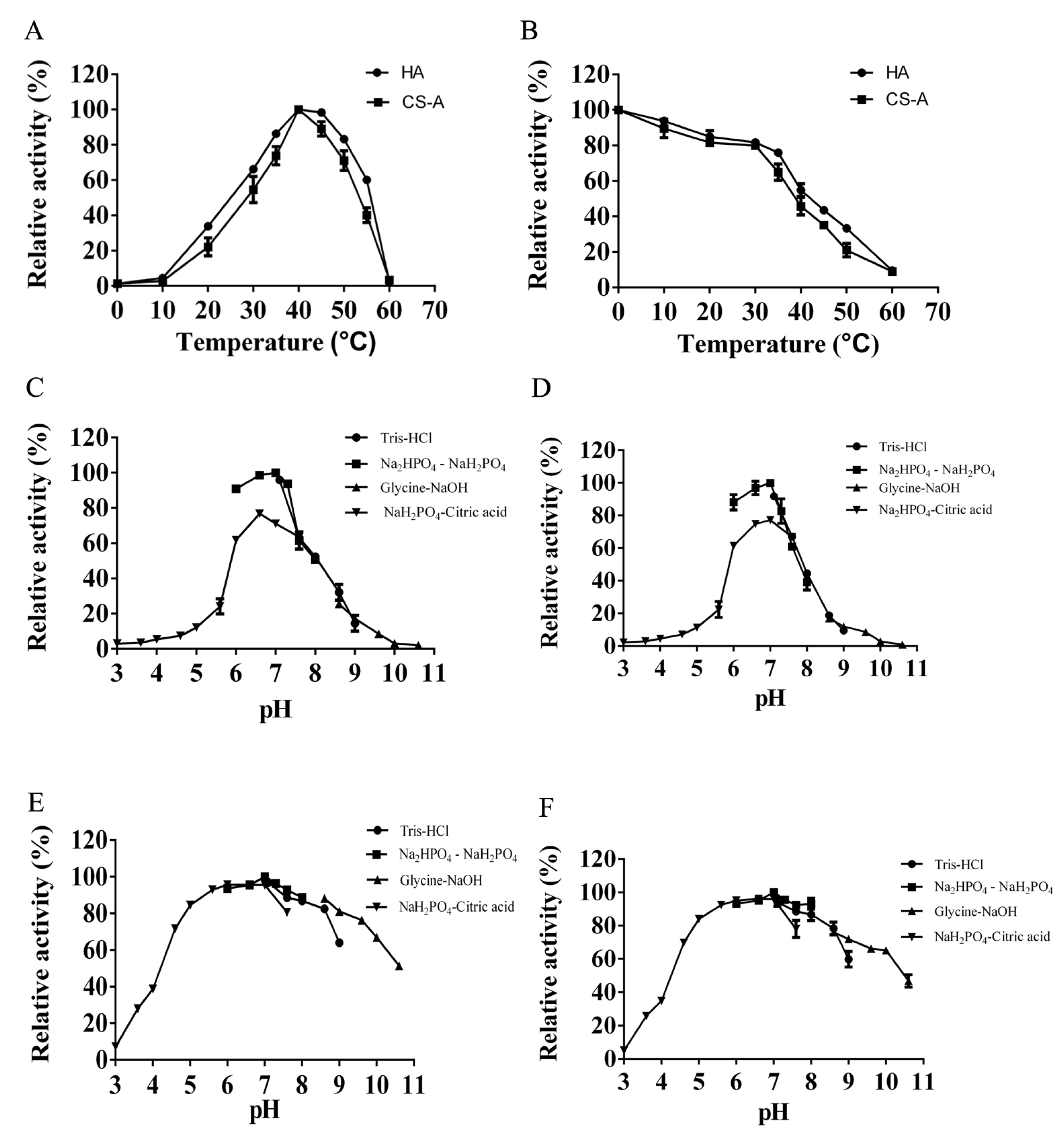
2.2. Purification and Biochemical Characterization of HCLaseM
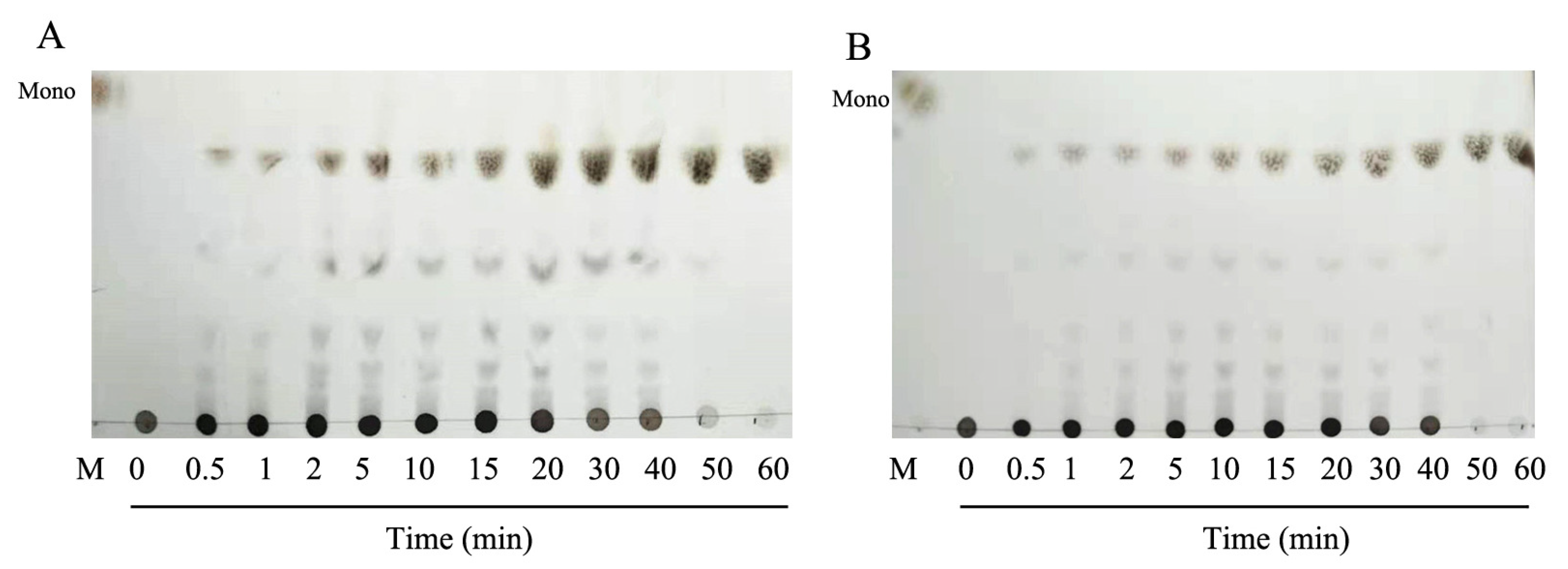
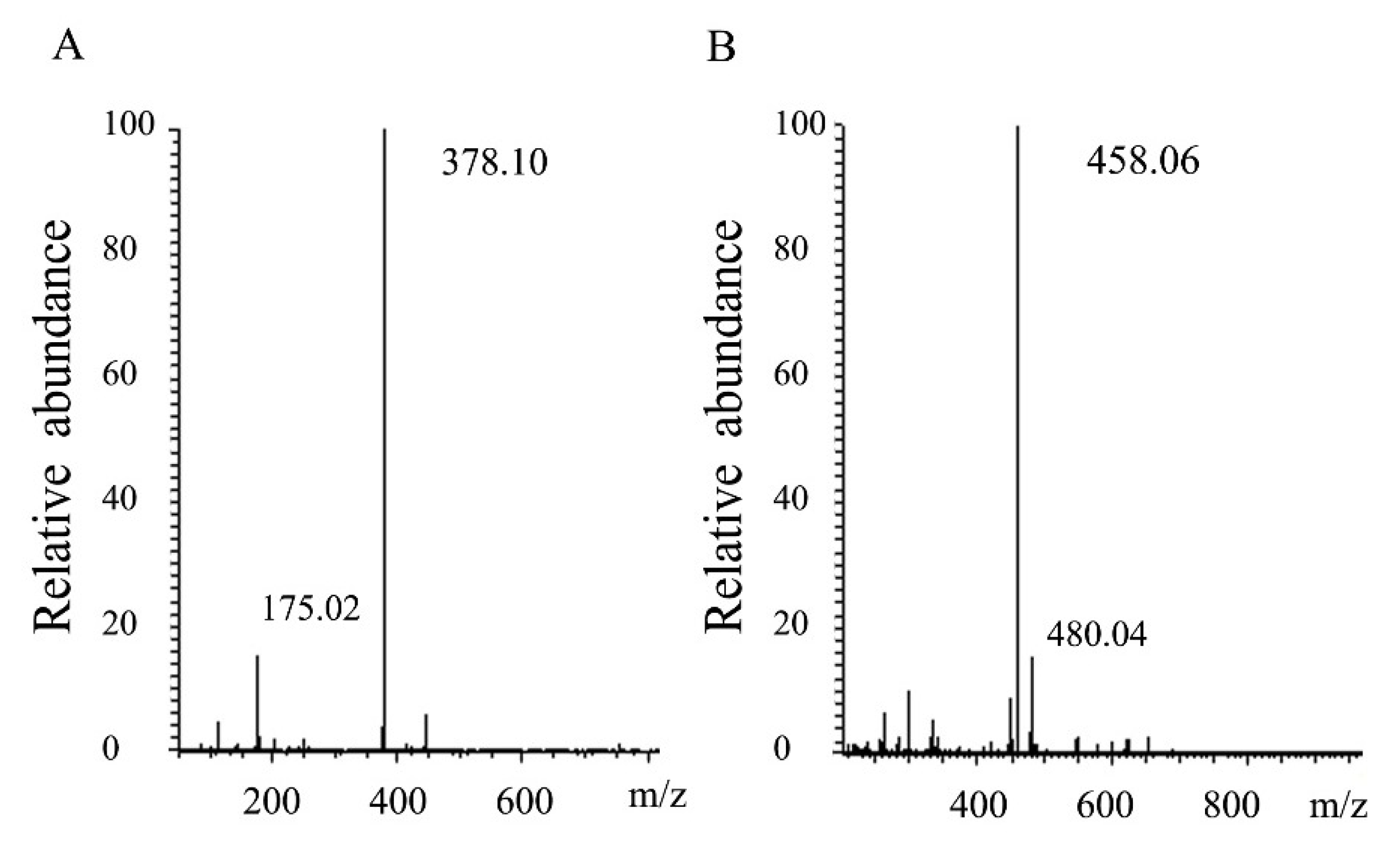
2.3. Degradation Pattern and End Production of HCLaseM
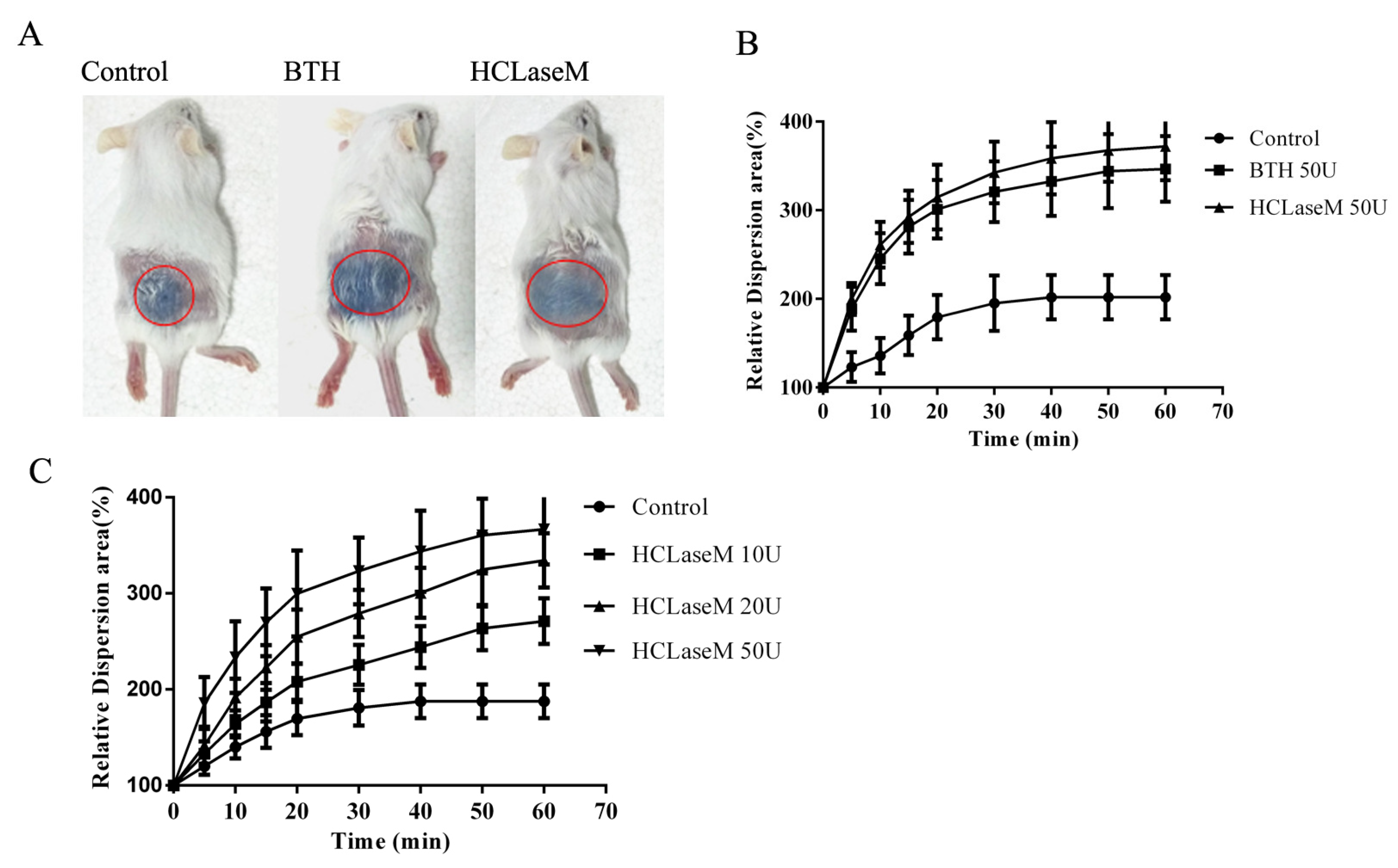
2.4. Effect of Hyaluronidase on the Diffusion of Trypan Blue
3. Discussion
4. Materials and Methods
4.1. Material and Animals
4.2. Cloning and Sequence Analysis of GAG Lyase Gene
4.3. Expression and Purification of Recombinant HCLaseM
4.4. Assay of HCLaseM Activity
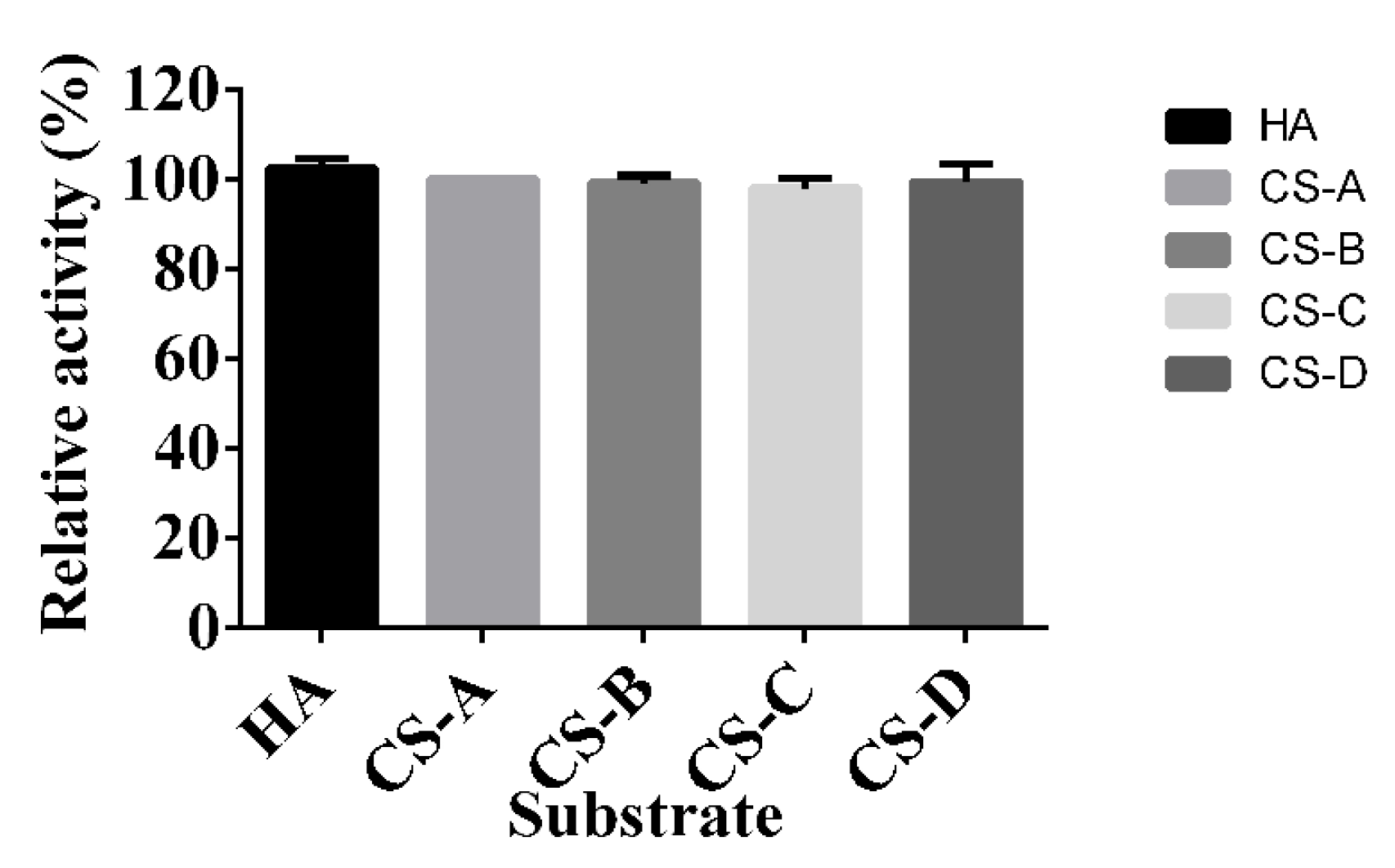
4.5. Substrate Specificity of HCLaseM
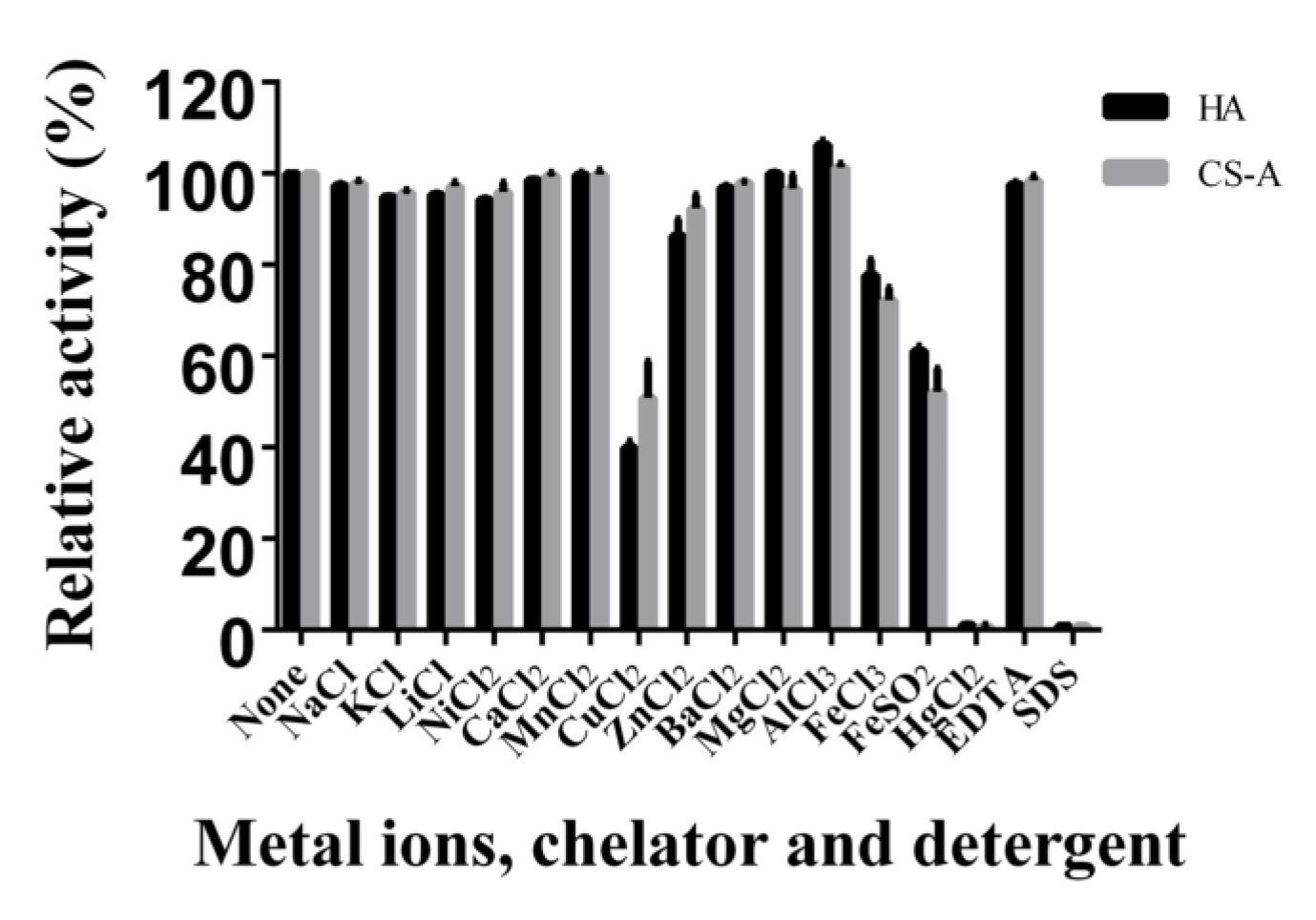
4.6. Effects of Temperature, pH, Metal Ions, Chelators, and Detergents on HCLaseM
4.7. Enzymatic Reaction Kinetics of HCLaseM Towards HA and CS-A
4.8. Degradation Pattern of HCLaseM
4.9. End Products of HCLaseM
4.10. Effect of Hyaluronidase on the Diffusion of Trypan Blue
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Linhardt, R.J.; Toida, T. Role of glycosaminoglycans in cellular communication. Acc. Chem. Res. 2004, 37, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Karbownik, M.S.; Nowak, J.Z. Hyaluronan: Towards novel anti-cancer therapeutics. Pharmacol. Rep. 2013, 65, 1056–1074. [Google Scholar] [CrossRef]
- Varki, A.J.G. Biological roles of oligosaccharides: All of the theories are correct. Glycobiology 1993, 3, 97–130. [Google Scholar] [CrossRef] [PubMed]
- Vlodavsky, I.; Fuks, Z.; Ishai-Michaeli, R.; Bashkin, P.; Levi, E.; Korner, G.; Bar-Shavit, R.; Klagsbrun, M. Extracellular matrix-resident basic fibroblast growth factor: Implication for the control of angiogenesis. J. Cell Biochem. 1991, 45, 167–176. [Google Scholar] [CrossRef]
