Conotoxin Diversity in the Venom Gland Transcriptome of the Magician’s Cone, Pionoconus magus
Abstract
1. Introduction
2. Results and Discussion
2.1. Quality of the Assembly
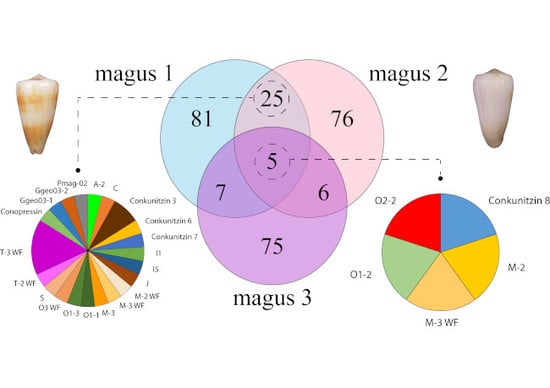
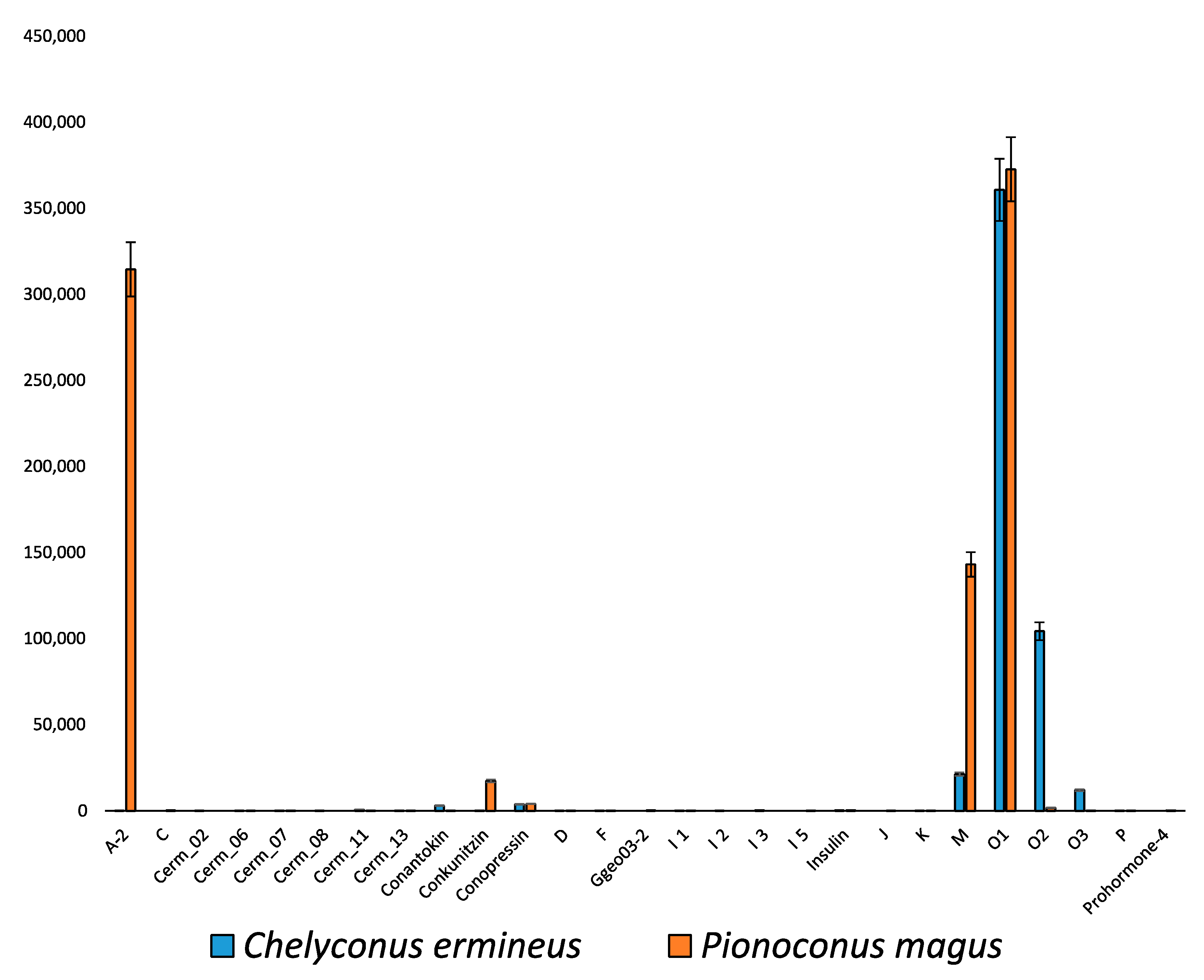
2.2. Diversity of Conotoxin Precursors and Hormones in P. magus
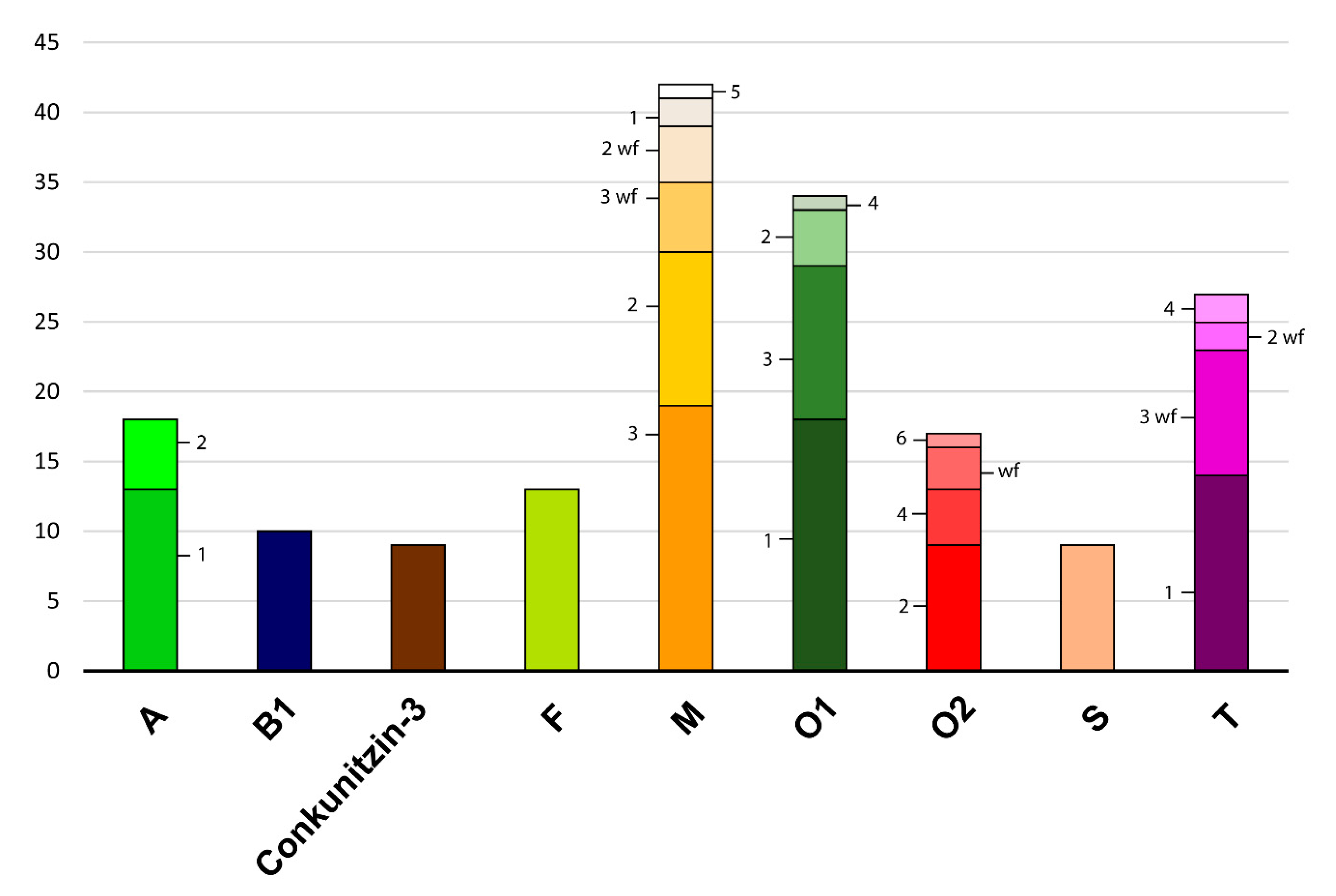
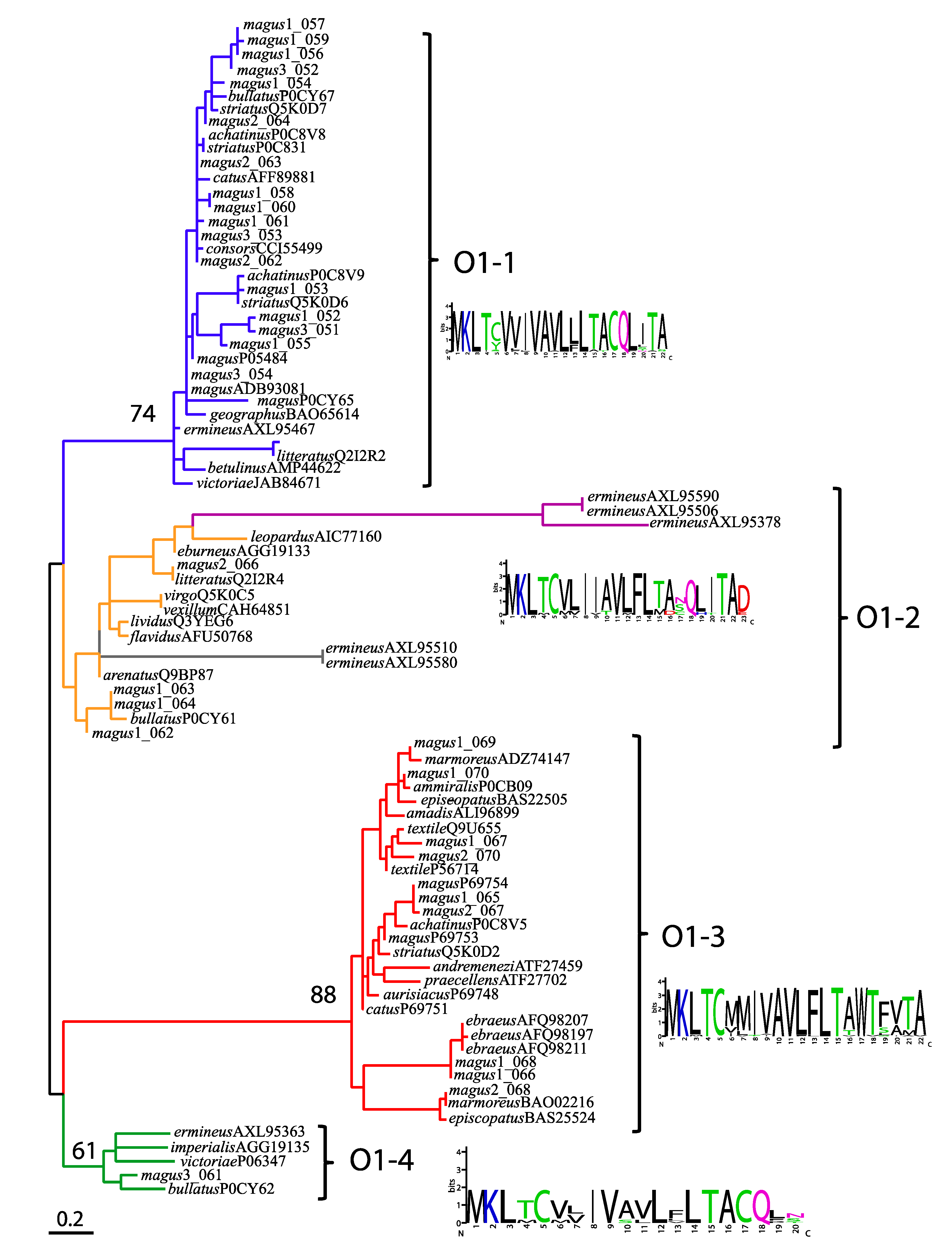
2.3. Diversity within Superfamilies
2.4. Conserved Conotoxin Precursors
2.5. Other Proteins Identified in the Venom Gland Transcriptome
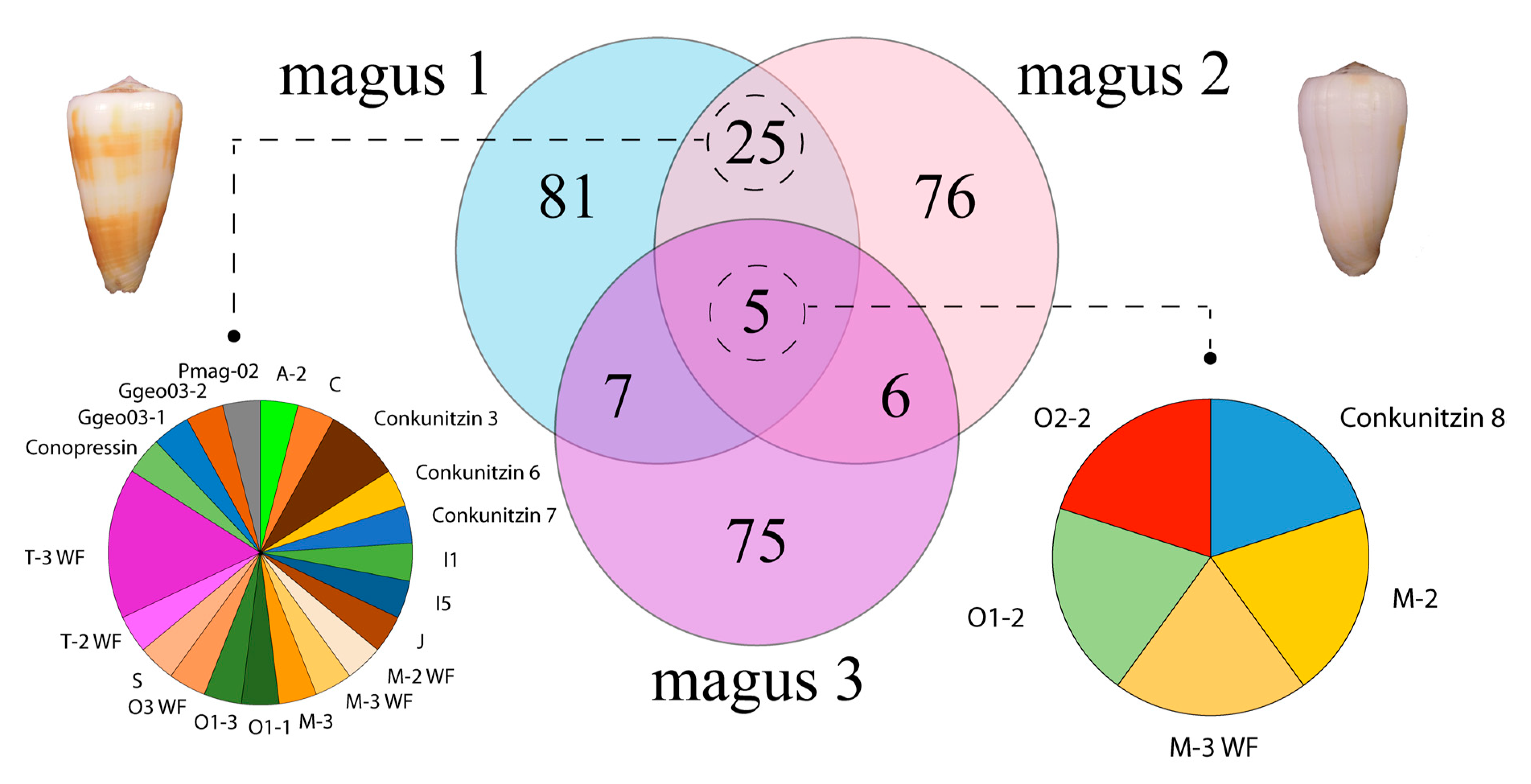
2.6. Relative Transcript Abundance and Differential Expression Analysis
3. Materials and Methods
3.1. Sampling and RNA Extraction
3.2. Library Preparation and Sequencing
3.3. Transcriptome Assembly
3.4. Prediction and Annotation of Conotoxin Precursors and Associated Proteins
3.5. Phylogenetic Analyses of the O1 Superfamily
3.6. Transcript Abundance and Differential Expression Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tucker, J.K.; Tenorio, M.J. Illustrated Catalog of the Living Cone Shells; MDM Publishing: Wellington, FL, USA, 2013; p. 517. [Google Scholar]
- Robinson, S.D.; Li, Q.; Bandyopadhyay, P.K.; Gajewiak, J.; Yandell, M.; Papenfuss, A.T.; Purcell, A.W.; Norton, R.S.; Safavi-Hemami, H. Hormone-like peptides in the venoms of marine cone snails. Gen. Comp. Endocrinol. 2017, 244 (Suppl. C), 11–18. [Google Scholar] [CrossRef]
- Robinson, S.D.; Norton, R.S. Conotoxin gene superfamilies. Mar. Drugs 2014, 12, 6058–6101. [Google Scholar] [CrossRef]
- Terlau, H.; Olivera, B.M. Conus venoms: A rich source of novel ion channel-targeted peptides. Physiol. Rev. 2004, 84, 41–68. [Google Scholar] [CrossRef]
- Kaas, Q.; Westermann, J.-C.; Craik, D.J. Conopeptide characterization and classifications: An analysis using ConoServer. Toxicon 2010, 55, 1491–1509. [Google Scholar] [CrossRef]
- Mansbach, A.R.; Travers, T.; McMahon, H.B.; Fair, M.J.; Gnanakaran, S. Snails in silico: A review of computational studies on the conopeptides. Mar. Drugs 2019, 17, 145. [Google Scholar] [CrossRef]
- Abalde, S.; Tenorio, M.J.; Afonso, C.M.L.; Zardoya, R. Conotoxin diversity in Chelyconus ermineus (Born, 1778) and the convergent origin of piscivory in the Atlantic and Indo-Pacific cones. Genome Biol. Evol. 2018, 10, 2643–2662. [Google Scholar] [CrossRef]
