Exploiting the Feedstock Flexibility of the Emergent Synthetic Biology Chassis Vibrio natriegens for Engineered Natural Product Production
Abstract
1. Introduction
2. Results
2.1. Investigating V. natriegens Feedstock Flexibility
2.2. Heterologous Production of Beta-carotene in V. natriegens
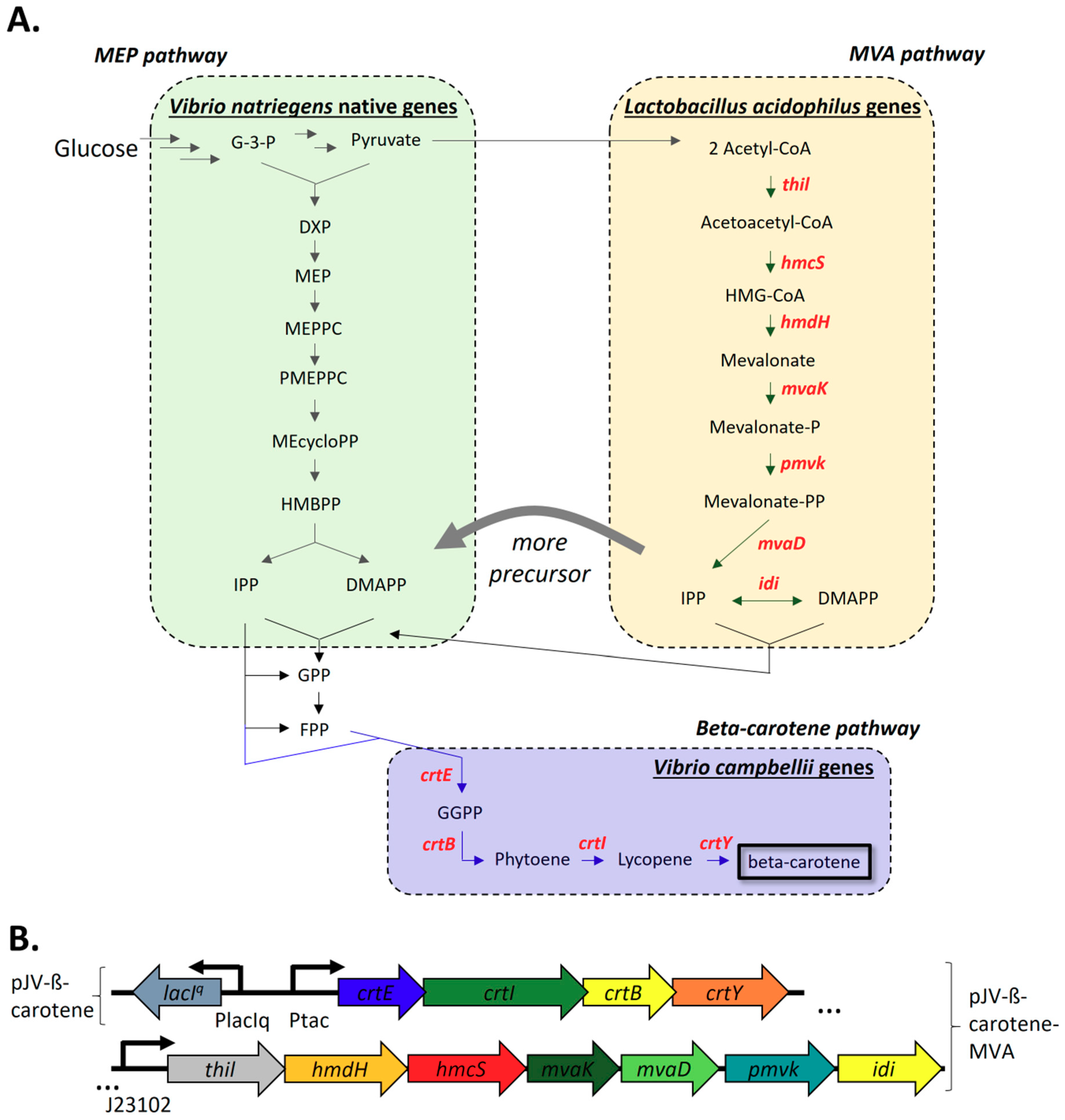
2.2.1. Construction and Expression of The V. Campbellii Beta-Carotene Biosynthesis Pathway and L. acidophilus Mevalonate Pathway
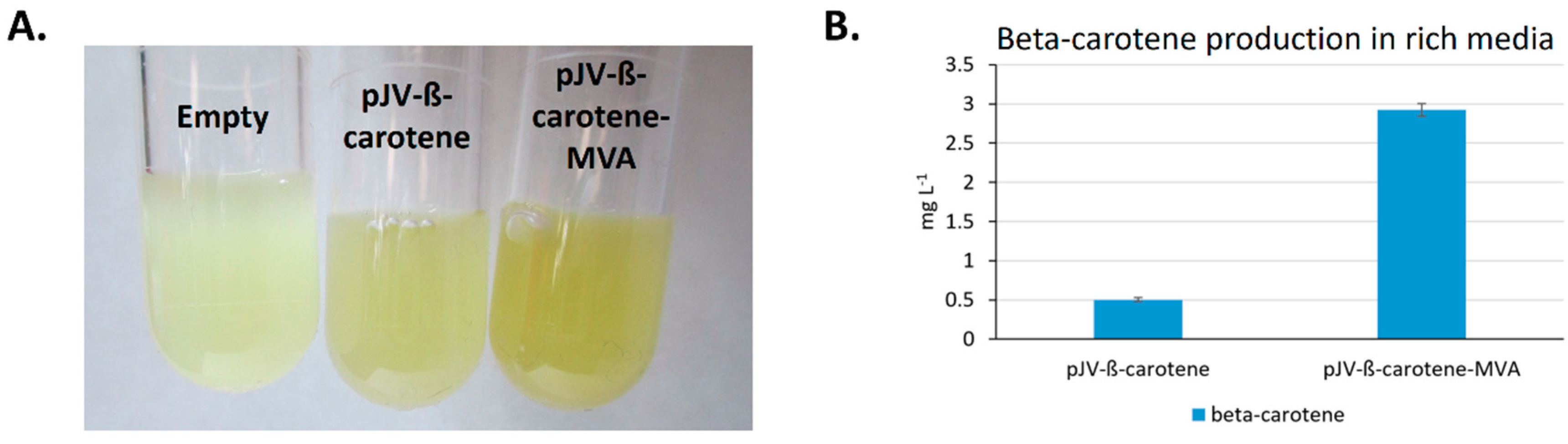
2.2.2. Heterologous Production of Beta-Carotene in Nutrient Rich Medium
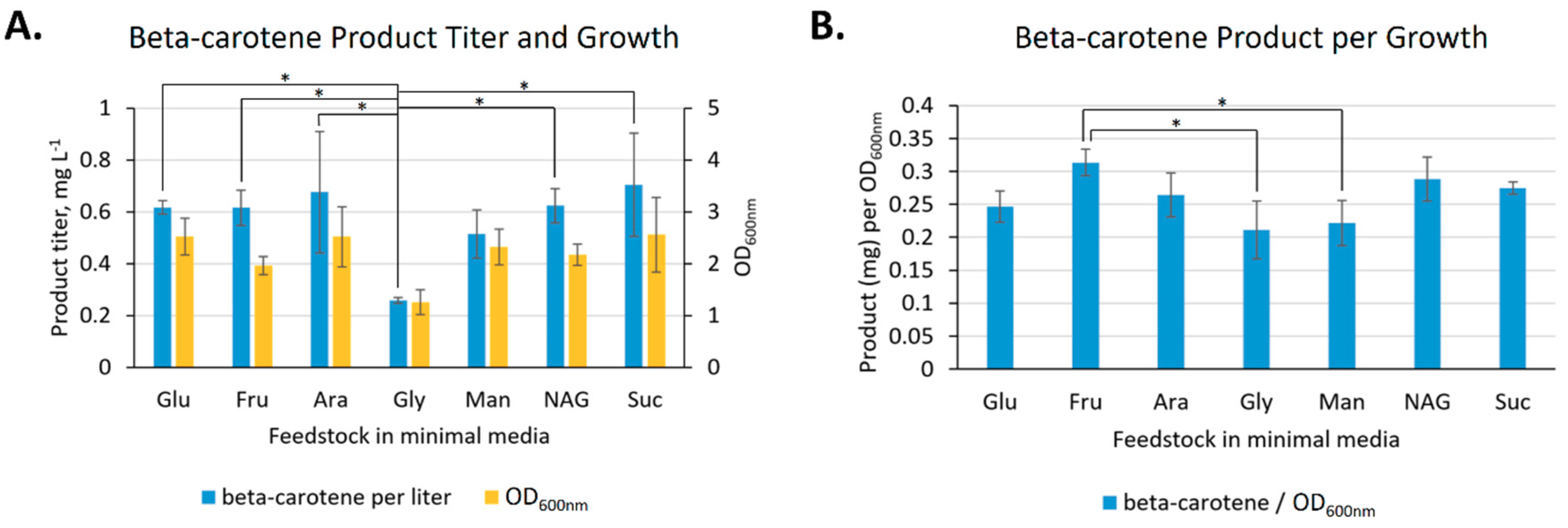
2.2.3. Feedstock Flexibility of V. natriegens for Producing Beta-carotene
2.3. Heterologous Production of Violacein in V. natriegens
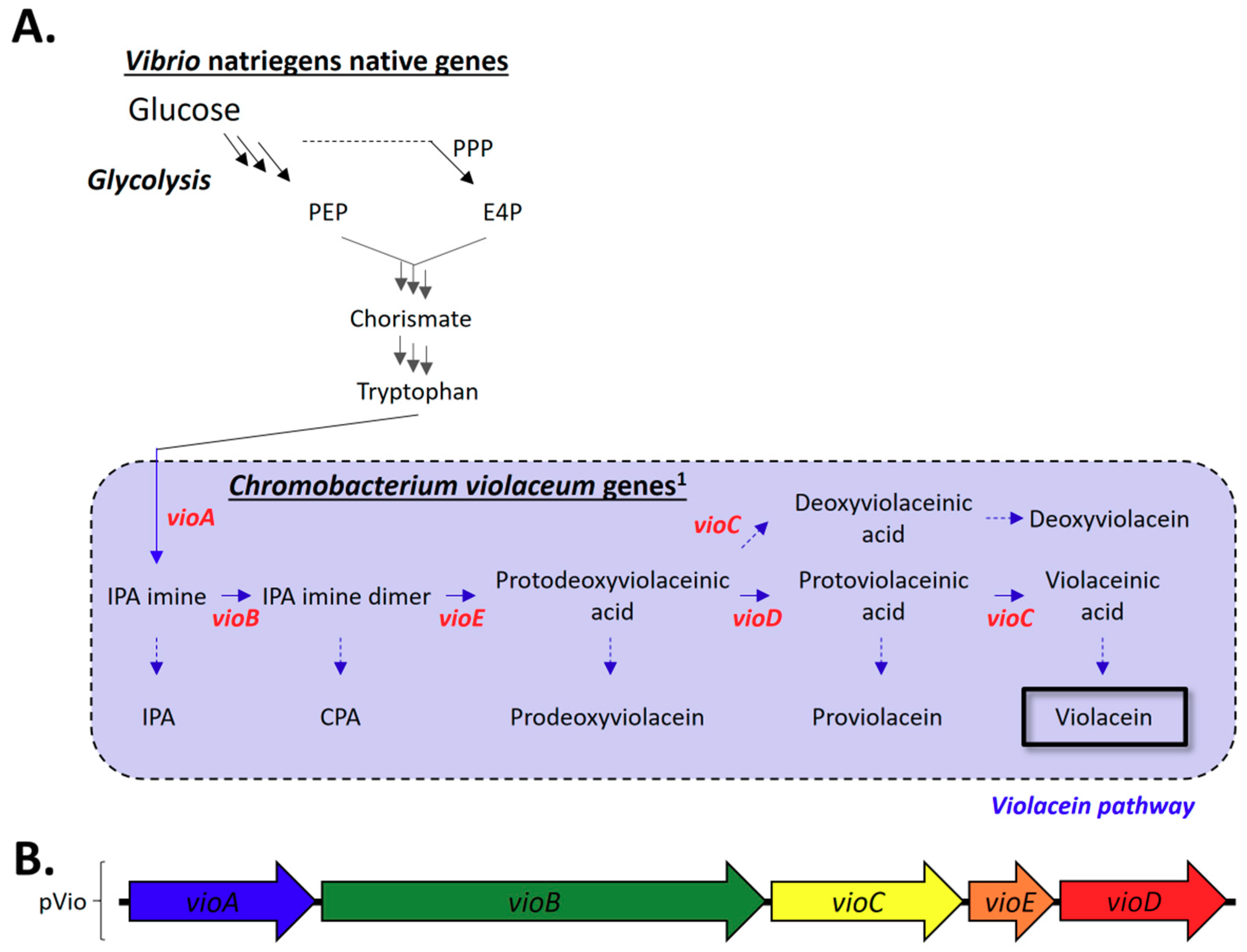
2.3.1. Construction of the C. violaceum Violacein Biosynthesis Pathway in V. natriegens
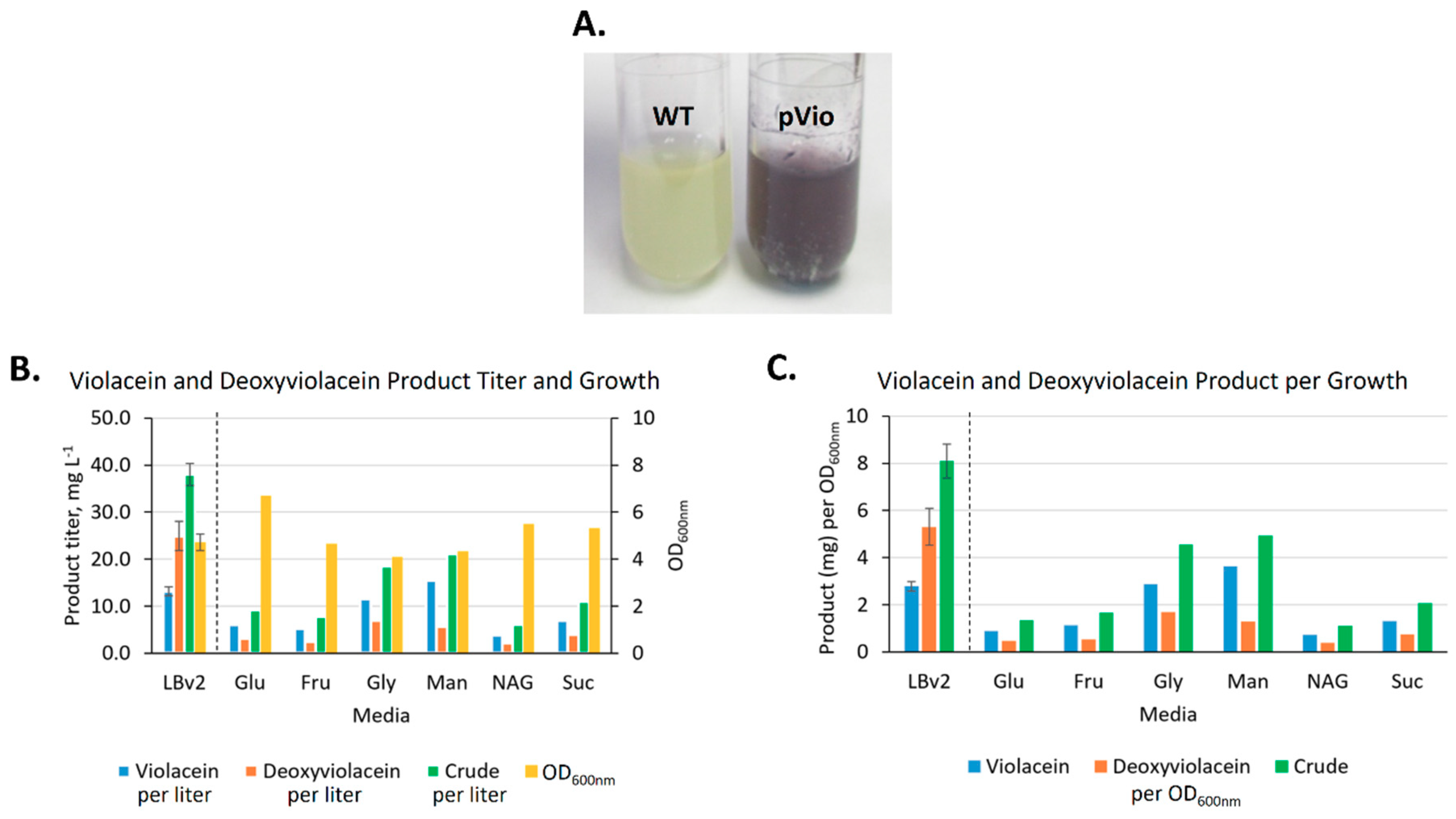
2.3.2. Heterologous Production and Feedstock Flexibility of Violacein Production
3. Discussion
3.1. Heterologous Production of Beta-carotene
3.2. Heterologous Production of Violacein
4. Materials and Methods
4.1. Materials and Nomenclature
4.2. BioLog Phenotype MicroArrays™
4.3. Cloning of the Beta-Carotene Pathway
4.4. Cloning of the MVA Pathway
| # | Name | Features | Refs. |
|---|---|---|---|
| 1 | pJV298 | oriT, p15A ori, lacIq, gfp under lacIq control, CmR | [35] |
| 2 | pA5D5GFP | J23102 promoter, B0032 RBS, gfp, and B0015 terminator assembled in the DVK_AE vector from the CIDAR MoClo kit | [10,82] |
| 3 | TOPO-ß-carotene | ß-carotene in pCR™4Blunt-TOPO® | This study |
| 4 | pJV-ß-carotene | pJV298 with crtY, crtB, crtI, crtE under lacIq control | This study |
| 5 | TOPO-MVA1 | MVA1 cluster in pCR™4Blunt-TOPO® | This study |
| 6 | TOPO-MVA2 | MVA2 cluster in pCR™4Blunt-TOPO® | This study |
| 7 | pJV-ß-carotene-A5D5-MVA2 | pJV-ß-carotene; J23102 promoter, B0032 RBS, mvaK, mvaD, pmvk, idi | This study |
| 8 | pJV-ß-carotene-MVA | pJV-ß-carotene; J23102 promoter, B0032 RBS, thil, hmdH, hmcS, MVA2 | This study |
| 9 | Bba_J72214-BBa_J72090 | P15A ori, CmR, araBAD promoter, araC, vioA, vioB, vioC, vioE | [43,83] |
| 10 | Synthesized vioD 1 | Recoded; from C. violaceum ATCC 12472 | This study |
| 11 | pVio | Bba_J72214-BBa_J72090 with vioD | This study |
| # | Name | Sequence |
|---|---|---|
| 1 | CrtY_F | TCATCTCATCATTCCGATAGCGGCACT |
| 2 | CrtE_R | ATGAACAGTTCTTCTCGAAGTAAAGCCAG |
| 3 | pJV-ß-car-R | ATGTATATCTCCTTAAGCTTACGCC |
| 4 | pJV-ß-car-F | TGAGGATCCGGTGATTGATTG |
| 5 | Bcar-4pJV-F | gcttaaggagatatacatATGAACAGTTCTTCTCGAAG |
| 6 | Bcar-4pJV-R | aatcaccggatcctcaTCATCTCATCATTCCGATAG |
| 7 | Cluster 1 F | ATGAAAGATATTTATATTGTCGCTGC |
| 8 | Cluster 1 R | TTATTTAACTTTGTATTGACGAACATGGC |
| 9 | Cluster 2 F | ATGAAAAGTAGTTTTTTAGCTCATGG |
| 10 | Cluster 2 R | TTATTTAATTAACTGATCAATTTGATTTTTTAGTGGC |
| 11 | Clust1_ck-F | CGTTGTCGGTGGTTCGATTA |
| 12 | Clust1-ck-R | ACGAGCAACCCAACCTTATC |
| 13 | Clust2-ck-F | CTTAGGCGAACTGGCAGATATTA |
| 14 | Clust2-ck-R | TGAGGTTGGCACGTGATTAG |
| 15 | A5D5-F | tgccgctatcggaatgatgagatgaggagTTGACAGCTAGCTCAGTC |
| 16 | A5D5-R | gcagcgacaatataaatatctttcatcattTAGTACTTTCCTGTGTGACTC |
| 17 | Cluster2-SacI-R | actacttttcatgagctcTTATTTAACTTTGTATTGACGAACATG |
| 18 | Cluster2-SacI-F | caaagttaaataagagctcATGAAAAGTAGTTTTTTAGCTC |
| 19 | MVA2-ovrlp-R | caatcaatcaccggatcctcaTTATTTAATTAACTGATCAATTTGATTTTTTAG |
| 20 | pJV-bcar-n-R | TCATCTCATCATTCCGATAGCG |