- Takahashi, Y.; Li, L.L.; Kamiryo, M.; Asteriou, T.; Moustakas, A.; Yamashita, H.; Heldin, P. Hyaluronan fragments induce endothelial cell differentiation in a CD44- and CXCL1/GRO1-dependent manner. J. Biol. Chem. 2005, 280, 24195–24204. [Google Scholar] [CrossRef]
- Rooney, P.; Wang, M.; Kumar, P.; Kumar, S. Angiogenic oligosaccharides of hyaluronan enhance the production of collagens by endothelial cells. J. Cell Sci. 1993, 105, 213–218. [Google Scholar] [PubMed]
- West, D.C.; Kumar, S. The effect of hyaluronate and its oligosaccharides on endothelial cell proliferation and monolayer integrity. Exp. Cell. Res. 1989, 183, 179–196. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, J.; Liu, C.; Song, C.; Li, P.; Yin, F.; Xiao, Y.; Li, J.; Jiang, W.; Zong, A.; et al. Protective effects of low molecular weight chondroitin sulfate on amyloid beta (Aβ)-induced damage in vitro and in vivo. Neuroscience. 2015, 305, 169–182. [Google Scholar] [CrossRef]
- Ma, Q.; Dudas, B.; Hejna, M.; Cornelli, U.; Lee, J.M.; Lorens, S.; Mervis, R.; Hanin, I.; Fareed, J. The blood-brain barrier accessibility of a heparin-derived oligosaccharides C3. Thromb. Res. 2002, 105, 447–453. [Google Scholar] [CrossRef]
- Zhu, B.; Chen, M.; Yin, H.; Du, Y.; Ning, L. Enzymatic hydrolysis of alginate to produce oligosaccharides by a new purified endo-type alginate lyase. Mar. Drugs. 2016, 14, 108. [Google Scholar] [CrossRef]
- Bradbury, E.J.; Moon, L.D.F.; Popat, R.J.; King, V.R.; Bennett, G.S.; Patel, P.N.; Fawcett, J.W.; Mcmahon, S.B. Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature 2002, 416, 636–640. [Google Scholar] [CrossRef] [PubMed]