- Li, Q.; Barghi, N.; Lu, A.; Fedosov, A.E.; Bandyopadhyay, P.K.; Lluisma, A.O.; Concepcion, G.P.; Yandell, M.; Olivera, B.M.; Safavi-Hemami, H. Divergence of the venom exogene repertoire in two sister species of Turriconus. Genome Biol. Evol. 2017, 9, 2211–2225. [Google Scholar] [CrossRef]
- Peng, C.; Yao, G.; Gao, B.-M.; Fan, C.-X.; Bian, C.; Wang, J.; Cao, Y.; Wen, B.; Zhu, Y.; Ruan, Z.; et al. High-throughput identification of novel conotoxins from the Chinese tubular cone snail (Conus betulinus) by multi-transcriptome sequencing. GigaScience 2016, 5, 17. [Google Scholar] [CrossRef]
- Dutertre, S.; Biass, D.; Stöcklin, R.; Favreau, P. Dramatic intraspecimen variations within the injected venom of Conus consors: An unsuspected contribution to venom diversity. Toxicon 2010, 55, 1453–1462. [Google Scholar] [CrossRef]
- Dutertre, S.; Jin, A.-H.; Vetter, I.; Hamilton, B.; Sunagar, K.; Lavergne, V.; Dutertre, V.; Fry, B.G.; Antunes, A.; Venter, D.J.; et al. Evolution of separate predation-and defence-evoked venoms in carnivorous cone snails. Nat. Commun. 2014, 5, 3521. [Google Scholar] [CrossRef]
- Phuong, M.A.; Mahardika, G.N. Targeted sequencing of venom genes from cone snail genomes improves understanding of conotoxin molecular evolution. Mol. Biol. Evol. 2018, 35, 1210–1224. [Google Scholar] [CrossRef] [PubMed]
- Duda, T.F.; Kohn, A.J.; Palumbi, S.R. Origins of diverse feeding ecologies within Conus, a genus of venomous marine gastropods. Biol. J. Linn. Soc. 2001, 73, 391–409. [Google Scholar] [CrossRef]
- Duda, T.F.; Palumbi, S.R. Molecular genetics of ecological diversification: Duplication and rapid evolution of toxin genes of the venomous gastropod Conus. Proc. Natl. Acad. Sci. USA 1999, 96, 6820–6823. [Google Scholar] [CrossRef] [PubMed]
- Phuong, M.A.; Alfaro, M.E.; Mahardika, G.N.; Marwoto, R.M.; Prabowo, R.E.; von Rintelen, T.; Vogt, P.W.H.; Hendricks, J.R.; Puillandre, N. Lack of signal for the impact of conotoxin gene diversity on speciation rates in cone snails. Syst. Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Phuong, M.A.; Mahardika, G.N.; Alfaro, M.E. Dietary breadth is positively correlated with venom complexity in cone snails. BMC Genom. 2016, 17, 401. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.J.; Dutertre, S.; Vetter, I.; Christie, M.J. Conus venom peptide pharmacology. Pharmacol. Rev. 2012, 64, 259–298. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, D.; Gonzalez, W.; Fissore, R.A.; Carvacho, I. Conotoxins as tools to understand the physiological function of voltage-gated calcium (CaV) channels. Mar. Drugs 2017, 15, 313. [Google Scholar] [CrossRef]
