Double the Chemistry, Double the Fun: Structural Diversity and Biological Activity of Marine-Derived Diketopiperazine Dimers
Abstract
1. General Considerations
2. Chemistry and Biological Properties of Marine Diketopiperazine Dimers
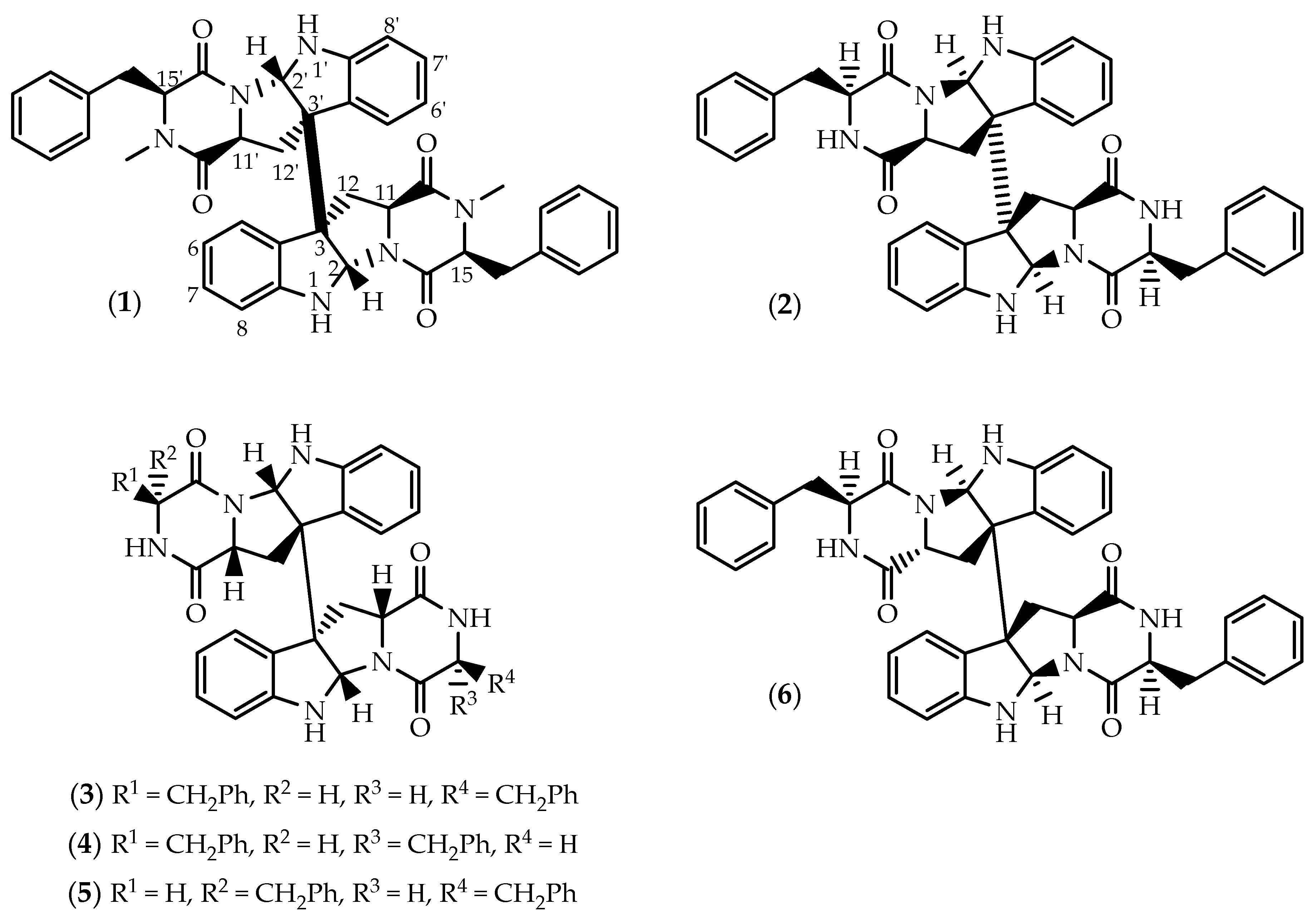
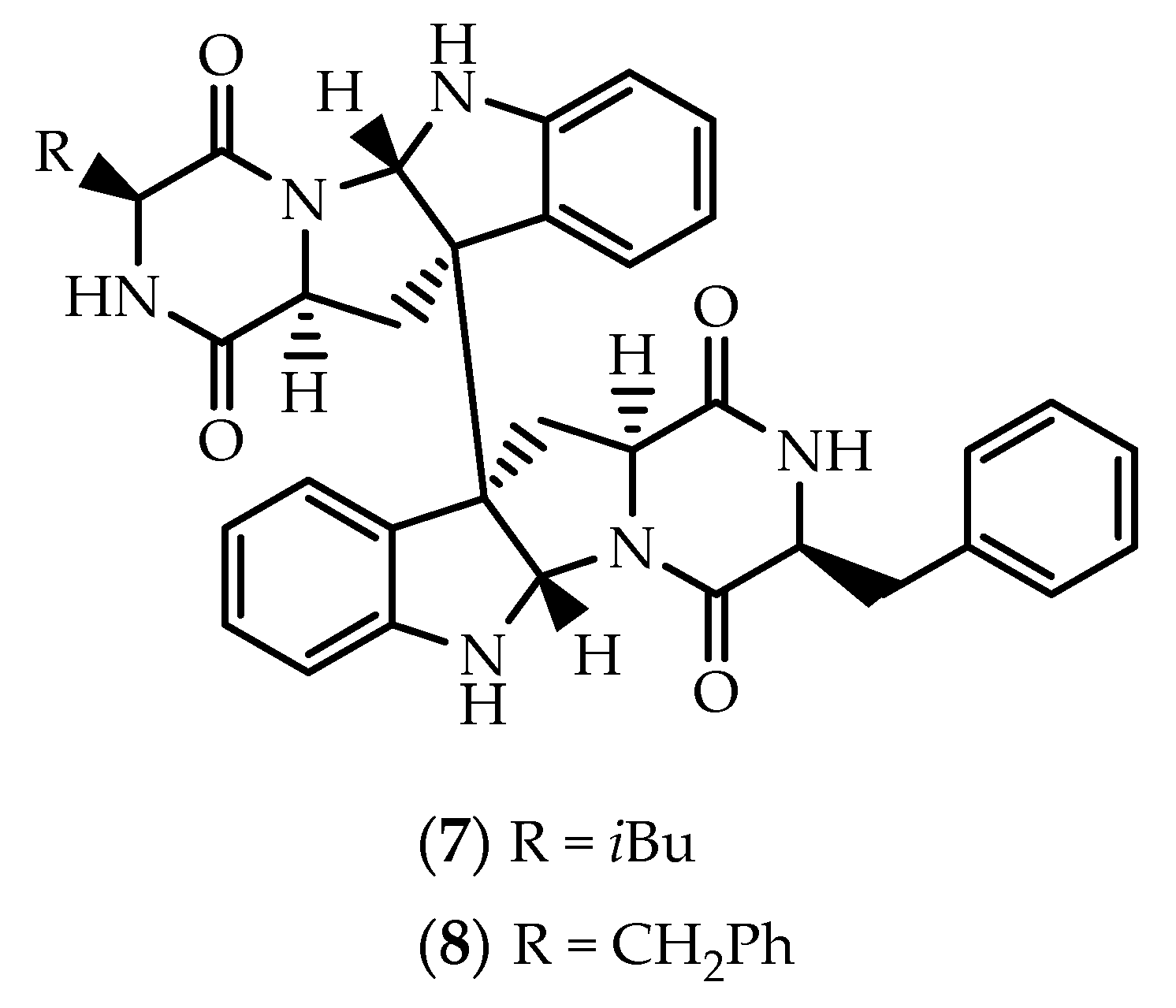
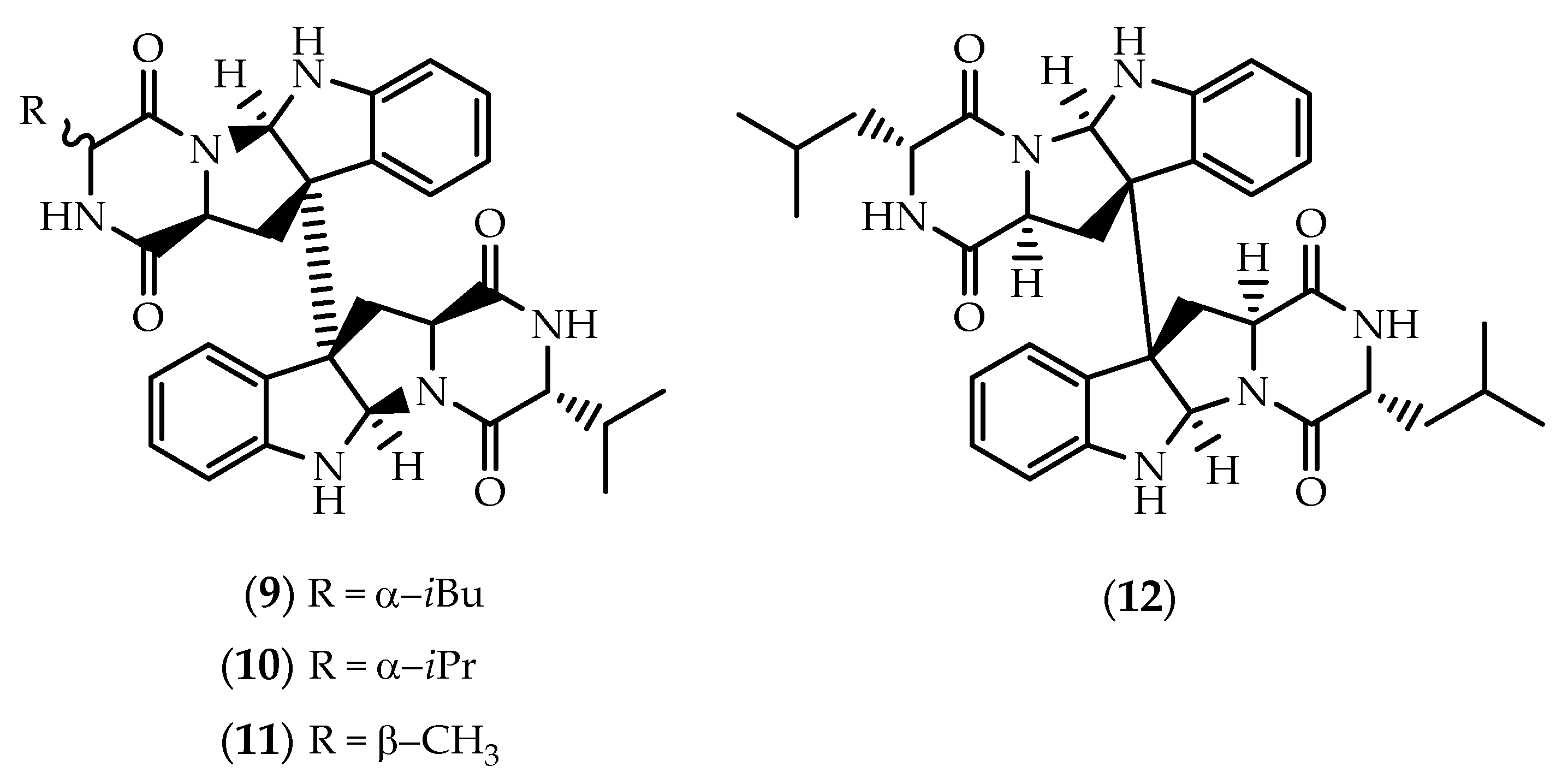
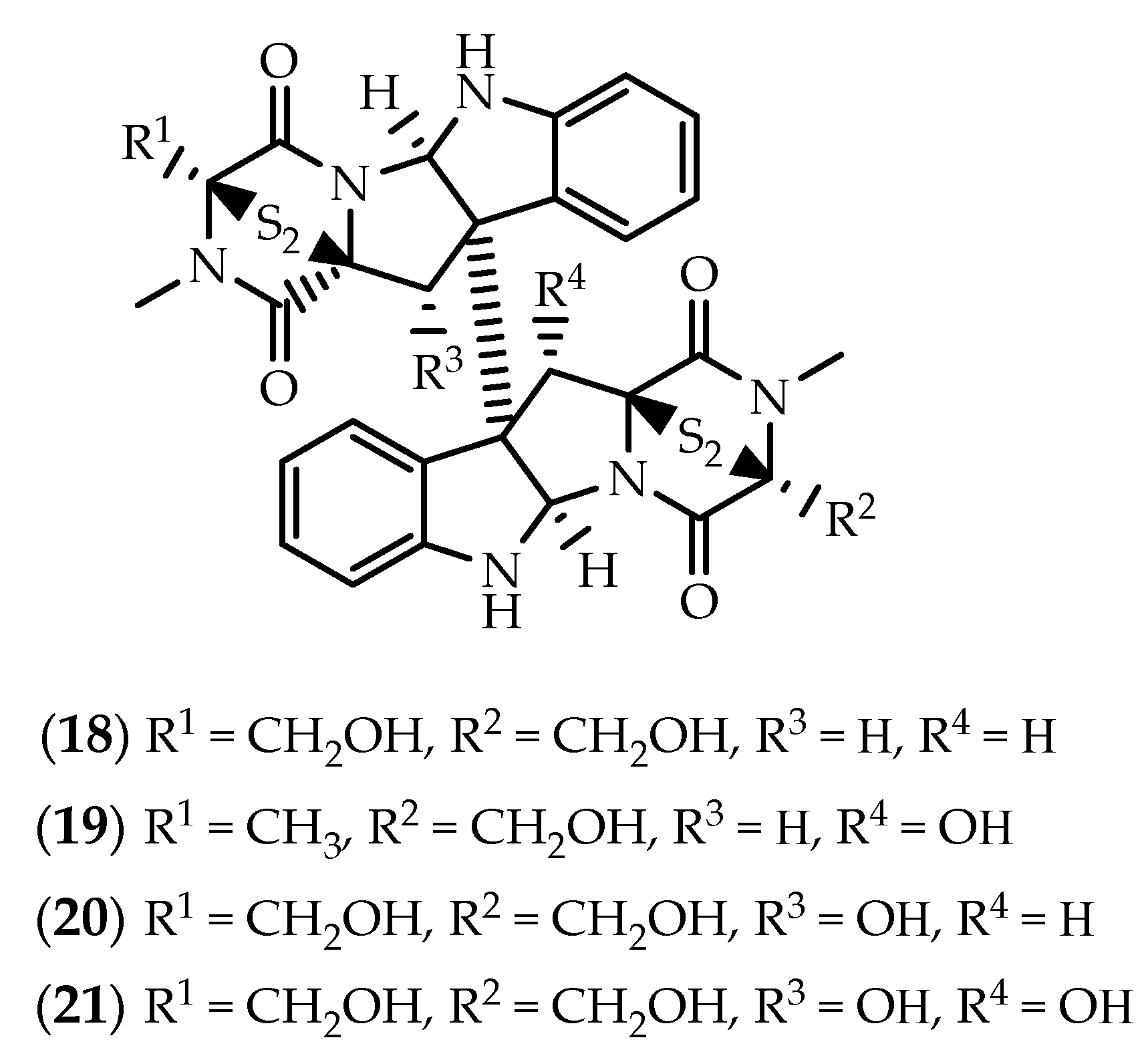
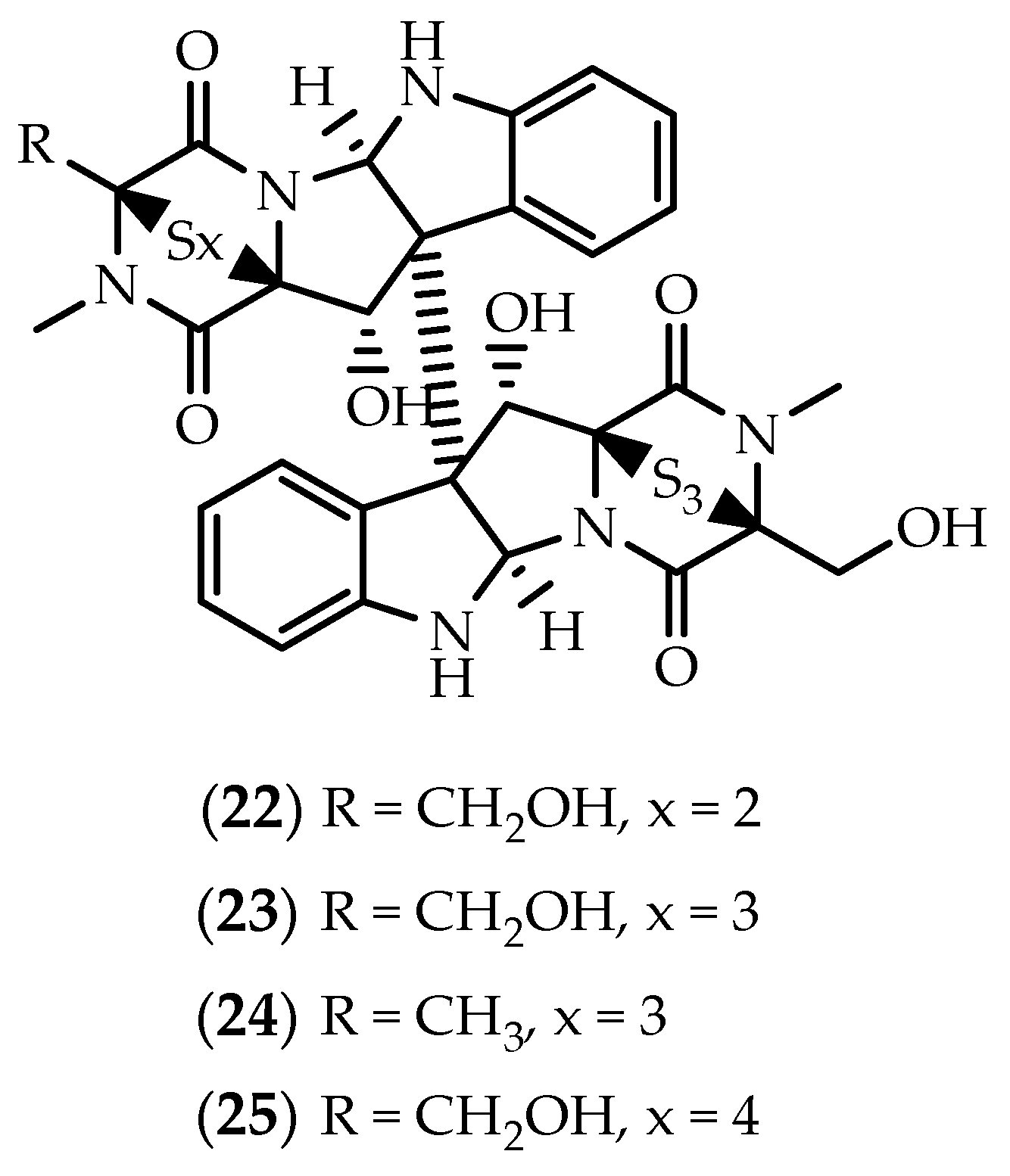
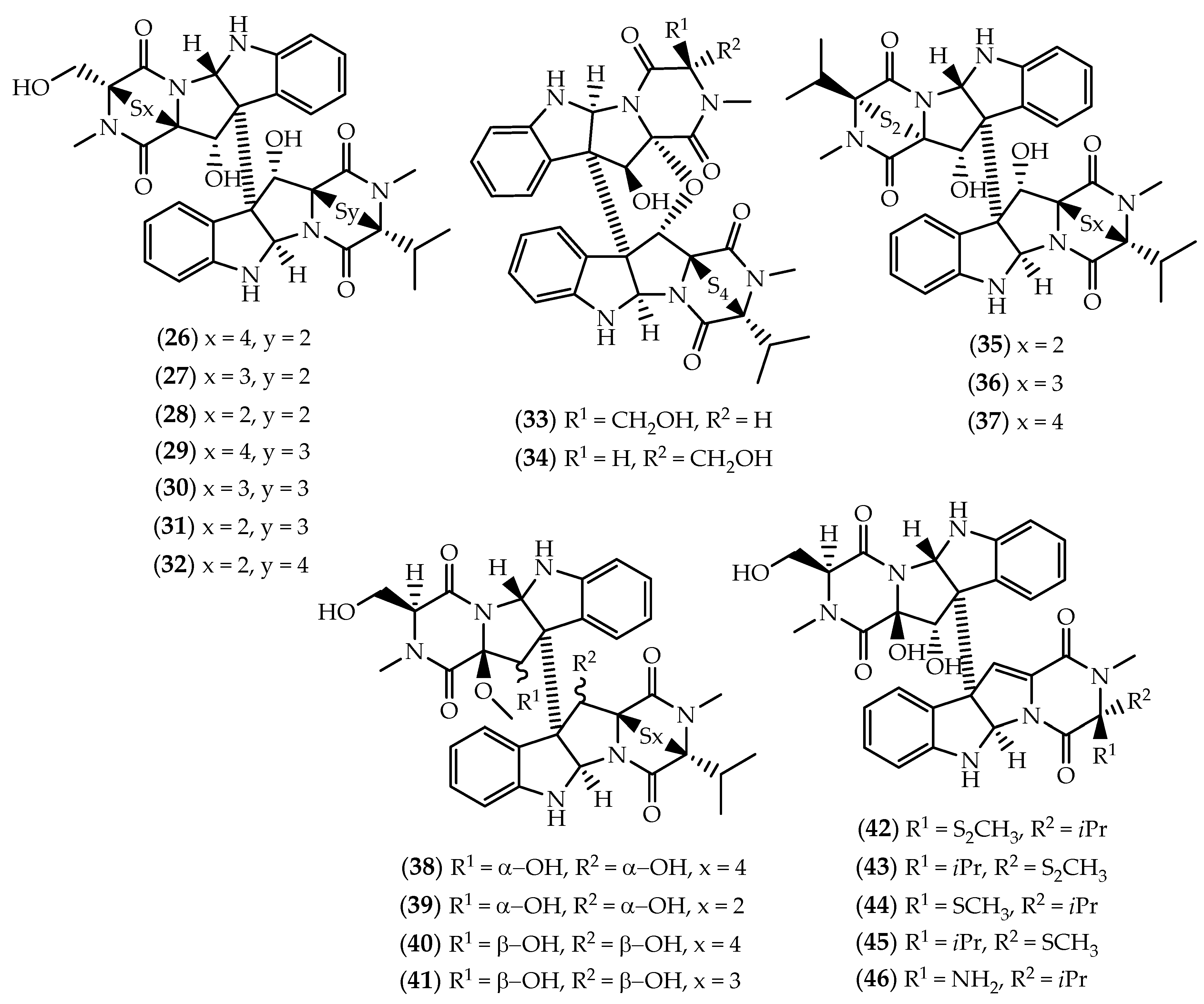
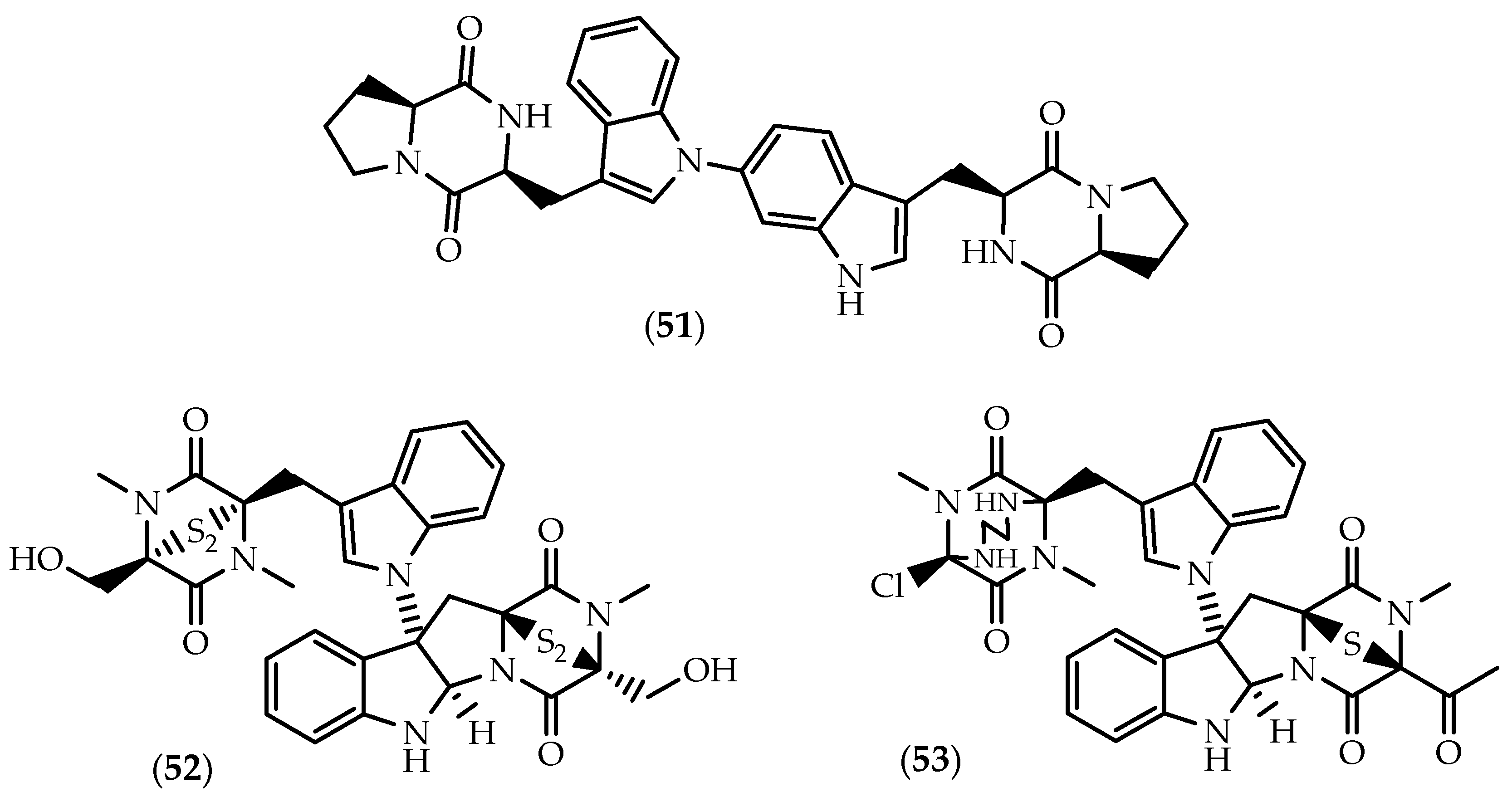
2.1. Dimers Joined Through C3-C3’ Bond
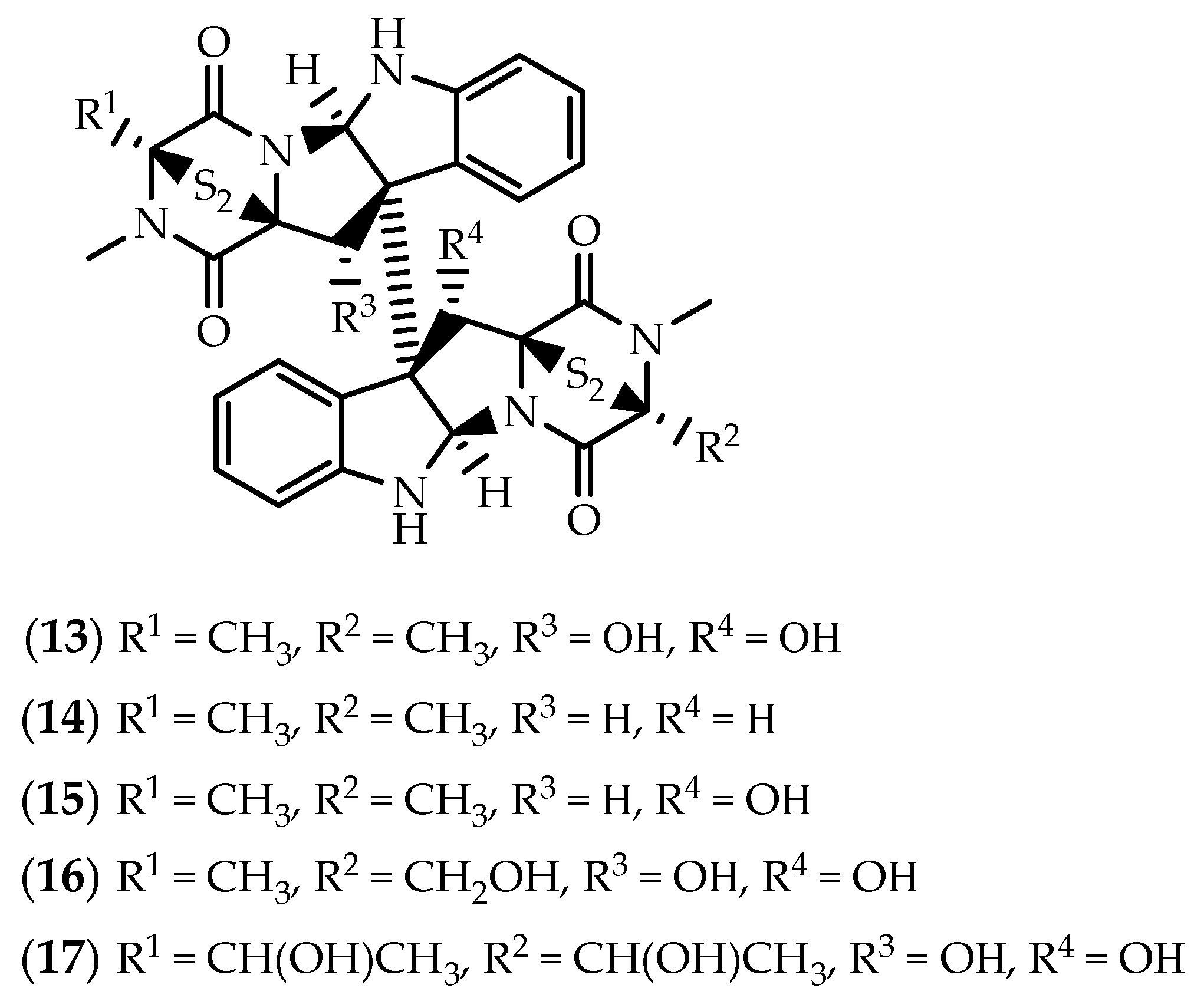
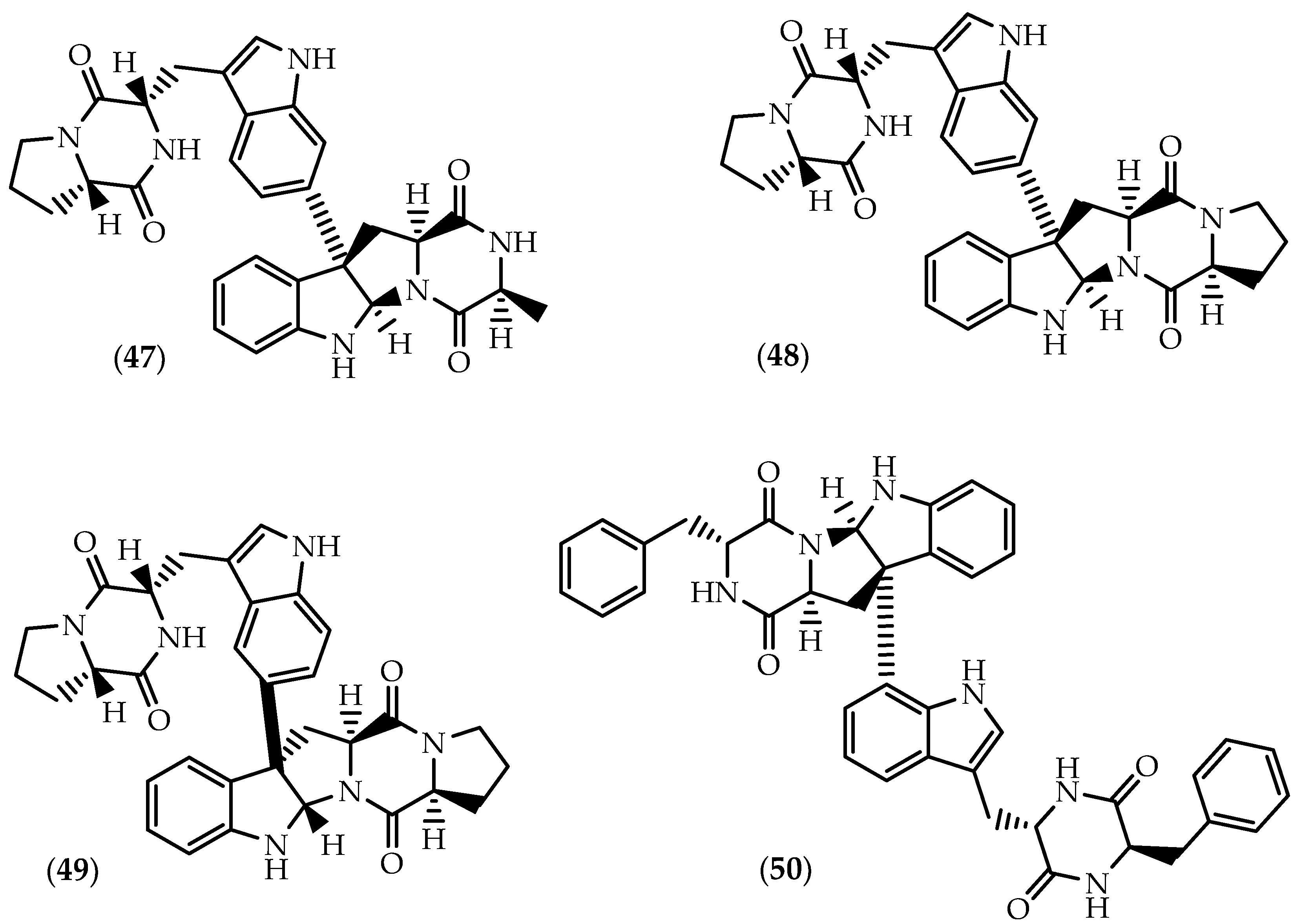
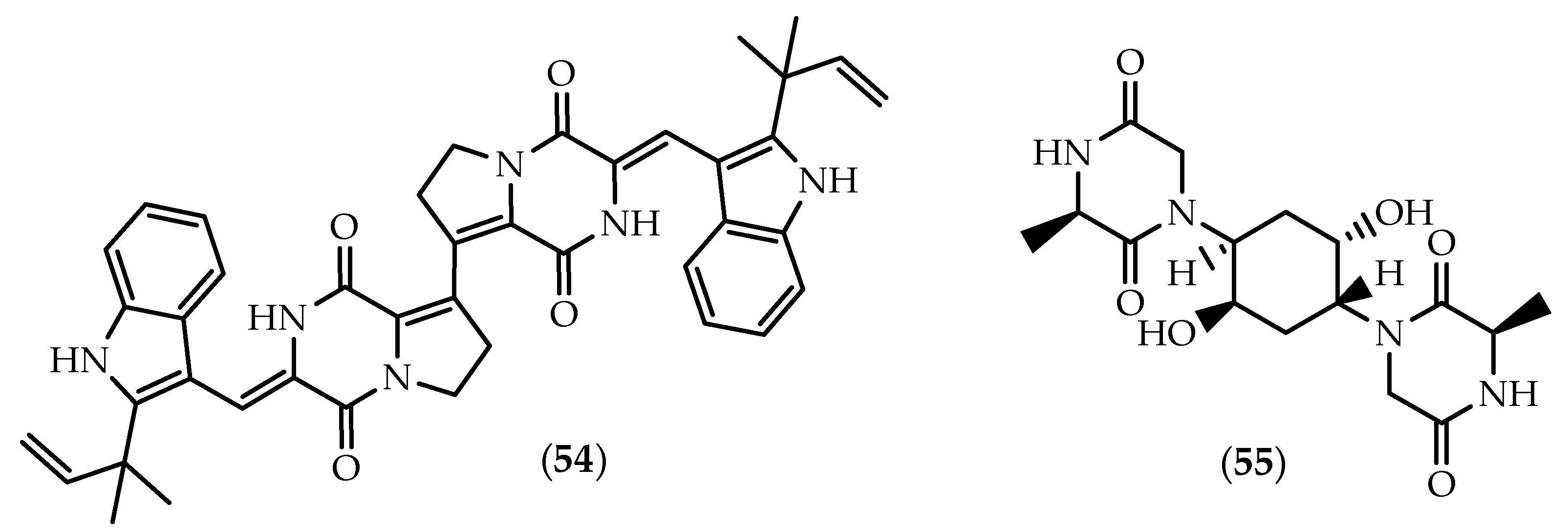
2.2. Dimers Joined Through Distinct Linkages
3. Discussion
3.1. Considerations on the Occurrence of Marine Diketopiperazine Dimers
3.2. Pharmacological Highlights and the Importance of Being Dimeric
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2019, 36, 122–173. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.B.; Evdokimov, N.M.; Lefranc, F.; Valentão, P.; Kornienko, A.; Pereira, D.M.; Andrade, P.B.; Gomes, N.G.M. Marine-derived anticancer agents: Clinical benefits, innovative mechanisms, and new targets. Mar. Drugs 2019, 17, 329. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.B.; Dasari, R.; Lefranc, F.; Kornienko, A.; Kiss, R.; Gomes, N.G.M. Targeting enzymatic pathways with marine-derived clinical agents. In Natural Products Targeting Clinically Relevant Enzymes, 1st ed.; Andrade, P.B., Valentão, P., Pereira, D.M., Eds.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2017; pp. 255–275. [Google Scholar]
- Huang, R.M.; Yi, X.X.; Zhou, Y.; Su, X.; Peng, Y.; Gao, C.H. An update on 2,5 diketopiperazines from marine organisms. Mar. Drugs 2014, 12, 6213–6235. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Zhou, X.; Xu, T.; Yang, X.; Liu, Y. Diketopiperazines from marine organisms. Chem. Biodivers. 2010, 7, 2809–2829. [Google Scholar] [CrossRef] [PubMed]
- Sano, S.; Nakao, M. Chemistry of 2,5-diketopiperazine and its bis-lactim ether: A brief review. Heterocycles 2015, 91, 1349–1375. [Google Scholar] [CrossRef]
- Borthwick, A.D. 2,5-Diketopiperazines: Synthesis, reactions, medicinal chemistry, and bioactive natural products. Chem. Rev. 2012, 112, 3641–3716. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.M.; Liang, X.A.; Kong, Y.; Jia, B. Structural diversity and biological activities of indole diketopiperazine alkaloids from fungi. J. Agric. Food Chem. 2016, 64, 6659–6671. [Google Scholar] [CrossRef] [PubMed]
- Ressureição, A.S.M.; Delatouche, R.; Gennari, C.; Piarulli, U. Bifunctional 2,5-diketopiperazines as rigid three-dimensional scaffolds in receptors and peptidomimetics. Eur. J. Org. Chem. 2011, 2011, 217–228. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, P.; Ma, H.; Zhu, W. Developments around the bioactive diketopiperazines: A patent review. Expert Opin. Ther. Pat. 2013, 23, 1415–1433. [Google Scholar] [CrossRef]
- Sung, B.J.; Hwang, K.Y.; Jeon, Y.H.; Lee, J.I.; Heo, Y.S.; Kim, J.H.; Moon, J.; Yoon, J.M.; Hyun, Y.L.; Kim, E.; et al. Structure of the catalytic domain of human phosphodiesterase 5 with bound drug molecules. Nature 2003, 425, 98–102. [Google Scholar] [CrossRef]
- Borthwick, A.D. Oral oxytocin antagonists. J. Med. Chem. 2010, 53, 6525–6538. [Google Scholar] [CrossRef] [PubMed]
- Gomes, N.G.M.; Lefranc, F.; Kijjoa, A.; Kiss, R. Can some marine-derived fungal metabolites become actual anticancer agents? Mar. Drugs 2015, 13, 3950–3991. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Yamazaki-Nakamura, Y.; Yakushiji, F. Medicinal chemistry and chemical biology of diketopiperazine-type antimicrotubule and vascular-disrupting agents. Chem. Pharm. Bull. 2013, 61, 889–901. [Google Scholar] [CrossRef] [PubMed]
- Mohanlal, R.W.; Lloyd, K.; Huang, L. Plinabulin, a novel small molecule clinical stage IO agent with anti-cancer activity, to prevent chemo–induced neutropenia and immune related AEs. J. Clin. Oncol. 2018, 36, 126. [Google Scholar] [CrossRef]