- Henke, C.A.; Roongta, U.; Mickelson, D.J.; Knutson, J.R.; Mccarthy, J.B. CD44-related chondroitin sulfate proteoglycan, a cell surface receptor implicated with tumor cell invasion, mediates endothelial cell migration on fibrinogen and invasion into a fibrin matrix. J. Clin. Invest. 1996, 97, 2541. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Grummer, S.E.; Kriegel, D.; Marmur, E. Hyaluronidase. Drematol. Surg. 2010, 36, 1071–1077. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xie, B.; Wei, W.; Hui, M.; Su, Z. Design and preparation of chimeric hyaluronidase as a chaperone for the subcutaneous administration of biopharmaceuticals. Biochem. Eng. J. 2016, 112, 32–41. [Google Scholar] [CrossRef]
- Yin, F.X.; Wang, F.S.; Sheng, J.Z. Uncovering the catalytic direction of chondroitin AC exolyase: From the reducing end towards the non-reducing end. J. Biol. Chem. 2016, 291, 4399–4406. [Google Scholar] [CrossRef]
- Kale, V.; Fridjonsson, O.; Jonsson, J.O.; Kristinsson, H.G.; Omarsdottir, S.; Hreggvidsson, G.O. Chondroitin lyase from a marine Arthrobacter sp MAT3885 for the production of chondroitin sulfate disaccharides. Mar. Biotechnol. 2015, 17, 479–492. [Google Scholar] [CrossRef]
- Lunin, V.V.; Li, Y.; Linhardt, R.J.; Miyazono, H.; Kyogashima, M.; Kaneko, T.; Bell, A.W.; Cygler, M. High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: An enzyme–substrate complex defines the catalytic mechanism. J. Mol. Biol. 2004, 337, 367–386. [Google Scholar] [CrossRef]
- Hong, S.W.; Kim, B.T.; Shin, H.Y.; Kim, W.S.; Lee, K.S.; Kim, Y.S.; Kim, D.H. Purification and characterization of novel chondroitin ABC and AC lyases from Bacteroides stercoris HJ-15, a human intestinal anaerobic bacterium. Eur. J. Biochem. 2002, 269, 2934–2940. [Google Scholar] [CrossRef]
- Kurata, A.; Matsumoto, M.; Kobayashi, T.; Deguchi, S.; Kishimoto, N. Hyaluronate Lyase of a Deep-Sea Bacillus niacini. Mar. Biotechnol. 2015, 17, 277–284. [Google Scholar] [CrossRef]