- Tikhonov, D.B.; Zhorov, B.S. Predicting structural details of the sodium channel pore basing on animal toxin studies. Front. Pharmacol. 2018, 9, 880. [Google Scholar] [CrossRef]
- Gao, B.; Peng, C.; Yang, J.; Yi, Y.; Zhang, J.; Shi, Q. Cone snails: A big store of conotoxins for novel drug discovery. Toxins 2017, 9, 397. [Google Scholar] [CrossRef]
- Layer, T.R.; McIntosh, M.J. Conotoxins: Therapeutic potential and application. Mar. Drugs 2006, 4, 119–142. [Google Scholar] [CrossRef]
- Olivera, B.M.; Rivier, J.; Clark, C.; Ramilo, C.A.; Corpuz, G.P.; Abogadie, F.C.; Mena, E.E.; Woodward, S.R.; Hillyard, D.R.; Cruz, L.J. Diversity of Conus neuropeptides. Science 1990, 249, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Barghi, N.; Concepcion, G.P.; Olivera, B.M.; Lluisma, A.O. High conopeptide diversity in Conus tribblei revealed through analysis of venom duct transcriptome using two high-throughput sequencing platforms. Mar. Biotechnol. 2015, 17, 81–98. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Li, C.; Dong, S.; Wu, Y.; Zhangsun, D.; Luo, S. Discovery methodology of novel conotoxins from Conus species. Mar. Drugs 2018, 16, 417. [Google Scholar] [CrossRef] [PubMed]
- Prashanth, J.R.; Lewis, R.J.; Dutertre, S. Towards an integrated venomics approach for accelerated conopeptide discovery. Toxicon 2012, 60, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Himaya, S.W.A.; Jin, A.-H.; Dutertre, S.; Giacomotto, J.; Mohialdeen, H.; Vetter, I.; Alewood, P.F.; Lewis, R.J. Comparative venomics reveals the complex prey capture strategy of the piscivorous cone snail Conus catus. J. Proteome Res. 2015, 14, 4372–4381. [Google Scholar] [CrossRef] [PubMed]
- Jin, A.-H.; Dutertre, S.; Dutt, M.; Lavergne, V.; Jones, A.; Lewis, R.J.; Alewood, P.F. Transcriptomic-proteomic correlation in the predation-evoked venom of the cone snail, Conus imperialis. Mar. Drugs 2019, 17, 177. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.D.; Li, Q.; Lu, A.; Bandyopadhyay, P.K.; Yandell, M.; Olivera, B.M.; Safavi-Hemami, H. The venom repertoire of Conus gloriamaris (Chemnitz, 1777), the glory of the sea. Mar. Drugs 2017, 15, 145. [Google Scholar] [CrossRef]
- Endean, R.; Duchemin, C. The venom apparatus of Conus magus. Toxicon 1967, 4, 275–284. [Google Scholar] [CrossRef]