- Cornacchia, C.; Cacciatore, I.; Baldassarre, L.; Mollica, A.; Feliciani, F.; Pinnen, F. 2,5 Diketopiperazines as neuroprotective agents. Mini Rev. Med. Chem. 2012, 12, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Teixidó, M.; Zurita, E.; Malakoutikhah, M.; Tarragó, T.; Giralt, E. Diketopiperazines as a tool for the study of transport across the blood-brain barrier (BBB) and their potential use as BBB-Shuttles. J. Am. Chem. Soc. 2007, 129, 11802–11813. [Google Scholar] [CrossRef]
- Kogut, E.F.; Thoen, J.C.; Lipton, M.A. Examination and enhancement of enantioselective autoinduction in cyanohydrin formation by cyclo[(R)-His-(R)-Phe]. J. Org. Chem. 1998, 63, 4604–4610. [Google Scholar] [CrossRef]
- Oku, J.I.; Inoue, S. Asymmetric cyanohydrin synthesis catalysed by a synthetic cyclic dipeptide. J. Chem. Soc. Chem. Commun. 1981, 229–230. [Google Scholar] [CrossRef]
- Martins, M.B.; Carvalho, I. Diketopiperazines: Biological activity and synthesis. Tetrahedron 2007, 63, 9923–9932. [Google Scholar] [CrossRef]
- Jia, B.; Ma, Y.; Chen, D.; Chen, P.; Hu, Y. Studies on structure and biological activity of indole diketopiperazine alkaloids. Prog. Chem. 2018, 30, 1067–1081. [Google Scholar]
- Li, S.M. Prenylated indole derivatives from fungi: Structure diversity, biological activities, biosynthesis and chemoenzymatic synthesis. Nat. Prod. Rep. 2010, 27, 57–78. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Sanchis, P.; Savina, S.A.; Albericio, F.; Álvarez, M. Structure, bioactivity and synthesis of natural products with hexahydropyrrolo [2,3-b] indole. Chem. Eur. J. 2011, 17, 1388–1408. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Y.; Zhang, X.; Lai, D.; Zhou, L. Structural diversity and biological activities of the cyclodipeptides from fungi. Molecules 2017, 22, 2026. [Google Scholar] [CrossRef] [PubMed]
- Li, S.M. Applications of dimethylallyltryptophan synthases and other indole prenyltransferases for structural modification of natural products. Appl. Microbiol. Biotechnol. 2009, 84, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Belin, P.; Moutiez, M.; Lautru, S.; Seguin, J.; Pernodet, J.L.; Gondry, M. The nonribosomal synthesis of diketopiperazines in tRNA-dependent cyclodipeptide synthase pathways. Nat. Prod. Rep. 2012, 29, 961–979. [Google Scholar] [CrossRef] [PubMed]
- Giessen, T.W.; Marahiel, M.A. Ribosome-independent biosynthesis of biologically active peptides: Application of synthetic biology to generate structural diversity. FEBS Lett. 2012, 586, 2065–2075. [Google Scholar] [CrossRef] [PubMed]
- Payne, J.A.E.; Schoppet, M.; Hansen, M.H.; Cryle, M.J. Diversity of nature’s assembly lines—Recent discoveries in non-ribosomal peptide synthesis. Mol. BioSyst. 2017, 13, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Gondry, M.; Sauguet, L.; Belin, P.; Thai, R.; Amouroux, R.; Tellier, C.; Tuphile, K.; Jacquet, M.; Braud, S.; Courçon, M.; et al. Cyclodipeptide synthases are a family of tRNA-dependent peptide bond–forming enzymes. Nat. Chem. Biol. 2009, 5, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yu, H.; Li, S.M. Expanding tryptophan-containing cyclodipeptide synthase spectrum by identification of nine members from Streptomyces strains. Appl. Microbiol. Biotechnol. 2018, 102, 4435–4444. [Google Scholar] [CrossRef]
- Sauguet, L.; Moutiez, M.; Li, Y.; Belin, P.; Seguin, J.; Le Du, M.H.; Thai, R.; Masson, C.; Fonvielle, M.; Pernodet, J.L.; et al. Cyclodipeptide synthases, a family of class-I aminoacyl-tRNAsynthetase-like enzymes involved in non-ribosomal peptide synthesis. Nucleic Acids Res. 2011, 39, 4475–4489. [Google Scholar] [CrossRef]
- Borgman, P.; Lopez, R.D.; Lane, A.L. The expanding spectrum of diketopiperazine natural product biosynthetic pathways containing cyclodipeptide synthases. Org. Biomol. Chem. 2019, 17, 2305–2314. [Google Scholar] [CrossRef] [PubMed]
- Moutiez, M.; Schmitt, E.; Seguin, J.; Thai, R.; Favry, E.; Belin, P.; Mechulam, Y.; Gondry, M. Unravelling the mechanism of non-ribosomal peptide synthesis by cyclodipeptide synthases. Nat. Commun. 2014, 5, 5141. [Google Scholar] [CrossRef] [PubMed]