| 21 | pJV-b-m2-4m1-F | CATGTTCGTCAATACAAAGTTAAATAAGAGC |
| 22 | pJV-b-m2-4m1-R | GCAGCGACAATATAAATATCTTTCATCAT |
| 23 | pJV-empty-F | GATCCGGTGATTGATTGAG |
| 24 | pJV-empty-R | CTTACATTAATTGCGTTGCGC |
| 25 | VioD-F | AAAATTCTGGTGATTGGCGCGGGC |
| 26 | VioD-R | ttaCTCGAGGCGCTGCAGCGC |
| 27 | Bba-4VioD-F | cgagtaaGGATCCGAGGCTTGGATTCTCA |
| 28 | Bba-4vioD-R | ttttttacctccttaaggatcTTAGCGCTTGGCCGCGAAA |
| 29 | vioD-inschk-F | GCGGTTTTCGCGGCCAAGCG |
| 30 | vioD-inschk-R | GGCAGGGCGGGGCGTAATTTGAT |
| Name | Sequence |
|---|---|
| Ptac promoter | ttgacaattaatcatcggctcgtataatg |
| J23102 promoter | ttgacagctagctcagtcctaggtactgtgctagc |
| B0032m RBS | agagtcacacaggaaagtacta |
4.5. Cloning of the Empty Control pJV298 Plasmid
4.6. Cloning of the Violacein Pathway
4.7. Heterologous Production of Beta-carotene
4.8. Heterologous Production of Violacein
4.9. Statistical Significance Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A: Additional BioLog Results
| Compound | Response | Compound | Response |
|---|---|---|---|
| Carbohydrates | Carboxylic acids, cont. | ||
| D-Glucosamine | 159 | Monomethyl succinate | 82 |
| D-Ribose | 154 | Butyric acid | 81 |
| Inosine | 145 | α-Ketobutyric acid | 80 |
| L-Arabinose | 145 | 4-Hydroxybenzoic acid | 73 |
| Uridine | 133 | 5-Keto-D-gluconic acid | 70 |
| 2′-Deoxyadenosine | 131 | Formic acid | 69 |
| D-Fructose | 131 | Oxalomalic acid | 54 |
| Adenosine | 128 | D-Malic acid | 50 |
| D-Galactose | 123 | Amino acids | |
| Glycerol | 122 | Gly-Asp | 149 |
| ß-Methyl-D-glucoside | 116 | L-Glutamic acid | 149 |
| Thymidine | 116 | L-Asparagine | 148 |
| L-Rhamnose | 106 | L-Aspartic acid | 148 |
| D-Mannitol | 105 | D-Alanine | 147 |
| Arbutin | 100 | L-Glutamine | 140 |
| D-Cellobiose | 95 | Ala-Gly | 137 |
| Maltose | 92 | L-Proline | 136 |
| Maltotrioise | 88 | L-Serine | 136 |
| Sucrose | 88 | L-Threonine | 129 |
| α-D-Glucose | 87 | L-Alanine | 128 |
| D-Trehalose | 82 | Gly-Pro | 117 |
| N-Acetyl-D-Glucosamine | 79 | Hydroxy-L-proline | 113 |
| D-Arabinose | 78 | L-Histidine | 108 |
| Gentiobiose | 73 | L-Arginine | 98 |
| L-Lyxose | 70 | Glycine | 96 |
| D,L-α-Glycerol phosphate | 65 | Gly-Glu | 94 |
| D-Arabitol | 56 | L-Ornithine | 80 |
| Carboxylic acids | L-Pyroglutamic acid | 79 | |
| L-Malic acid | 145 | D-Serine | 52 |
| Succinic acid | 135 | Polymers | |
| Fumaric acid | 134 | Dextrin | 112 |
| Pyruvic acid | 132 | Pectin | 110 |
| D-Gluconic acid | 129 | Laminarin | 95 |
| L-Lactic acid | 128 | Gamma-cyclodextrin | 62 |
| Acetic acid | 119 | Fatty acids | |
| D,L-Malic acid | 119 | Tween 20 | 109 |
| Citric acid | 116 | Tween 40 | 108 |
| Bromosuccinic acid | 104 | Tween 80 | 92 |
| Propionic acid | 102 | Esters | |
| Malonic acid | 97 | Methylpyruvate | 107 |
| Quinic acid | 97 | Amines | |
| γ-amino-N-butyric acid | 92 | Putrescine | 67 |
| ß-hydroxybutyric acid | 90 | Alcohols | |
| α-Hydroxybutric acid | 87 | Dihydroxyacetone | 58 |
Appendix B: Additional Information Regarding Genes used in this Study
| Protein | Gene | Gene ID | #bp 3 | Notes 4 | Source/Ref 5 |
|---|---|---|---|---|---|
| Lycopene cyclase | crtY1 | 5556089 | 1158 | crtB(14) | Vc |
| Phytoene/squalene synthase family protein | crtB1 | 5556092 | 915 | crtY(14), crtI(20) | Vc |
| Phytoene desaturase | crtI1 | 5556083 | 1614 | crtB(20), crtE(11) | Vc |
| Polyprenyl synthetase | crtE1 | 5556114 | 864 | crtI(11) | Vc |