- Li, Q.; Watkins, M.; Robinson, D.S.; Safavi-Hemami, H.; Yandell, M. Discovery of novel conotoxin candidates using machine learning. Toxins 2018, 10, 503. [Google Scholar] [CrossRef]
- McGivern, J.G. Ziconotide: A review of its pharmacology and use in the treatment of pain. Neuropsychiatr. Dis. Treat. 2007, 3, 69–85. [Google Scholar] [CrossRef]
- Olivera, B.M.; Gray, W.R.; Zeikus, R.; McIntosh, J.M.; Varga, J.; Rivier, J.; de Santos, V.; Cruz, L.J. Peptide neurotoxins from fish-hunting cone snails. Science 1985, 230, 1338–1343. [Google Scholar] [CrossRef] [PubMed]
- Röckel, D.; Korn, W.; Kohn, A.J. Manual of the Living Conidae. Vol. I, Indo-Pacific; Christa Hemmen Verlag: Wiesbaden, Germany, 1995. [Google Scholar]
- Tucker, J.K.; Tenorio, M.J. Systematic Classification of Recent and Fossil Conoidean Gastropods: With Keys to the Genera of Cone Shells; Conchbooks: Hackenheim, Germany, 2009. [Google Scholar]
- Perron, F.E.; Kohn, A.J. Larval dispersal and geographic distribution in coral reef gastropods of the genus Conus. In Proceedings of the Fifth International Coral Reef Congress, Tahiti, French Polynesian, 27 May–1 June 1985; Volume 4, pp. 67–72. [Google Scholar]
- Nybakken, J.; Perron, F. Ontogenetic change in the radula of Conus magus (Gastropoda). Mar. Biol. 1988, 98, 239–242. [Google Scholar] [CrossRef]
- Olivera, B.M.; Seger, J.; Horvath, M.P.; Fedosov, A.E. Prey-capture strategies of fish-hunting cone snails: Behavior, neurobiology and evolution. Brain Behav. Evol. 2015, 86, 58–74. [Google Scholar] [CrossRef]
- Kaas, Q.; Yu, R.; Jin, A.-H.; Dutertre, S.; Craik, D.J. ConoServer: Updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res. 2012, 40, D325–D330. [Google Scholar] [CrossRef] [PubMed]
- Kapono, C.A.; Thapa, P.; Cabalteja, C.C.; Guendisch, D.; Collier, A.C.; Bingham, J.-P. Conotoxin truncation as a post-translational modification to increase the pharmacological diversity within the milked venom of Conus magus. Toxicon 2013, 70, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Terrat, Y.; Biass, D.; Dutertre, S.; Favreau, P.; Remm, M.; Stöcklin, R.; Piquemal, D.; Ducancel, F. High-resolution picture of a venom gland transcriptome: Case study with the marine snail Conus consors. Toxicon 2012, 59, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Pi, C.; Liu, Y.; Peng, C.; Jiang, X.; Liu, J.; Xu, B.; Yu, X.; Yu, Y.; Jiang, X.; Wang, L.; et al. Analysis of expressed sequence tags from the venom ducts of Conus striatus: Focusing on the expression profile of conotoxins. Biochimie 2006, 88, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Puillandre, N.; Watkins, M.; Olivera, B.M. Evolution of Conus peptide genes: Duplication and positive selection in the A-superfamily. J. Mol. Evol. 2010, 70, 190–202. [Google Scholar] [CrossRef]