- Gondry, M.; Jacques, I.B.; Thai, R.; Babin, M.; Canu, N.; Seguin, J.; Belin, P.; Pernodet, J.L.; Moutiez, M. A comprehensive overview of the cyclodipeptide synthase family enriched with the characterization of 32 new enzymes. Front. Microbiol. 2018, 9, 46. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.K.; Choi, J.; Choi, S.J.; Baek, K.H. Cyclodipeptides: An overview of their biosynthesis and biological activity. Molecules 2017, 22, 1796. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, S.; Sato, M.; Tsunematsu, Y.; Watanabe, K. Evaluation of biosynthetic pathway and engineered biosynthesis of alkaloids. Molecules 2016, 21, 1078. [Google Scholar] [CrossRef]
- Giessen, T.W.; von Tesmar, A.M.; Marahiel, M.A. A tRNA-Dependent two-enzyme pathway for the generation of singly and doubly methylated ditryptophan 2,5-diketopiperazines. Biochemistry 2013, 52, 4274–4283. [Google Scholar] [CrossRef]
- Ortiz, A.; Sansinenea, E. Cyclic dipeptides: Secondary metabolites isolated from different microorganisms with diverse biological activities. Curr. Med. Chem. 2017, 24, 2773–2780. [Google Scholar] [CrossRef]
- Prasad, C. Bioactive cyclic dipeptides. Peptides 1995, 16, 151–164. [Google Scholar] [CrossRef]
- Gomes, N.G.M.; Pereira, D.M.; Valentão, P.; Andrade, P.B. Hybrid MS/NMR methods on the prioritization of natural products: Applications in drug discovery. J. Pharm. Biomed. Anal. 2018, 147, 234–249. [Google Scholar] [CrossRef]
- Bérubé, G. Natural and synthetic biologically active dimeric molecules: Anticancer agents, anti-HIV agents, steroid derivatives and opioid antagonists. Curr. Med. Chem. 2006, 13, 131–154. [Google Scholar] [CrossRef]
- Springer, J.P.; Büchi, G.; Kobbe, B.; Demain, A.L.; Clardy, J. The structure of ditryptophenaline—A new metabolite of Aspergillus flavus. Tetrahedron Lett. 1977, 18, 2403–2406. [Google Scholar] [CrossRef]
- Lin, A.Q.; Du, L.; Fang, Y.C.; Wang, F.Z.; Zhu, T.J.; Gu, Q.Q.; Zhu, W.M. iso-α-Cyclopiazonic acid, a new natural product isolated from the marine-derived fungus Aspergillus flavus C-F-3. Chem. Nat. Compd. 2009, 45, 677–680. [Google Scholar] [CrossRef]
- Sun, K.; Li, Y.; Guo, L.; Wang, Y.; Liu, P.; Zhu, W. Indole diterpenoids and isocoumarin from the fungus, Aspergillus flavus, isolated from the prawn, Penaeus vannamei. Mar. Drugs 2014, 12, 3970–3981. [Google Scholar] [CrossRef]
- Zhang, J.; Peng, J.; Liu, T.; Xin, Z. Isolation and identification of endophyte and its secondary metabolites from Salicornia bigelovii Torr. based on type I polyketide synthase (PKS I) gene. Shipin Kexue (Food Sci.) 2016, 37, 114–119. [Google Scholar]
- Nakagawa, M.; Sugumi, H.; Kodato, S.; Hino, T. Oxidative dimerization of Nb-acyltryptophans total synthesis and absolute configuration of ditryptophenaline. Tetrahedron Lett. 1981, 22, 5323–5326. [Google Scholar] [CrossRef]
- Maes, C.M.; Potgieter, M.; Steyn, P.S. N.m.r. assignments, conformation, and absolute configuration of ditryptophenaline and model dioxopiperazines. J. Chem. Soc. Perkin Trans. 1 1986, 861–866. [Google Scholar] [CrossRef]
- Saruwatari, T.; Yagishita, F.; Mino, T.; Noguchi, H.; Hotta, K.; Watanabe, K. Cytochrome P450 as dimerization catalyst in diketopiperazine alkaloid biosynthesis. ChemBioChem 2014, 15, 656–659. [Google Scholar] [CrossRef]
- Barrow, C.J.; Sedlock, S.M. 1’-(2-Phenyl-ethylene)-ditryptophenaline, a new dimeric diketopiperazine from Aspergillus flavus. J. Nat. Prod. 1994, 57, 1239–1244. [Google Scholar] [CrossRef]
- Barrow, C.J.; Cai, P.; Snyder, J.K.; Sedlock, D.M.; Sun, H.H.; Cooper, R. WIN 64821, a new competitive antagonist to substance P, isolated from an Aspergillus species: Structure determination and solution conformation. J. Org. Chem. 1993, 58, 6016–6021. [Google Scholar] [CrossRef]
- Sedlock, D.M.; Barrow, C.J.; Brownell, J.E.; Hong, A.; Gillum, A.M.; Houck, D.R. WIN 64821, a novel neurokinin antagonist produced by an Aspergillus sp. I. Fermentation and isolation. J. Antibiot. 1994, 47, 391–398. [Google Scholar] [CrossRef]
- Ding, L.; Li, F.C.; Qin, M.; Qin, S.; Kelter, G.; Fiebig, H.H.; Laatsch, H. Anti-tumor compounds isolated from marine Aspergillus sp. Chin. J. Nat. Med. 2008, 6, 421–424. [Google Scholar] [CrossRef]