| Acetyl-CoA acetyltransferase | thil | 3252766 | 1158(1169) | hmdH(1) | La |
| Hydroxymethyl glutaryl-CoA reductase | hmdH | 3252708 | 1212 | thil(1) | La |
| Hydroxymethyl glutaryl-CoA synthase | hmcS | 3252698 | 1164 | La | |
| Mevalonate kinase | mvaK | 3253078 | 909(923) | La | |
| Mevalonate diphosphate decarboxylate | mvaD | 3253057 | 963 | La | |
| Phosphomevalonate kinase | pmvk2 | 3253097 | 1083(1097) | La | |
| Isopentenyl pyrophosphate isomerase | idi2 | 3253102 | 1020(1034) | La | |
| L-tryptophan synthase | vioA | 24947400 | 1257 | Cv | |
| Iminophenyl-pyruvate dimer synthase | vioB | 24945600 | 2997 | Cv | |
| Protodeoxyviolaceinate monooxygenase | vioE | 24949508 | 576 | Cv | |
| Tryptophan hydroxylase | vioD | 24947515 | 1122 | Cv | |
| Violacein synthase | vioC | 24948167 | 1290 | Cv |
Appendix C: Plasmid Maps
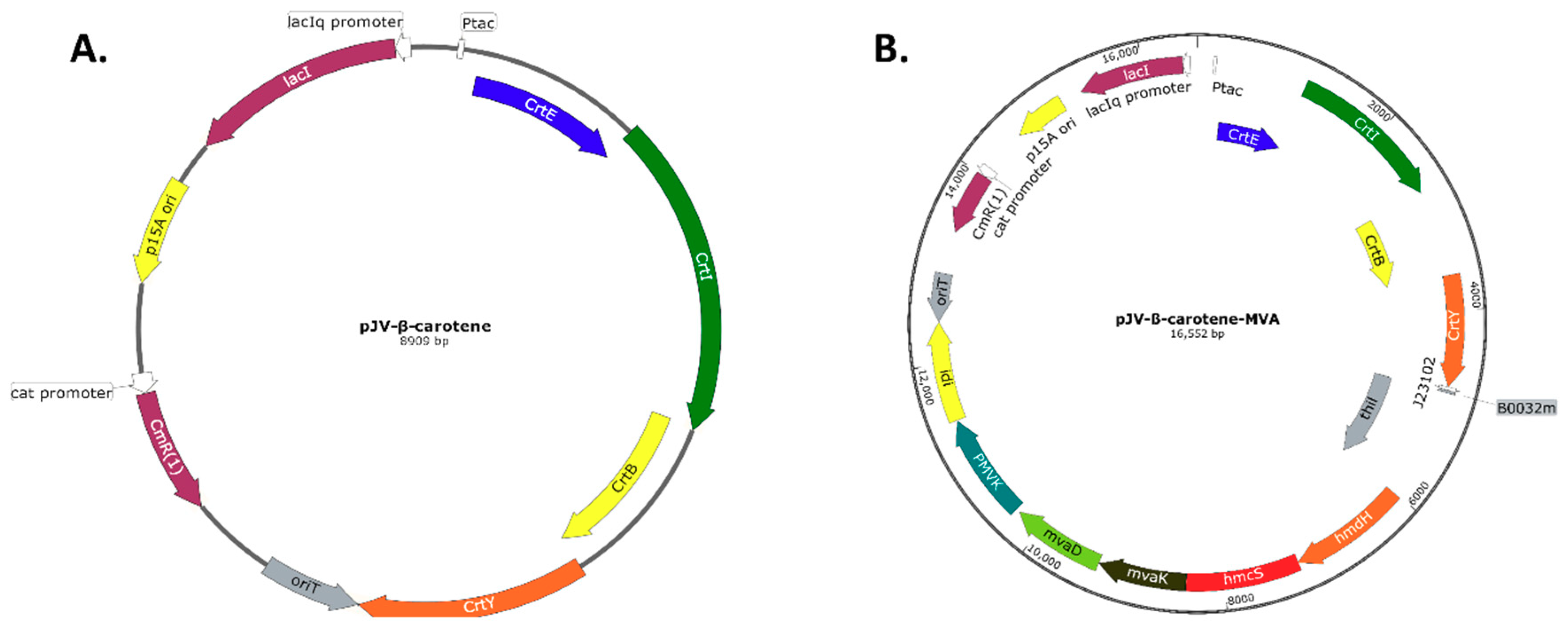
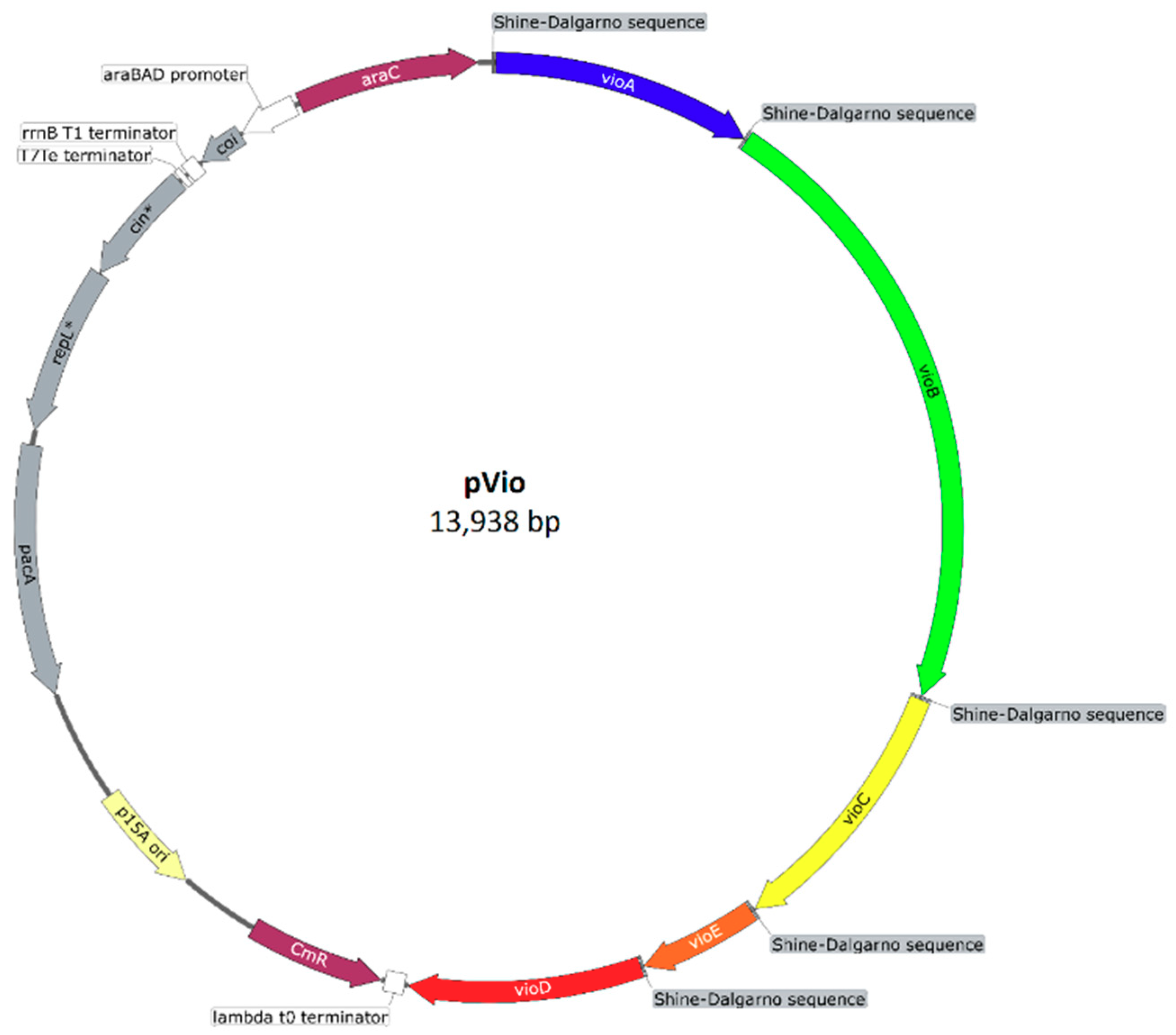
References
- Adams, B.L. The next generation of synthetic biology chassis: Moving synthetic biology from the laboratory to the field. ACS Synth. Biol. 2016, 5, 1328–1330. [Google Scholar] [CrossRef] [PubMed]
- Calero, P.; Nikel, P.I. Chasing bacterial chassis for metabolic engineering: A perspective review from classical to non-traditional microorganisms. Microb. Biotechnol. 2019, 12, 98–124. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.J.; Theriot, J.A.; Sood, P.; Marshall, W.F.; Landweber, L.F.; Fritz-Laylin, L.; Polka, J.K.; Oliferenko, S.; Gerbich, T.; Gladfelter, A.; et al. Non-model model organisms. BMC Biol. 2017, 15, 55. [Google Scholar] [CrossRef] [PubMed]
- Payne, W.J.; Eagon, R.G.; Williams, A.K. Some observations on the physiology of Pseudomonas natriegens nov. spec. Antonie Van Leeuwenhoek 1961, 27, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Dalia, T.N.; Hayes, C.A.; Stolyar, S.; Marx, C.J.; McKinlay, J.B.; Dalia, A.B. Multiplex genome editing by natural transformation (MuGENT) for synthetic biology in Vibrio natriegens. ACS Synth. Biol. 2017, 6, 1650–1655. [Google Scholar] [CrossRef] [PubMed]
- Eagon, R.G. Pseudomonas natriegens, a marine bacterium with a generation time of less than 10 minutes. J. Bacteriol. 1962, 83, 736–737. [Google Scholar] [PubMed]
- Hoffart, E.; Grenz, S.; Lange, J.; Nitschel, R.; Muller, F.; Schwentner, A.; Feith, A.; Lenfers-Lucker, M.; Takors, R.; Blombach, B. High substrate uptake rates empower Vibrio natriegens as production host for industrial biotechnology. Appl. Environ. Microbiol. 2017, 83, e01614-17. [Google Scholar] [CrossRef]
- Lee, H.H.; Ostrov, N.; Wong, B.G.; Gold, M.A.; Khalil, A.S.; Church, G.M. Functional genomics of the rapidly replicating bacterium Vibrio natriegens by CRISPRi. Nat. Microbiol. 2019, 4, 1105–1113. [Google Scholar] [CrossRef]
- Long, C.P.; Gonzalez, J.E.; Cipolla, R.M.; Antoniewicz, M.R. Metabolism of the fast-growing bacterium Vibrio natriegens elucidated by (13)C metabolic flux analysis. Metab. Eng. 2017, 44, 191–197. [Google Scholar] [CrossRef]
- Tschirhart, T.; Shukla, V.; Kelly, E.E.; Schultzhaus, Z.; NewRingeisen, E.; Erickson, J.S.; Wang, Z.; Garcia, W.; Curl, E.; Egbert, R.G.; et al. Synthetic biology parts and tools for the fast-growing marine bacterium Vibrio natriegens. ACS Synth. Biol. 2019, 8, 2069–2079. [Google Scholar] [CrossRef]
- Wang, Z.; Lin, B.; Hervey, W.J.t.; Vora, G.J. Draft genome sequence of the fast-growing marine bacterium Vibrio natriegens strain ATCC 14048. Genome Announc. 2013, 1, e00589-13. [Google Scholar] [CrossRef] [PubMed]