- Carruthers, M.; Yurchenko, A.A.; Augley, J.J.; Adams, C.E.; Herzyk, P.; Elmer, K.R. De novo transcriptome assembly, annotation and comparison of four ecological and evolutionary model salmonid fish species. BMC Genom. 2018, 19, 32. [Google Scholar]
- Holding, M.L.; Margres, M.J.; Mason, A.J.; Parkinson, C.L.; Rokyta, D.R. Evaluating the performance of de novo assembly methods for venom-gland transcriptomics. Toxins 2018, 10, 249. [Google Scholar] [CrossRef]
- Slatkin, M. Isolation by distance in equilibrium and non-equilibrium populations. Evolution 1993, 47, 264–279. [Google Scholar] [CrossRef] [PubMed]
- Duda, T.F., Jr.; Chang, D.; Lewis, B.D.; Lee, T. Geographic variation in venom allelic composition and diets of the widespread predatory marine gastropod Conus ebraeus. PLoS ONE 2009, 4, e6245. [Google Scholar] [CrossRef] [PubMed]
- Barghi, N.; Concepcion, G.P.; Olivera, B.M.; Lluisma, A.O. Comparison of the venom peptides and their expression in closely related Conus species: Insights into adaptive post-speciation evolution of Conus exogenomes. Genome Biol. Evol. 2015, 7, 1797–1814. [Google Scholar] [CrossRef] [PubMed]
- Jin, A.-H.; Dutertre, S.; Kaas, Q.; Lavergne, V.; Kubala, P.; Lewis, R.J.; Alewood, P.F. Transcriptomic messiness in the venom duct of Conus miles contributes to conotoxin diversity. Mol. Cell. Proteom. 2013, 12, 3824–3833. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Bandyopadhyay, P.K.; Olivera, B.M.; Yandell, M. Elucidation of the molecular envenomation strategy of the cone snail Conus geographus through transcriptome sequencing of its venom duct. BMC Genom. 2012, 13, 284. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Bandyopadhyay, P.K.; Olivera, B.M.; Yandell, M. Characterization of the Conus bullatus genome and its venom-duct transcriptome. BMC Genom. 2011, 12, 60. [Google Scholar] [CrossRef]
- Azam, L.; McIntosh, J.M. Alpha-conotoxins as pharmacological probes of nicotinic acetylcholine receptors. Acta Pharmacol. Sin. 2009, 30, 771. [Google Scholar] [CrossRef]