- Barrow, C.J.; Musza, L.L.; Cooper, R. Structure-activity studies of the natural product substance P antagonist WIN 64821. Bioorg. Med. Chem. Lett. 1995, 5, 377–380. [Google Scholar] [CrossRef]
- Popp, J.L.; Musza, L.L.; Barrow, C.J.; Rudewicz, P.J.; Houck, D.R. WIN 64821, a novel neurokinin antagonist produced by an Aspergillus sp. III. Biosynthetic analogs. J. Antibiot. 1994, 47, 411–419. [Google Scholar] [CrossRef]
- Oleynek, J.J.; Sedlock, D.M.; Barrow, C.J.; Appell, K.C.; Casiano, F.; Haycock, D.; Ward, S.J.; Kaplita, P.; Gillum, A.M. WIN 64821, a novel neurokinin antagonist produced by an Aspergillus sp. II. Biological activity. J. Antibiot. 1994, 47, 399–410. [Google Scholar] [CrossRef]
- Overman, L.E.; Paone, D.V. Enantioselective total syntheses of ditryptophenaline and ent-WIN 64821. J. Am. Chem. Soc. 2001, 123, 9465–9467. [Google Scholar] [CrossRef]
- Xu, J.; Hu, Q.; Ding, W.; Wang, P.; Di, Y. New asymmetrical bispyrrolidinoindoline diketopiperazines from the marine fungus Aspergillus sp. DX4H. Nat. Prod. Res. 2018, 32, 815–820. [Google Scholar] [CrossRef]
- Gu, B.B.; Gui, Y.H.; Liu, L.; Su, Z.Y.; Jiao, W.H.; Li, L.; Sun, F.; Wang, S.P.; Yang, F.; Lin, H.W. A new asymmetric diketopiperazine dimer from the sponge-associated fungus Aspergillus versicolor 16F-11. Magn. Reson. Chem. 2019, 57, 49–54. [Google Scholar] [CrossRef]
- Cho, K.H.; Sohn, J.H.; Oh, H. Isolation and structure determination of a new diketopiperazine dimer from marine-derived fungus Aspergillus sp. SF-5280. Nat. Prod. Res. 2018, 32, 214–221. [Google Scholar] [CrossRef]
- Kaur, A.; Raja, H.A.; Darveaux, B.A.; Chen, W.L.; Swanson, S.M.; Pearce, C.J.; Oberlies, N.H. New diketopiperazine dimer from a filamentous fungal isolate of Aspergillus sydowii. Magn. Reson. Chem. 2015, 53, 616–619. [Google Scholar] [CrossRef]
- Ovenden, S.P.; Sberna, G.; Tait, R.M.; Wildman, H.G.; Patel, R.; Li, B.; Steffy, K.; Nguyen, N.; Meurer-Grimes, B.M. A diketopiperazine dimer from a marine-derived isolate of Aspergillus niger. J. Nat. Prod. 2004, 67, 2093–2095. [Google Scholar] [CrossRef]
- Pérez-Balado, C.; Rodríguez-Graña, P.; de Lera, A.R. Stereocontrolled and versatile total synthesis of bispyrrolidinoindoline diketopiperazine alkaloids: Structural revision of the fungal isolate (+)-asperdimin. Chemistry 2009, 15, 9928–9937. [Google Scholar] [CrossRef]
- Gomes, N.M.; Dethoup, T.; Singburaudom, N.; Gales, L.; Silva, A.M.S.; Kijjoa, A. Eurocristatine, a new diketopiperazine dimer from the marine sponge-associated fungus Eurotium cristatum. Phytochem. Lett. 2012, 5, 717–720. [Google Scholar] [CrossRef]
- Li, Y.; Sun, K.L.; Wang, Y.; Fu, P.; Liu, P.P.; Wang, C.; Zhu, W.M. A cytotoxic pyrrolidinoindoline diketopiperazine dimer from the algal fungus Eurotium herbariorum HT-2. Chin. Chem. Lett. 2013, 24, 1049–1052. [Google Scholar] [CrossRef]
- May Zin, W.W.; Buttachon, S.; Dethoup, T.; Pereira, J.A.; Gales, L.; Inácio, Â.; Costa, P.M.; Lee, M.; Sekeroglu, N.; Silva, A.M.S.; et al. Antibacterial and antibiofilm activities of the metabolites isolated from the culture of the mangrove-derived endophytic fungus Eurotium chevalieri KUFA 0006. Phytochemistry 2017, 141, 86–97. [Google Scholar] [CrossRef]
- Tadano, S.; Sugimachi, Y.; Sumimoto, M.; Tsukamoto, S.; Ishikawa, H. Collective synthesis and biological evaluation of tryptophan-based dimeric diketopiperazine alkaloids. Chemistry 2016, 22, 1277–1291. [Google Scholar] [CrossRef]
- Du, F.Y.; Li, X.M.; Li, C.S.; Shang, Z.; Wang, B.G. Cristatumins A-D, new indole alkaloids from the marine-derived endophytic fungus Eurotium cristatum EN-220. Bioorg. Med. Chem. Lett. 2012, 22, 4650–4653. [Google Scholar] [CrossRef]
- Lorenzo, P.; Álvarez, R.; de Lera, Á.R. Total synthesis and structural revision of (+)-cristatumin C. J. Nat. Prod. 2014, 77, 421–423. [Google Scholar] [CrossRef]
- Liu, J.; Gu, B.; Yang, L.; Yang, F.; Lin, H. New anti-inflammatory cyclopeptides from a sponge-derived fungus Aspergillus violaceofuscus. Front. Chem. 2018, 6, 226. [Google Scholar] [CrossRef]
- Weindling, R.; Emerson, O.H. The isolation of a toxic substance from the culture filtrate of Trichoderma. Phytopathology 1936, 26, 1068–1070. [Google Scholar]