- Guo, X.; Shi, Y.; Sheng, J.; Wang, F. A novel hyaluronidase produced by Bacillus sp. A50. PLoS ONE 2014, 9, e94156. [Google Scholar] [CrossRef]
- Han, W.; Wang, W.; Zhao, M.; Sugahara, K.; Li, F. A novel eliminase from a marine bacterium that degrades hyaluronan and chondroitin sulfate. J. Biol. Chem. 2014, 289, 27886–27898. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Zhang, J.; Zhang, J.; Jiang, Y.; Shen, Z.; Guan, H.; Jiang, X. Purification and characterization of chondroitinase ABC from Acinetobacter sp. C26. Int. J. Biol. Macromol. 2017, 95, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Yang, S.; Fu, X.; Xie, W.; Li, L.; Liu, Z.; Mou, H.; Zhu, C. Expression, Purification and Characterization of Chondroitinase AC II from marine bacterium Arthrobacter sp. CS01. Mar. Drugs. 2019, 17, 185. [Google Scholar] [CrossRef]
- Tork, S.E.; Shahein, Y.E.; El-Hakim, A.E.; Abdel-Aty, A.M.; Aly, M.M. Purification and partial characterization of serine-metallokeratinase from a newly isolated Bacillus pumilus NRC21. Int. J. Biol. Macromol. 2016, 86, 189–196. [Google Scholar] [CrossRef]
- Muchmore, D.B.; Vaughn, D.E. Review of the mechanism of action and clinical efficacy of recombinant human hyaluronidase coadministration with current prandial insulin formulations. J. Diabetes Sci. Technol. 2010, 4, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Jedrzejas, M.; Mello, L.; de, G.B.; Li, S. Mechanism of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase. Structure of complexes with the substrate. J. Biol. Chem. 2002, 277, 28287–28297. [Google Scholar] [CrossRef] [PubMed]