- Ellison, M.; Olivera, B.M. α4/3 Conotoxins: Phylogenetic distribution, functional properties, and structure–function insights. Chem. Rec. 2007, 7, 341–353. [Google Scholar] [CrossRef]
- Kohn, A.J.; Hunter, C. The feeding process in Conus imperialis. Veliger 2001, 44, 232–234. [Google Scholar]
- Prashanth, J.R.; Dutertre, S.; Jin, A.H.; Lavergne, V.; Hamilton, B.; Cardoso, F.C.; Griffin, J.; Venter, D.J.; Alewood, P.F.; Lewis, R.J. The role of defensive ecological interactions in the evolution of conotoxins. Mol. Ecol. 2016, 25, 598–615. [Google Scholar] [CrossRef]
- Jacob, R.B.; McDougal, O.M. The M-superfamily of conotoxins: A review. Cell. Mol. Life Sci. 2010, 67, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Loughnan, M.; Nicke, A.; Jones, A.; Schroeder, C.I.; Nevin, S.T.; Adams, D.J.; Alewood, P.F.; Lewis, R.J. Identification of a novel class of nicotinic receptor antagonists: Dimeric conotoxins VxXIIA, VxXIIB, and VxXIIC from Conus vexillum. J. Biol. Chem. 2006, 281, 24745–24755. [Google Scholar] [CrossRef] [PubMed]
- Loughnan, M.L.; Nicke, A.; Lawrence, N.; Lewis, R.J. Novel αD-conopeptides and their precursors Identified by cDNA cloning define the D-conotoxin superfamily. Biochemistry 2009, 48, 3717–3729. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Zhang, T.; Kompella, S.N.; Yan, M.; Lu, A.; Wang, Y.; Shao, X.; Chi, C.; Adams, D.J.; Ding, J.; et al. Conotoxin αD-GeXXA utilizes a novel strategy to antagonize nicotinic acetylcholine receptors. Sci. Rep. 2015, 5, 14261. [Google Scholar] [CrossRef] [PubMed]
- Jin, A.-H.; Vetter, I.; Himaya, S.W.A.; Alewood, P.F.; Lewis, R.J.; Dutertre, S. Transcriptome and proteome of Conus planorbis identify the nicotinic receptors as primary target for the defensive venom. Proteomics 2015, 15, 4030–4040. [Google Scholar] [CrossRef]
- Hernández-Sámano, C.A.; Falcón, A.; Zamudio, F.; Batista, V.C.; Michel-Morfín, E.J.; Landa-Jaime, V.; López-Vera, E.; Jeziorski, C.M.; Aguilar, B.M. αD-conotoxins in species of the Eastern Pacific: The case of Conus princeps from Mexico. Toxins 2019, 11, 405. [Google Scholar] [CrossRef]
- Liu, Z.; Li, H.; Liu, N.; Wu, C.; Jiang, J.; Yue, J.; Jing, Y.; Dai, Q. Diversity and evolution of conotoxins in Conus virgo, Conus eburneus, Conus imperialis and Conus marmoreus from the South China Sea. Toxicon 2012, 60, 982–989. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, X.; Ren, Z.; Tang, W.; Zou, Q.; Wang, J.; Chen, S.; Zhang, H.; Xu, A. Oxidative folding of conopeptides modified by Conus protein disulfide isomerase. Protein J. 2017, 36, 407–416. [Google Scholar] [CrossRef]