- Weinstock, M.T.; Hesek, E.D.; Wilson, C.M.; Gibson, D.G. Vibrio natriegens as a fast-growing host for molecular biology. Nat. Methods 2016, 13, 849–851. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.G.; Kwak, D.H.; Park, S.; Woo, S.; Yang, J.S.; Kang, C.W.; Kim, B.; Noh, M.H.; Seo, S.W.; Jung, G.Y. Vibrio sp. dhg as a platform for the biorefinery of brown macroalgae. Nat. Commun. 2019, 10, 2486. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.D.; Finnigan, W.; Wolde-Michael, E.; Kelly, P.; Blaker, J.J.; Hay, S.; Breitling, R.; Takano, E.; Scrutton, N.S. Synthetic biology for fibers, adhesives, and active camouflage materials in protection and aerospace. MRS Commun. 2019, 9, 486–504. [Google Scholar] [CrossRef]
- Tao, W.; Lv, L.; Chen, G.Q. Engineering Halomonas species TD01 for enhanced polyhydroxyalkanoates synthesis via CRISPRi. Microb. Cell Fact. 2017, 16, 48. [Google Scholar] [CrossRef]
- Wendisch, V.F.; Brito, L.F.; Gil Lopez, M.; Hennig, G.; Pfeifenschneider, J.; Sgobba, E.; Veldmann, K.H. The flexible feedstock concept in industrial biotechnology: Metabolic engineering of Escherichia coli, Corynebacterium glutamicum, Pseudomonas, Bacillus and yeast strains for access to alternative carbon sources. J. Biotechnol. 2016, 234, 139–157. [Google Scholar] [CrossRef]
- Harrison, R.G.; Todd, P.W.; Rudge, S.R.; Petrides, D.P. Bioseparations Science and Engineering, 2nd ed.; Oxford University Press: Oxford, UK, 2015; p. 432. [Google Scholar]
- Sabri, S.; Nielsen, L.K.; Vickers, C.E. Molecular control of sucrose utilization in Escherichia coli W, an efficient sucrose-utilizing strain. Appl. Environ. Microbiol. 2013, 79, 478–487. [Google Scholar] [CrossRef]
- Choi, S.Y.; Yoon, K.H.; Lee, J.I.; Mitchell, R.J. Violacein: Properties and production of a versatile bacterial pigment. Biomed. Res. Int. 2015, 2015, 465056. [Google Scholar] [CrossRef]
- Fang, M.Y.; Zhang, C.; Yang, S.; Cui, J.Y.; Jiang, P.X.; Lou, K.; Wachi, M.; Xing, X.H. High crude violacein production from glucose by Escherichia coli engineered with interactive control of tryptophan pathway and violacein biosynthetic pathway. Microb. Cell Fact. 2015, 14, 8. [Google Scholar] [CrossRef]
- Jeschek, M.; Gerngross, D.; Panke, S. Rationally reduced libraries for combinatorial pathway optimization minimizing experimental effort. Nat. Commun. 2016, 7, 11163. [Google Scholar] [CrossRef]
- Jiang, P.X.; Wang, H.S.; Zhang, C.; Lou, K.; Xing, X.H. Reconstruction of the violacein biosynthetic pathway from Duganella sp. B2 in different heterologous hosts. Appl. Microbiol. Biotechnol. 2010, 86, 1077–1088. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.A.; Vernacchio, V.R.; Lachance, D.M.; Lebovich, M.; Fu, L.; Shirke, A.N.; Schultz, V.L.; Cress, B.; Linhardt, R.J.; Koffas, M.A. ePathOptimize: A combinatorial approach for transcriptional balancing of metabolic pathways. Sci. Rep. 2015, 5, 11301. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kim, S.-W.; Nguyen, D.Q.A.; Li, H.; Kim, S.B.; Seo, Y.-G.; Yang, J.-K.; Chung, I.-Y.; Kim, D.H.; Kim, C.-J. Production of β-carotene by recombinant Escherichia coli with engineered whole mevalonate pathway in batch and fed-batch cultures. Biotechnol. Bio process Eng. 2009, 14, 559–564. [Google Scholar] [CrossRef]