- Bell, M.R.; Johnson, J.R.; Wildi, B.S.; Woodward, R.B. The structure of gliotoxin. J. Am. Chem. Soc. 1958, 80, 1001. [Google Scholar] [CrossRef]
- Gardiner, M.D.; Waring, P.; Howlett, B.J. The epipolythiodioxopiperazine (ETP) class of fungal toxins: Distribution, mode of action, functions and biosynthesis. Microbiology 2005, 151, 1021–1032. [Google Scholar] [CrossRef]
- Welch, T.R.; Williams, R.M. Epidithiodioxopiperazines: Occurrence, synthesis and biogenesis. Nat. Prod. Rep. 2014, 31, 1376–1404. [Google Scholar] [CrossRef]
- Chai, C.L.; Waring, P. Redox sensitive epidithiodioxopiperazines in biological mechanisms of toxicity. Redox Rep. 2000, 5, 257–264. [Google Scholar] [CrossRef]
- Waring, P.; Chai, C.L.L. The multiple properties of gliotoxin and other epipolythiodioxopiperazine metabolites. Aust. J. Chem. 2015, 68, 178–183. [Google Scholar] [CrossRef]
- Iwasa, E.; Hamashima, Y.; Sodeoka, M. Epipolythiodiketopiperazine alkaloids: Total syntheses and biological activities. Isr. J. Chem. 2011, 51, 420–433. [Google Scholar] [CrossRef]
- Jiang, C.S.; Guo, Y.W. Epipolythiodioxopiperazines from fungi: Chemistry and bioactivities. Mini Rev. Med. Chem. 2011, 11, 728–745. [Google Scholar] [CrossRef]
- Son, B.W.; Jensen, P.R.; Kauffman, C.A.; Fenical, W. New cytotoxic epidithiodioxopiperazines related to verticillin A from a marine isolate of the fungus Penicillium. Nat. Prod. Lett. 1999, 13, 213–222. [Google Scholar] [CrossRef]
- Figueroa, M.; Graf, T.N.; Ayers, S.; Adcock, A.F.; Kroll, D.J.; Yang, J.; Swanson, S.M.; Munoz-Acuna, U.; Carcache de Blanco, E.J.; Agrawal, R.; et al. Cytotoxic epipolythiodioxopiperazine alkaloids from filamentous fungi of the Bionectriaceae. J. Antibiot. 2012, 65, 559–564. [Google Scholar] [CrossRef]
- Dong, J.Y.; He, H.P.; Shen, Y.M.; Zhang, K.Q. Nematicidal epipolysulfanyldioxopiperazines from Gliocladium roseum. J. Nat. Prod. 2005, 68, 1510–1513. [Google Scholar] [CrossRef]
- Katagiri, K.; Sato, K.; Hayakawa, S.; Matsushima, T.; Minato, H. Verticillin A, a new antibiotic from Verticillium sp. J. Antibiot. 1970, 23, 420–422. [Google Scholar] [CrossRef]
- Minato, H.; Matsumoto, M.; Katayama, T. Verticillin A, a new antibiotic from Verticillium sp. J. Chem. Soc. D 1971, 44–45. [Google Scholar] [CrossRef]
- Paschall, A.V.; Yang, D.; Lu, C.; Choi, J.H.; Li, X.; Liu, F.; Figueroa, M.; Oberlies, N.H.; Pearce, C.; Bollag, W.B.; et al. H3K9 trimethylation silences Fas expression to confer colon carcinoma immune escape and 5-fluorouracil chemoresistance. J. Immunol. 2015, 195, 1868–1882. [Google Scholar] [CrossRef]
- Lu, C.; Paschall, A.V.; Shi, H.; Savage, N.; Waller, J.L.; Sabbatini, M.E.; Oberlies, N.H.; Pearce, C.; Liu, K. The MLL1-H3K4me3 axis-mediated PD-L1 expression and pancreatic cancer immune evasion. J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef]
- Chu, M.; Truumees, I.; Rothofsky, M.L.; Patel, M.G.; Gentile, F.; Das, P.R.; Puar, M.S.; Lin, S.L. Inhibition of c-fos proto-oncogene induction by Sch 52900 and Sch 52901, novel diketopiperazine produced by Gliocladium sp. J. Antibiot. 1995, 48, 1440–1445. [Google Scholar] [CrossRef][Green Version]
- Liu, F.; Liu, Q.; Yang, D.; Bollag, W.B.; Robertson, K.; Wu, P.; Liu, K. Verticillin A overcomes apoptosis resistance in human colon carcinoma through DNA methylation-dependent upregulation of BNIP3. Cancer Res. 2011, 71, 6807–6816. [Google Scholar] [CrossRef]
- Lu, C.; Yang, D.; Sabbatini, M.E.; Colby, A.H.; Grinstaff, M.W.; Oberlies, N.H.; Pearce, C.; Liu, K. Contrasting roles of H3K4me3 and H3K9me3 in regulation of apoptosis and gemcitabine resistance in human pancreatic cancer cells. BMC Cancer 2018, 18, 149. [Google Scholar] [CrossRef]
- Zewdu, A.; Lopez, G.; Braggio, D.; Kenny, C.; Constantino, D.; Bid, H.K.; Batte, K.; Iwenofu, O.H.; Oberlies, N.H.; Pearce, C.J.; et al. Verticillin A inhibits leiomyosarcoma and malignant peripheral nerve sheath tumor growth via induction of apoptosis. Clin. Exp. Pharmacol. 2016, 6, 221. [Google Scholar]