- Safavi-Hemami, H.; Hu, H.; Gorasia, D.G.; Bandyopadhyay, P.K.; Veith, P.D.; Young, N.D.; Reynolds, E.C.; Yandell, M.; Olivera, B.M.; Purcell, A.W. Combined proteomic and transcriptomic interrogation of the venom gland of Conus geographus uncovers novel components and functional compartmentalization. Mol. Cell. Proteom. 2014, 13, 938–953. [Google Scholar] [CrossRef]
- Jouiaei, M.; Yanagihara, A.A.; Madio, B.; Nevalainen, T.J.; Alewood, P.F.; Fry, B.G. Ancient venom systems: A review on Cnidaria toxins. Toxins 2015, 7, 2251–2271. [Google Scholar] [CrossRef]
- Tasoulis, T.; Isbister, G.K. A review and database of snake venom proteomes. Toxins 2017, 9, 290. [Google Scholar] [CrossRef] [PubMed]
- Lavergne, V.; Dutertre, S.; Jin, A.-h.; Lewis, R.J.; Taft, R.J.; Alewood, P.F. Systematic interrogation of the Conus marmoreus venom duct transcriptome with ConoSorter reveals 158 novel conotoxins and 13 new gene superfamilies. BMC Genom. 2013, 14, 708. [Google Scholar] [CrossRef] [PubMed]
- Rucavado, A.; Nicolau, C.A.; Escalante, T.; Kim, J.; Herrera, C.; Gutiérrez, J.M.; Fox, J.W. Viperid envenomation wound exudate contributes to increased vascular permeability via a DAMPs/TLR-4 mediated pathway. Toxins 2016, 8, 349. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, S.H.; Leipold, E. Conotoxins of the O-superfamily affecting voltage-gated sodium channels. Cell. Mol. Life Sci. 2007, 64, 1329–1340. [Google Scholar] [CrossRef] [PubMed]
- Craig, A.G.; Zafaralla, G.; Cruz, L.J.; Santos, A.D.; Hillyard, D.R.; Dykert, J.; Rivier, J.E.; Gray, W.R.; Imperial, J.; DelaCruz, R.G.; et al. An O-glycosylated neuroexcitatory Conus peptide. Biochemistry 1998, 37, 16019–16025. [Google Scholar] [CrossRef] [PubMed]
- López-Vera, E.; Jacobsen, R.B.; Ellison, M.; Olivera, B.M.; Teichert, R.W. A novel alpha conotoxin (α-PIB) isolated from C. purpurascens is selective for skeletal muscle nicotinic acetylcholine receptors. Toxicon 2007, 49, 1193–1199. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.D.; McIntosh, J.M.; Hillyard, D.R.; Cruz, L.J.; Olivera, B.M. The A-superfamily of Conotoxins: Structural and functional divergence. J. Biol. Chem. 2004, 279, 17596–17606. [Google Scholar] [CrossRef] [PubMed]
- Abalde, S.; Tenorio, M.J.; Uribe, J.E.; Zardoya, R. Conidae phylogenomics and evolution. Zool. Scr. 2019, 48, 194–214. [Google Scholar] [CrossRef]