- Moore, S.J.; Lai, H.E.; Kelwick, R.J.; Chee, S.M.; Bell, D.J.; Polizzi, K.M.; Freemont, P.S. EcoFlex: A multifunctional MoClo kit for E. coli synthetic biology. ACS Synth. Biol. 2016, 5, 1059–1069. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Guo, L. Biosynthesis of β-carotene in engineered E. coli using the MEP and MVA pathways. Microb. Cell Fact. 2014, 13, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Zhang, C.; Bi, C.; Li, Q.; Zhang, X. Combinatory optimization of chromosomal integrated mevalonate pathway for beta-carotene production in Escherichia coli. Microb. Cell Fact. 2016, 15, 202. [Google Scholar] [CrossRef]
- Yoon, S.H.; Lee, S.H.; Das, A.; Ryu, H.K.; Jang, H.J.; Kim, J.Y.; Oh, D.K.; Keasling, J.D.; Kim, S.W. Combinatorial expression of bacterial whole mevalonate pathway for the production of β-carotene in E. coli. J. Biotechnol. 2009, 140, 218–226. [Google Scholar] [CrossRef]
- Yoon, S.-H.Y.; Park, H.-M.; Kim, J.-E.K.; Lee, S.-H.; Choi, M.-S.; Kim, J.-Y.K.; Oh, D.-K.O.; Keasling, J.D.; Kim, S.-W. Increased ß-carotene production in recombinant Escherichia coli harboring an engineered isoprenoid precursor pathway with mevalonate addition. Biotechnol. Prog. 2007, 23, 599–605. [Google Scholar] [CrossRef]
- Flesch, G.; Rohmer, M. Prokaryotic hopanoids: The biosynthesis of the bacteriohopane skeleton. Eur. J. Biochem. 1988, 175, 405–411. [Google Scholar] [CrossRef]
- Rodríguez-Concepción, M.; Boronat, A. Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 2002, 130, 1079–1089. [Google Scholar] [CrossRef]
- Wang, Z.; Lin, B.; Mostaghim, A.; Rubin, R.A.; Glaser, E.R.; Mittraparp-Arthorn, P.; Thompson, J.R.; Vuddhakul, V.; Vora, G.J. Vibrio campbellii hmgA-mediated pyomelanization impairs quorum sensing, virulence, and cellular fitness. Front. Microbiol. 2013, 4, 379. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; O’Shaughnessy, T.J.; Soto, C.M.; Rahbar, A.M.; Robertson, K.L.; Lebedev, N.; Vora, G.J. Function and regulation of Vibrio campbellii proteorhodopsin: Acquired phototrophy in a classical organoheterotroph. PLoS ONE 2012, 7, e38749. [Google Scholar] [CrossRef] [PubMed]
- Dunn, A.K.; Millikan, D.S.; Adin, D.M.; Bose, J.L.; Stabb, E.V. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl. Environ. Microbiol. 2006, 72, 802–810. [Google Scholar] [CrossRef] [PubMed]
- van Kessel, J.C.; Rutherford, S.T.; Cong, J.P.; Quinodoz, S.; Healy, J.; Bassler, B.L. Quorum sensing regulates the osmotic stress response in Vibrio harveyi. J. Bacteriol. 2015, 197, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Altermann, E.; Russell, W.M.; Azcarate-Peril, M.A.; Barrangou, R.; Buck, B.L.; McAuliffe, O.; Souther, N.; Dobson, A.; Duong, T.; Callanan, M.; et al. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 2005, 102, 3906–3912. [Google Scholar] [CrossRef] [PubMed]
- Balibar, C.J.; Walsh, C.T. In vitro biosynthesis of violacein from L-tryptophan by the enzymes VioA-E from Chromobacterium violaceum. Biochemistry 2006, 45, 15444–15457. [Google Scholar] [CrossRef]
- Dantas, C.; Volpe, P.L.O.; Durán, N.; Ferreira, M.M.C. The violacein biosynthesis monitored by multi-wavelength fluorescence spectroscopy and by the PARAFAC method. J. Braz. Chem. Soc. 2012, 23, 2054–2064. [Google Scholar] [CrossRef][Green Version]
- iGEM. Available online: https://2016.igem.org/Team:Washington/Project1 (accessed on 23 July 2019).
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef]