- Nukui, M.; Taylor, K.B.; McPherson, D.T.; Shigenaga, M.K.; Jedrzejas, M. The function of hydrophobic residues in the catalytic cleft of Streptococcus pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms. J. Biol. Chem. 2003, 278, 3079–3088. [Google Scholar] [CrossRef]
- Li, S.; Jedrzejas, M. Hyaluronan binding and degradation by Streptococcus agalactiae hyaluronate lyase. J. Biol. Chem. 2001, 276, 41407–41416. [Google Scholar] [CrossRef]
- Allen, A.G.; Lindsay, H.; Seilly, D.; Bolitho, S.; Peters, S.E.; Maskell, D.J. Identification and characterisation of hyaluronate lyase from Streptococcus suis. Microb. Pathog. 2004, 36, 327–335. [Google Scholar] [CrossRef]
- Linn, S.; Chan, T.; Lipeski, L.; Salyers, A.A. Isolation and characterization of two chondroitin lyases from Bacteroides thetaiotaomicron. J. Bacteriol. 1983, 156, 859–866. [Google Scholar]
- Prabhakar, V.; Capila, I.; Bosques, C.J.; Pojasek, K.; Sasisekharan, R. Chondroitinase ABC I from Proteus vulgaris: Cloning, recombinant expression and active site identification. Biochem. J. 2005, 386, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Hamai, A.; Hashimoto, N.; Mochizuki, H.; Kato, F.; Makiguchi, Y.; Horie, K.; Suzuki, S. Two distinct chondroitin sulfate ABC lyases. An endoeliminase yielding tetrasaccharides and an exoeliminase preferentially acting on oligosaccharides. J. Biol. Chem. 1997, 272, 9123–9130. [Google Scholar] [CrossRef] [PubMed]
- Shaya; Hahn, D.; Park, B.S.; Sim, N.Y.; Kim, J.S.; Cygler, Y.S. Characterization of chondroitin sulfate lyase ABC from Bacteroides thetaiotaomicron WAL2926. Biochemistry 2008, 47, 6650–6661. [Google Scholar] [CrossRef] [PubMed]
- Gu, K.F.; Linhardt, R.J.; Laliberté, M.; Zimmermann, J. Purification, characterization and specificity of chondroitin lyases and glycuronidase from Flavobacterium heparinum. Biochem. J. 1996, 312, 569–577. [Google Scholar] [CrossRef]