- Andrews, S. FastQC. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 24 September 2019).
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Smith-Unna, R.; Boursnell, C.; Patro, R.; Hibberd, J.M.; Kelly, S. TransRate: Reference-free quality assessment of de novo transcriptome assemblies. Genome Res. 2016, 26, 1134–1144. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Wheeler, D.L. GenBank. Nucleic Acids Res. 2005, 33, D34–D38. [Google Scholar] [CrossRef] [PubMed]
- Uniprot Consortium T. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357. [Google Scholar] [CrossRef] [PubMed]
- Leng, N.; Dawson, J.A.; Thomson, J.A.; Ruotti, V.; Rissman, A.I.; Smits, B.M.G.; Haag, J.D.; Gould, M.N.; Stewart, R.M.; Kendziorski, C. EBSeq: An empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 2013, 29, 1035–1043. [Google Scholar] [CrossRef]

| Sample | Specimen No. | Voucher ID MNCN | Location | SRA Accesion No. | Raw Reads | Clean Reads | Contigs | Conotoxin Reads | BUSCO | Transrate |
|---|---|---|---|---|---|---|---|---|---|---|
| magus1 | OK194 | 15.05/87629 | Ishigaki, Japan | SRR9831243 | 31,530,877 | 30,998,130 | 61.842 | 10,294,213 | 34.7% | 0.33 |
| magus2 | OK206 | 15.05/87641 | Ishigaki, Japan | SRR9831255 | 41,380,216 | 40,552,301 | 73.039 | 13,566,387 | 34.0% | 0.30 |
| magus3 | – | – | Central Philippines | SRX5015024 | 16,303,626 | 15,925,034 | 129.180 | 4,045,608 | 72.4% | 0.34 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pardos-Blas, J.R.; Irisarri, I.; Abalde, S.; Tenorio, M.J.; Zardoya, R. Conotoxin Diversity in the Venom Gland Transcriptome of the Magician’s Cone, Pionoconus magus. Mar. Drugs 2019, 17, 553. https://doi.org/10.3390/md17100553
Pardos-Blas JR, Irisarri I, Abalde S, Tenorio MJ, Zardoya R. Conotoxin Diversity in the Venom Gland Transcriptome of the Magician’s Cone, Pionoconus magus. Marine Drugs. 2019; 17(10):553. https://doi.org/10.3390/md17100553
Chicago/Turabian StylePardos-Blas, José R., Iker Irisarri, Samuel Abalde, Manuel J. Tenorio, and Rafael Zardoya. 2019. "Conotoxin Diversity in the Venom Gland Transcriptome of the Magician’s Cone, Pionoconus magus" Marine Drugs 17, no. 10: 553. https://doi.org/10.3390/md17100553
APA StylePardos-Blas, J. R., Irisarri, I., Abalde, S., Tenorio, M. J., & Zardoya, R. (2019). Conotoxin Diversity in the Venom Gland Transcriptome of the Magician’s Cone, Pionoconus magus. Marine Drugs, 17(10), 553. https://doi.org/10.3390/md17100553