- Lee, M.E.; Aswani, A.; Han, A.S.; Tomlin, C.J.; Dueber, J.E. Expression-level optimization of a multi-enzyme pathway in the absence of a high-throughput assay. Nucleic Acids Res. 2013, 41, 10668–10678. [Google Scholar] [CrossRef]
- Brazilian National Genome Project, C. The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc. Natl. Acad. Sci. USA 2003, 100, 11660–11665. [Google Scholar]
- Kittleson, J.T.; DeLoache, W.; Cheng, H.Y.; Anderson, J.C. Scalable plasmid transfer using engineered P1-based phagemids. ACS Synth. Biol. 2012, 1, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Sarovich, D.S.; Pemberton, J.M. pPSX: A novel vector for the cloning and heterologous expression of antitumor antibiotic gene clusters. Plasmid 2007, 57, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, E.; Michniewski, S.; Gätgens, C.; Münch, E.; Müller, F.; Polen, T.; Millard, A.; Blombach, B.; Frunzke, J. Generation of a prophage-free variant of the fast-growing bacterium Vibrio natriegens. Appl. Environ. Microbiol. 2019, 85, e00853-19. [Google Scholar] [CrossRef] [PubMed]
- Failmezger, J.; Scholz, S.; Blombach, B.; Siemann-Herzberg, M. Cell-free protein synthesis from fast-growing Vibrio natriegens. Front. Microbiol. 2018, 9, 1146. [Google Scholar] [CrossRef]
- Darlington, A.P.S.; Kim, J.; Jimenez, J.I.; Bates, D.G. Dynamic allocation of orthogonal ribosomes facilitates uncoupling of co-expressed genes. Nat. Commun. 2018, 9, 695. [Google Scholar] [CrossRef]
- Gyorgy, A.; Jimenez, J.I.; Yazbek, J.; Huang, H.H.; Chung, H.; Weiss, R.; Del Vecchio, D. Isocost Lines Describe the Cellular Economy of Genetic Circuits. Biophys. J. 2015, 109, 639–646. [Google Scholar] [CrossRef]
- Aiyar, S.E.; Gaal, T.; Gourse, R.L. rRNA promoter activity in the fast-growing bacterium Vibrio natriegens. J. Bacteriol. 2002, 184, 1349–1358. [Google Scholar] [CrossRef]
- European Commission. Available online: https://cordis.europa.eu/project/rcn/199219/factsheet/en (accessed on 23 July 2019).
- Metgen. Available online: http://www.metgen.com/retapp/ (accessed on 23 July 2019).
- Steinbach, D.; Kruse, A.; Sauer, J.; Vetter, P. Sucrose is a promising feedstock for the synthesis of the platform chemical hydroxymethylfurfural. Energies 2018, 11, 645. [Google Scholar] [CrossRef]
- Enquist-Newman, M.; Faust, A.M.; Bravo, D.D.; Santos, C.N.; Raisner, R.M.; Hanel, A.; Sarvabhowman, P.; Le, C.; Regitsky, D.D.; Cooper, S.R.; et al. Efficient ethanol production from brown macroalgae sugars by a synthetic yeast platform. Nature 2014, 505, 239–243. [Google Scholar] [CrossRef]
- Lee, H.H.; Ostrov, N.; Wong, B.G.; Gold, M.A.; Khalil, A.; Church, G.M. Vibrio natriegens, a new genomic powerhouse. bioRxiv 2016, 058487. [Google Scholar]
- International Energy Agency (IEA). Biofuels for Transport: An International Perspective; IEA: Paris, France, 2004. [Google Scholar]
- Bruschi, M.; Boyes, S.J.; Sugiarto, H.; Nielsen, L.K.; Vickers, C.E. A transferable sucrose utilization approach for non-sucrose-utilizing Escherichia coli strains. Biotechnol. Adv. 2012, 30, 1001–1010. [Google Scholar] [CrossRef] [PubMed]
- Arifin, Y.; Sabri, S.; Sugiarto, H.; Kromer, J.O.; Vickers, C.E.; Nielsen, L.K. Deletion of cscR in Escherichia coli W improves growth and poly-3-hydroxybutyrate (PHB) production from sucrose in fed batch culture. J. Biotechnol. 2011, 156, 275–278. [Google Scholar] [CrossRef] [PubMed]