- Sun, X.; Wang, Z.; Bi, Y.; Wang, Y.; Liu, H. Genetic and functional characterization of the hyaluronate lyase HylB and the beta-N-acetylglucosaminidase HylZ in Streptococcus zooepidemicus. Curr. Microbiol. 2015, 70, 35–42. [Google Scholar] [CrossRef]
- Zhu, C.; Zhang, J.; Li, L.; Zhang, J.; Jiang, Y.; Shen, Z.; Guan, H.; Jiang, X. Biotechnology, Purification and Characterization of Hyaluronate Lyase from Arthrobacter globiformis A152. Appl. Biochem. Biotechnol. 2017, 182, 216–228. [Google Scholar] [CrossRef]
- Fu, J.; Jiang, Z.; Chang, J.; Han, B.; Liu, W.; Peng, Y. Purification, characterization of Chondroitinase ABC from Sphingomonas paucimobilis and in vitro cardiocytoprotection of the enzymatically degraded CS-A. Int. J. Biol. Macromol. 2018, 115, 737–745. [Google Scholar] [CrossRef]
- Benitez, A.; Yates, T.J.; Lopez, L.E.; Cerwinka, W.H.; Bakkar, A.; Lokeshwar, V.B. Targeting hyaluronidase for cancer therapy: Antitumor activity of sulfated hyaluronic acid in prostate cancer cells. Cancer Res. 2011, 71, 4085–4095. [Google Scholar] [CrossRef]
- Vogel, H.G.; Vogel, W.H.; Schölkens, B.A.; Sandow, J.; Müller, G.; Vogel, W.F.; Chapter, Q. Guidelines for the Care and Use of Laboratory Animals; Springer: Heidelberg, Germany, 2002; pp. 1369–1382. [Google Scholar]
- Yamagata, T.; Saito, H.; Habuchi, O.; Suzuki, S. Purification and properties of bacterial chondroitinases and chondrosulfatases. J. Biol. Chem. 1968, 243, 1523–1535. [Google Scholar]
- Frost, G.I.; Stern, R. A microtiter-based Assay for hyaluronidase activity not requiring specialized reagents. Anal. Biochem. 1997, 251, 263–269. [Google Scholar] [CrossRef]

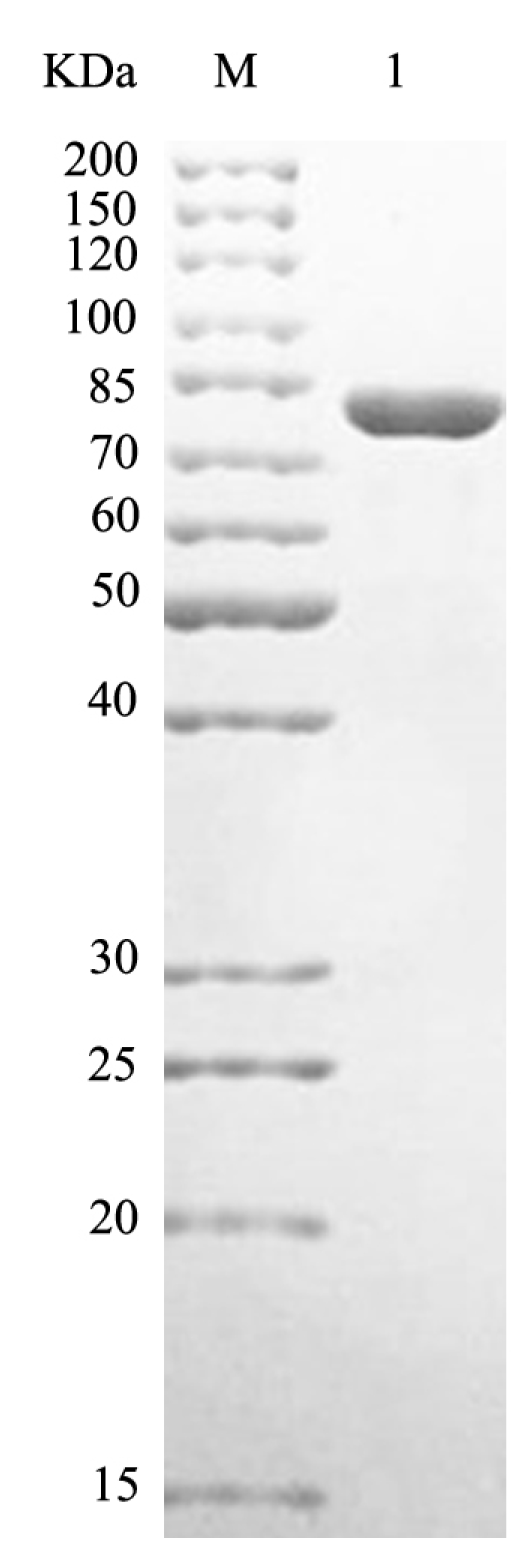