- Zhang, Y.X.; Chen, Y.; Guo, X.N.; Zhang, X.W.; Zhao, W.M.; Zhong, L.; Zhou, J.; Xi, Y.; Lin, L.P.; Ding, J. 11,11′-dideoxy-verticillin: A natural compound possessing growth factor receptor tyrosine kinase-inhibitory effect with anti-tumor activity. Anticancer Drugs 2005, 16, 515–524. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, Y.X.; Li, M.H.; Zhao, W.M.; Shi, Y.H.; Miao, Z.H.; Zhang, X.W.; Lin, L.P.; Ding, J. Antiangiogenic activity of 11,11′-dideoxyverticillin, a natural product isolated from the fungus Shiraia bambusicola. Biochem. Biophys. Res. Commun. 2005, 329, 1334–1342. [Google Scholar] [CrossRef]
- Watts, K.R.; Ratnam, J.; Ang, K.H.; Tenney, K.; Compton, J.E.; McKerrow, J.; Crews, P. Assessing the trypanocidal potential of natural and semisynthetic diketopiperazines from two deep water marine-derived fungi. Bioorg. Med. Chem. 2010, 18, 2566–2574. [Google Scholar] [CrossRef]
- Ebrahim, W.; Kjer, J.; Amrani, M.E.; Wray, V.; Lin, W.; Ebel, R.; Lai, D.; Proksch, P. Pullularins E and F, two new peptides from the endophytic fungus Bionectria ochroleuca isolated from the mangrove plant Sonneratia caseolaris. Mar. Drugs 2012, 10, 1081–1091. [Google Scholar] [CrossRef]
- Joshi, B.K.; Gloer, J.B.; Wicklow, D.T. New verticillin and glisoprenin analogues from Gliocladium catenulatum, a mycoparasite of Aspergillus flavus sclerotia. J. Nat. Prod. 1999, 62, 730–733. [Google Scholar] [CrossRef]
- Zheng, C.J.; Kim, C.J.; Bae, K.S.; Kim, Y.H.; Kim, W.G. Bionectins A-C, epidithiodioxopiperazines with anti-MRSA activity, from Bionectra byssicola F120. J. Nat. Prod. 2006, 69, 1816–1819. [Google Scholar] [CrossRef]
- Zheng, C.J.; Park, S.H.; Koshino, H.; Kim, Y.H.; Kim, W.G. Verticillin G, a new antibacterial compound from Bionectra byssicola. J. Antibiot. 2007, 60, 61–64. [Google Scholar] [CrossRef]
- Hauser, D.; Weber, H.P.; Sigg, H.P. Isolierung and strukturaufklärung von Chaetocin. Helv. Chem. Acta 1970, 53, 1061–1073. [Google Scholar] [CrossRef]
- Weber, H.P. The molecular structure and absolute configuration of chaetocin. Acta Crystallogr. Sect. B Struct. Sci. 1972, 28, 2945–2951. [Google Scholar] [CrossRef]
- Greiner, D.; Bonaldi, T.; Eskeland, R.; Roemer, E.; Imhof, A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3–9. Nat. Chem. Biol. 2005, 1, 143–145. [Google Scholar] [CrossRef]
- Greiner, D.; Bonaldi, T.; Eskeland, R.; Roemer, E.; Imhof, A. Reply to “Chaetocin is a nonspecific inhibitor of histone lysine methyltransferases”. Nat. Chem. Biol. 2013, 9, 137. [Google Scholar] [CrossRef]
- Cherblanc, F.L.; Chapman, K.L.; Reid, J.; Borg, A.J.; Sundriyal, S.; Alcazar-Fuoli, L.; Bignell, E.; Demetriades, M.; Schofield, C.J.; DiMaggio, P.A., Jr.; et al. On the histone lysine methyltransferase activity of fungal metabolite chaetocin. J. Med. Chem. 2013, 56, 8616–8625. [Google Scholar] [CrossRef]
- Sodeoka, M.; Dodo, K.; Teng, Y.; Iuchi, K.; Hamashima, Y.; Iwasa, E.; Fujishiro, S. Synthesis and biological activities of chaetocin and its derivatives. Pure Appl. Chem. 2012, 84, 1369–1378. [Google Scholar] [CrossRef]
- Tran, H.T.; Kim, H.N.; Lee, I.K.; Nguyen-Pham, T.N.; Ahn, J.S.; Kim, Y.K.; Lee, J.J.; Park, K.S.; Kook, H.; Kim, H.J. Improved therapeutic effect against leukemia by a combination of the histone methyltransferase inhibitor chaetocin and the histone deacetylase inhibitor trichostatin A. J. Korean Med. Sci. 2013, 28, 237–246. [Google Scholar] [CrossRef]
- Lai, Y.S.; Chen, J.Y.; Tsai, H.J.; Chen, T.Y.; Hung, W.C. The SUV39H1 inhibitor chaetocin induces differentiation and shows synergistic cytotoxicity with other epigenetic drugs in acute myeloid leukemia cells. Blood Cancer J. 2015, 5, e313. [Google Scholar] [CrossRef]
- Liu, X.; Guo, S.; Liu, X.; Su, L. Chaetocin induces endoplasmic reticulum stress response and leads to death receptor 5-dependent apoptosis in human non-small cell lung cancer cells. Apoptosis 2015, 20, 1499–1507. [Google Scholar] [CrossRef]
- Mathison, A.; Salmonson, A.; Missfeldt, M.; Bintz, J.; Williams, M.; Kossak, S.; Nair, A.; de Assunção, T.M.; Christensen, T.; Buttar, N.; et al. Combined AURKA and H3K9 methyltransferase targeting inhibits cell growth by inducing mitotic catastrophe. Mol. Cancer Res. 2017, 15, 984–997. [Google Scholar] [CrossRef]