- Koutinas, A.A.; Wang, R.; Webb, C. Evaluation of wheat as generic feedstock for chemical production. Ind. Crop. Prod. 2004, 20, 75–88. [Google Scholar] [CrossRef]
- Jahrels, K.; Lengeler, J.W. Molecular analysis of two ScrR repressors and of a ScrR–FruR hybrid repressor for sucrose and D-fructose specific regulons from enteric bacteria. Mol. Microbiol. 1993, 9, 195–209. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, Q.; Sun, T.; Zhu, X.; Xu, H.; Tang, J.; Zhang, X.; Ma, Y. Engineering central metabolic modules of Escherichia coli for improving beta-carotene production. Metab. Eng. 2013, 17, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Bogacz-Radomska, L.; Harasym, J. β-Carotene—Properties and production methods. Food Qual. Saf. 2018, 2, 69–74. [Google Scholar] [CrossRef]
- Alonso-Gutierrez, J.; Chan, R.; Batth, T.S.; Adams, P.D.; Keasling, J.D.; Petzold, C.J.; Lee, T.S. Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metab. Eng. 2013, 19, 33–41. [Google Scholar] [CrossRef]
- Lemuth, K.; Steuer, K.; Albermann, C. Engineering of a plasmid-free Escherichia coli strain for improved in vivo biosynthesis of astaxanthin. Microb. Cell Fact. 2011, 10, 29–40. [Google Scholar] [CrossRef]
- Wang, C.-W.; Oh, M.-K.; Liao, J.C. Engineered isoprenoid pathway enhances astaxanthin production in Escherichia coli. Biotechnol. Bioeng. 1999, 62, 235–241. [Google Scholar] [CrossRef]
- Willrodt, C.; David, C.; Cornelissen, S.; Bühler, B.; Julsing, M.K.; Schmid, A. Engineering the productivity of recombinant Escherichia coli for limonene formation from glycerol in minimal media. Biotechnol. J. 2014, 9, 1000–1012. [Google Scholar] [CrossRef]
- Yang, J.; Zhao, G.; Sun, Y.; Zheng, Y.; Jiang, X.; Liu, W.; Xian, M. Bio-isoprene production using exogenous MVA pathway and isoprene synthase in Escherichia coli. Bioresour. Technol. 2012, 104, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Xian, M.; Su, S.; Zhao, G.; Nie, Q.; Jiang, X.; Zheng, Y.; Liu, W. Enhancing production of bio-isoprene using hybrid MVA pathway and isoprene synthase in E. coli. PLoS ONE 2012, 7, e33509. [Google Scholar] [CrossRef] [PubMed]
- Pitera, D.J.; Paddon, C.J.; Newman, J.D.; Keasling, J.D. Balancing a heterologous mevalonate pathway for improved isoprenoid production in Escherichia coli. Metab. Eng. 2007, 9, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Luo, Z.; Wang, Y.; Pham, N.T.; Tuck, L.; Perez-Pi, I.; Liu, L.; Shen, Y.; French, C.; Auer, M.; et al. Rapid pathway prototyping and engineering using in vitro and in vivo synthetic genome SCRaMbLE-in methods. Nat. Commun. 2018, 9, 1936. [Google Scholar] [CrossRef]
- Yang, C.; Jiang, P.; Xiao, S.; Zhang, C.; Lou, K.; Xing, X.-H. Fed-batch fermentation of recombinant Citrobacter freundii with expression of a violacein-synthesizing gene cluster for efficient violacein production from glycerol. Biochem. Eng. J. 2011, 57, 55–62. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Y.G.; Zhang, R.B.; Zhang, F.; Zhu, J. Glycerol/glucose co-fermentation: One more proficient process to produce propionic acid by Propionibacterium acidipropionici. Curr. Microbiol. 2011, 62, 152–158. [Google Scholar] [CrossRef]
- VanBriesen, J.M. Evaluation of methods to predict bacterial yield using thermodynamics. Biodegradation 2002, 13, 171–190. [Google Scholar] [CrossRef]