| Step | Total Activity (U) | Total Protein (mg) | Specific Activity (U/mg Protein) | Fold Purification | Yield (%) |
|---|---|---|---|---|---|
| Fermentation media | 7671 | 299.4 | 25.6 | 1 | 100 |
| Nickel column | 4290 | 15.4 | 278.3 | 10.9 | 55.9 |
| Substrate | Km (mg/mL) | Vmax (μM/min) |
|---|---|---|
| HA | 0.419 ± 0.019 | 0.0246 ± 0.0003 |
| CS-A | 0.478 ± 0.015 | 0.0264 ± 0.0002 |
| Substrate | ||||||
|---|---|---|---|---|---|---|
| Enzyme | Source | CS-A | CS-B | CS-C | HA | Reference |
| HCLaseM | Microbacterium sp. H14 | 100 | 99 | 98 | 102 | This study |
| ChSase ABC | BacteroidesStercoris1 | 100 | 32 | 40 | 0 | [18] |
| ChSase AC | B. Stercoris1 | 100 | 0 | 46 | 67 | [18] |
| ChSase ABC | FlavobacteriumHeparinum1 | 100 | 100 | 100 | 0 | [18] |
| ChSase AC | F. Heparinum1 | 100 | 0 | 110 | 107 | [18] |
| ChSase ABC | B.Thetaiotaomicron1 | 100 | 13–16 | 80–130 | 10–30 | [18] |
| ChSase ABC | Proteus. Vulgaris1 | 100 | 34 | 100 | 60 | [18] |
| BniHL | Bacillus niacin2 | 100 | 0 | 68 | 227 | [19] |
| HAase-B | Bacillus sp. A50 2 | 100 | 0 | 36 | 256 | [20] |
| HCLase | Vibrio sp. FC509 2 | 100 | 0 | 96 | 222 | [21] |
| ChSase ABC | Acinetobacter sp. C26 2 | 100 | 96 | 95 | 133 | [22] |
| ChSase AC II | Arthrobacter sp. CS01 2 | 100 | 0 | 86 | 255 | [23] |
| Enzyme | Source | Molecular Mass (kDa) | Optimal pH | Optimal Temperature (°C) | Action Pattern | References |
|---|---|---|---|---|---|---|
| HCLaseM | Microbacterium sp. H14 | 85.9 | 7 | 40 | Endo | This study |
| AsChnAC | Arthrobacter sp. | N/A | 7.2 | 37 | Exo | [15] |
| ChoA1 | Arthrobacter sp. MAT3885 | 83.4 | 6.0–7.5 | 40 | N/A | [16] |
| ChSase AC | Arthrobacter aurescens | N/A | N/A | N/A | Exo | [17] |
| ChSase ABC | Bacteroides stercoris | 116 | 7 | 40 | Exo | [18] |
| ChSase AC | Bacteroides stercoris | 84 | 5.7–6.0 | 45–50 | Endo | [18] |