- Isham, C.R.; Tibodeau, J.D.; Bossou, A.R.; Merchan, J.R.; Bible, K.C. The anticancer effects of chaetocin are independent of programmed cell death and hypoxia, and are associated with inhibition of endothelial cell proliferation. Br. J. Cancer 2012, 106, 314–323. [Google Scholar] [CrossRef]
- Jung, H.J.; Seo, I.; Casciello, F.; Jacquelin, S.; Lane, S.W.; Suh, S.I.; Suh, M.H.; Lee, J.S.; Baek, W.K. The anticancer effect of chaetocin is enhanced by inhibition of autophagy. Cell Death Dis. 2016, 7, e2098. [Google Scholar] [CrossRef]
- Chaib, H.; Nebbioso, A.; Prebet, T.; Castellano, R.; Garbit, S.; Restouin, A.; Vey, N.; Altucci, L.; Collette, Y. Anti-leukemia activity of chaetocin via death receptor-dependent apoptosis and dual modulation of the histone methyl-transferase SUV39H1. Leukemia 2012, 26, 662–674. [Google Scholar] [CrossRef]
- Tibodeau, J.D.; Benson, L.M.; Isham, C.R.; Owen, W.G.; Bible, K.C. The anticancer agent chaetocin is a competitive substrate and inhibitor of thioredoxin reductase. Antioxid. Redox Signal. 2009, 11, 1097–1106. [Google Scholar] [CrossRef]
- Isham, C.R.; Tibodeau, J.D.; Jin, W.; Xu, R.; Timm, M.M.; Bible, K.C. Chaetocin: A promising new antimyeloma agent with in vitro and in vivo activity mediated via imposition of oxidative stress. Blood 2007, 109, 2579–2588. [Google Scholar] [CrossRef]
- Han, X.; Han, Y.; Zheng, Y.; Sun, Q.; Ma, T.; Zhang, J.; Xu, L. Chaetocin induces apoptosis in human melanoma cells through the generation of reactive oxygen species and the intrinsic mitochondrial pathway, and exerts its antitumor activity in vivo. PLoS ONE 2017, 12, e0175950. [Google Scholar] [CrossRef]
- Dixit, D.; Ghildiyal, R.; Anto, N.P.; Sen, E. Chaetocin-induced ROS-mediated apoptosis involves ATM-YAP1 axis and JNK-dependent inhibition of glucose metabolism. Cell. Death Dis. 2014, 5, e1212. [Google Scholar] [CrossRef]
- Li, Z.; Huang, L.; Wei, L.; Hou, Z.; Ye, W.; Huang, S. Chaetocin induces caspase-dependent apoptosis in ovarian cancer cells via the generation of reactive oxygen species. Oncol. Lett. 2019, 18, 1915–1921. [Google Scholar] [CrossRef]
- He, J.; Chen, X.; Li, B.; Zhou, W.; Xiao, J.; He, K.; Zhang, J.; Xiang, G. Chaetocin induces cell cycle arrest and apoptosis by regulating the ROS-mediated ASK-1/JNK signaling pathways. Oncol. Rep. 2017, 38, 2489–2497. [Google Scholar] [CrossRef]
- Yang, G.; Weng, X.; Zhao, Y.; Zhang, X.; Hu, Y.; Dai, X.; Liang, P.; Wang, P.; Ma, L.; Sun, X.; et al. The histone H3K9 methyltransferase SUV39H links SIRT1 repression to myocardial infarction. Nat. Commun. 2017, 8, 14941. [Google Scholar] [CrossRef]
- Kung, A.L.; Zabludoff, S.D.; France, D.S.; Freedman, S.J.; Tanner, E.A.; Vieira, A.; Cornell-Kennon, S.; Lee, J.; Wang, B.; Wang, J.; et al. Small molecule blockade of transcriptional coactivation of the hypoxia-inducible factor pathway. Cancer Cell 2004, 6, 33–43. [Google Scholar] [CrossRef]
- Lee, Y.M.; Lim, J.H.; Yoon, H.; Chun, Y.S.; Park, J.W. Antihepatoma activity of chaetocin due to deregulated splicing of hypoxia-inducible factor 1alpha pre-mRNA in mice and in vitro. Hepatology 2011, 53, 171–180. [Google Scholar] [CrossRef]
- Reece, K.M.; Richardson, E.D.; Cook, K.M.; Campbell, T.J.; Pisle, S.T.; Holly, A.J.; Venzon, D.J.; Liewehr, D.J.; Chau, C.H.; Price, D.K.; et al. Epidithiodiketopiperazines (ETPs) exhibit in vitro antiangiogenic and in vivo antitumor activity by disrupting the HIF-1α/p300 complex in a preclinical model of prostate cancer. Mol. Cancer 2014, 13, 91. [Google Scholar] [CrossRef]
- Vo, M.C.; Nguyen-Pham, T.N.; Lee, H.J.; Jung, S.H.; Choi, N.R.; Hoang, M.D.; Kim, H.J.; Lee, J.J. Chaetocin enhances dendritic cell function via the induction of heat shock protein and cancer testis antigens in myeloma cells. Oncotarget 2017, 8, 46047–46056. [Google Scholar] [CrossRef][Green Version]
- Ono, T.; Kamimura, N.; Matsuhashi, T.; Nagai, T.; Nishiyama, T.; Endo, J.; Hishiki, T.; Nakanishi, T.; Shimizu, N.; Tanaka, H.; et al. The histone 3 lysine 9 methyltransferase inhibitor chaetocin improves prognosis in a rat model of high salt diet-induced heart failure. Sci. Rep. 2017, 7, 39752. [Google Scholar] [CrossRef]