- Fernández-Llamosas, H.; Castro, L.; Blázquez, M.L.; Díaz, E.; Carmona, M. Speeding up bioproduction of selenium nanoparticles by using Vibrio natriegens as microbial factory. Sci. Rep. 2017, 7, 16046. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2017, 45, D37–D42. [Google Scholar] [CrossRef]
- Brister, J.R.; Ako-Adjei, D.; Bao, Y.; Blinkova, O. NCBI viral genomes resource. Nucleic Acids Res. 2015, 43, D571–D577, (Database issue). [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef] [PubMed]
- Bochner, B.R.; Gadzinski, P.; Panomitros, E. Phenotype MicroArrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 2001, 11, 1246–1255. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Lei, X.H.; Bochner, B.R.; Wanner, B.L. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J. Bacteriol. 2003, 185, 4956–4972. [Google Scholar] [CrossRef] [PubMed]
- Spangler, J.R.; Caruana, J.C.; Phillips, D.A.; Walper, S.A. Broad range shuttle vector construction and promoter evaluation for the use of Lactobacillus plantarum WCFS1 as a microbial engineering platform. Synth. Biol. 2019, 4, ysz012. [Google Scholar] [CrossRef]
- Iverson, S.V.; Haddock, T.L.; Beal, J.; Densmore, D.M. CIDAR MoClo: Improved MoClo assembly standard and new E. coli part library enable rapid combinatorial design for synthetic and traditional biology. ACS Synth. Biol. 2016, 5, 99–103. [Google Scholar] [CrossRef]
- Addgene. Available online: https://www.addgene.org/40782/ (accessed on 9 July 2019).
- Paliakov, E.M.; Crow, B.S.; Bishop, M.J.; Norton, D.; George, J.; Bralley, J.A. Rapid quantitative determination of fat-soluble vitamins and coenzyme Q-10 in human serum by reversed phase ultra-high pressure liquid chromatography with UV detection. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2009, 877, 89–94. [Google Scholar] [CrossRef]
| Carbohydrates | ||
|---|---|---|
| N-Acetyl-D-glucosamine 1 | D-Galactose | Maltotriose |
| Adenosine | Gentiobiose | D-Mannitol 1 |
| D-Arabinose | Glycerol 1 | ß-Methyl-D-glucoside |
| L-Arabinose1 | D,L-α-Glycerol phosphate | L-Rhamnose |
| D-Arabitol | D-Glucosamine | D-Ribose |
| Arbutin | α-D-Glucose 1 | Sucrose1 |
| D-Cellubiose | Inosine | Thymidine |
| 2′-Deoxyadenosine | L-Lyxose | D-Trehalose |
| D-Fructose1 | Maltose | Uridine |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ellis, G.A.; Tschirhart, T.; Spangler, J.; Walper, S.A.; Medintz, I.L.; Vora, G.J. Exploiting the Feedstock Flexibility of the Emergent Synthetic Biology Chassis Vibrio natriegens for Engineered Natural Product Production. Mar. Drugs 2019, 17, 679. https://doi.org/10.3390/md17120679
Ellis GA, Tschirhart T, Spangler J, Walper SA, Medintz IL, Vora GJ. Exploiting the Feedstock Flexibility of the Emergent Synthetic Biology Chassis Vibrio natriegens for Engineered Natural Product Production. Marine Drugs. 2019; 17(12):679. https://doi.org/10.3390/md17120679
Chicago/Turabian StyleEllis, Gregory A., Tanya Tschirhart, Joseph Spangler, Scott A. Walper, Igor L. Medintz, and Gary J. Vora. 2019. "Exploiting the Feedstock Flexibility of the Emergent Synthetic Biology Chassis Vibrio natriegens for Engineered Natural Product Production" Marine Drugs 17, no. 12: 679. https://doi.org/10.3390/md17120679
APA StyleEllis, G. A., Tschirhart, T., Spangler, J., Walper, S. A., Medintz, I. L., & Vora, G. J. (2019). Exploiting the Feedstock Flexibility of the Emergent Synthetic Biology Chassis Vibrio natriegens for Engineered Natural Product Production. Marine Drugs, 17(12), 679. https://doi.org/10.3390/md17120679