| BniHL | Bacillus niacin | 120 | 6 | 45 | Endo | [19] |
| HAase-B | Bacillus sp. A50 | 116 | 6.5 | 44 | Endo | [20] |
| HCLase | Vibrio sp. FC509 | 90 | 8 | 30 | Endo | [21] |
| ChSase ABC | Acinetobacter sp. C26 | 76 | 6 | 42 | N/A | [22] |
| ChSase AC II | Arthrobacter sp. CS01 | 100 | 6.5 | 37 | Exo | [23] |
| cABC I | Proteus vulgaris | 112 | 8 | 37 | Endo | [31] |
| cABC II | Proteus vulgaris | 105 | 8 | 37 | Exo | [32] |
| ChonABC | Bacteroides thetaiotaomicron | 115 | 7.6 | 37 | Exo | [33] |
| ChSase AC | Flavobacterium heparinum | 74 | 6.8 | 40 | Endo | [34] |
| HylB | Streptococcus zooepidemicus | N/A | 6 | 37 | N/A | [35] |
| HAase | Arthrobacter globiformis | 74 | 6 | 42 | N/A | [36] |
| ChSase ABC | Sphingomonas paucimobilis | 82 | 6.5 | 40 | N/A | [37] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, J.; Han, X.; Song, G.; Gong, Q.; Yu, W. Cloning, Expression, and Characterization of a New Glycosaminoglycan Lyase from Microbacterium sp. H14. Mar. Drugs 2019, 17, 681. https://doi.org/10.3390/md17120681
Sun J, Han X, Song G, Gong Q, Yu W. Cloning, Expression, and Characterization of a New Glycosaminoglycan Lyase from Microbacterium sp. H14. Marine Drugs. 2019; 17(12):681. https://doi.org/10.3390/md17120681
Chicago/Turabian StyleSun, Junhao, Xu Han, Guanrui Song, Qianhong Gong, and Wengong Yu. 2019. "Cloning, Expression, and Characterization of a New Glycosaminoglycan Lyase from Microbacterium sp. H14" Marine Drugs 17, no. 12: 681. https://doi.org/10.3390/md17120681
APA StyleSun, J., Han, X., Song, G., Gong, Q., & Yu, W. (2019). Cloning, Expression, and Characterization of a New Glycosaminoglycan Lyase from Microbacterium sp. H14. Marine Drugs, 17(12), 681. https://doi.org/10.3390/md17120681