- Zuma, A.A.; Santos, J.O.; Mendes, I.; de Souza, W.; Machado, C.R.; Motta, M.C.M. Chaetocin-A histone methyltransferase inhibitor-impairs proliferation, arrests cell cycle and induces nucleolar disassembly in Trypanosoma cruzi. Acta Trop. 2017, 170, 149–160. [Google Scholar]
- Bae, J.S.; Han, M.; Yao, C.; Chung, J.H. Chaetocin inhibits IBMX-induced melanogenesis in B16F10 mouse melanoma cells through activation of ERK. Chem. Biol. Interact. 2016, 245, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Furuya, K.; Okudaira, M.; Shindo, T.; Sato, A. Corollospora pulchella, a marine fungus producing antibiotics, melinacidins III, IV and gancidin W. Annu. Rep. Sankyo Res. Lab. 1985, 37, 140–142. [Google Scholar]
- Argoudelis, A.D.; Mizsak, S.A. Melinacidins II, III and IV. Structural studies. J. Antibiot. 1977, 30, 468–473. [Google Scholar] [CrossRef] [PubMed]
- Ebead, G.A.; Overy, D.P.; Berrué, F.; Kerr, R.G. Westerdykella reniformis sp. nov., producing the antibiotic metabolites melinacidin IV and chetracin B. IMA Fungus 2012, 3, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, D.; Luan, Y.; Gu, Q.; Zhu, T. Cytotoxic metabolites from the Antarctic psychrophilic fungus Oidiodendron truncatum. J. Nat. Prod. 2012, 75, 920–927. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, Y.; Yu, R.; Feng, Y.; Wang, L.; Che, Q.; Gu, Q.; Li, D.; Li, J.; Zhu, T. Chetracins E and F, cytotoxic epipolythiodioxopiperazines from the marine-derived fungus Acrostalagmus luteoalbus HDN13–530. RSC Adv. 2018, 8, 53–58. [Google Scholar] [CrossRef]
- Song, X.; Zhao, Z.; Qi, X.; Tang, S.; Wang, Q.; Zhu, T.; Gu, Q.; Liu, M.; Li, J. Identification of epipolythiodioxopiperazines HDN-1 and chaetocin as novel inhibitor of heat shock protein 90. Oncotarget 2015, 6, 5263–5274. [Google Scholar] [CrossRef]
- Takahashi, C.; Numata, A.; Ito, Y.; Matsumura, E.; Araki, H.; Iwaki, H.; Kushida, K. Leptosins, antitumour metabolites of a fungus isolated from a marine alga. J. Chem. Soc. Perkin Trans. 1 1994, 1859–1864. [Google Scholar] [CrossRef]
- Takahashi, C.; Numata, A.; Matsumura, E.; Minoura, K.; Eto, H.; Shingu, T.; Ito, T.; Hasegawa, T. Leptosins I and J, cytotoxic substances produced by a Leptosphaeria sp. Physico-chemical properties and structures. J. Antibiot. 1994, 47, 1242–1249. [Google Scholar] [CrossRef]
- Takahashi, C.; Takai, Y.; Kimura, Y.; Numata, A.; Shigematsu, N.; Tanaka, H. Cytotoxic metabolites from a fungal adherent of a marine alga. Phytochemistry 1995, 38, 155–158. [Google Scholar] [CrossRef]
- Takahashi, C.; Minoura, K.; Yamada, T.; Numata, A.; Kushida, K.; Shingu, T.; Hagishita, S.; Nakai, H.; Sato, T.; Harada, H. Potent cytotoxic metabolites from a Leptosphaeria species. Structure determination and conformational analysis. Tetrahedron 1995, 51, 3483–3498. [Google Scholar] [CrossRef]
- Yamada, T.; Iwamoto, C.; Yamagaki, N.; Yamanouchi, T.; Minoura, K.; Yamori, T.; Uehara, Y.; Andoh, T.; Umemura, K.; Numata, A. Leptosins M-N1, cytotoxic metabolites from a Leptosphaeria species separated from a marine alga. Structure determination and biological activities. Tetrahedron 2002, 58, 479–487. [Google Scholar] [CrossRef]
- Yamada, T.; Iwamoto, C.; Yamagaki, N.; Yamanouchi, T.; Minoura, K.; Hagishita, S.; Numata, A. Leptosins O-S, cytotoxic metabolites from a strain of Leptosphaeria sp. isolated from a marine alga. Heterocycles 2004, 63, 641–653. [Google Scholar] [CrossRef]
- Du, L.; Robles, A.J.; King, J.B.; Mooberry, S.L.; Cichewicz, R.H. Cytotoxic dimeric epipolythiodiketopiperazines from the Ascomycetous fungus Preussia typharum. J. Nat. Prod. 2014, 77, 1459–1466. [Google Scholar] [CrossRef] [PubMed]
- Yanagihara, M.; Sasaki-Takahashi, N.; Suguhara, T.; Yamamoto, S.; Shinomi, M.; Yamashita, I.; Hayashida, M.; Yamanoha, B.; Numata, A.; Yamori, T.; et al. Leptosins isolated from marine fungus Leptoshaeria species inhibit DNA topoisomerases I and/or II and induce apoptosis by inactivation of Akt/protein kinase B. Cancer Sci. 2005, 96, 816–824. [Google Scholar] [CrossRef]
- Raju, R.; Piggott, A.M.; Conte, M.; Aalbersberg, W.G.; Feussner, K.; Capon, R.J. Naseseazines A and B: A new dimeric diketopiperazine framework from a marine-derived Actinomycete, Streptomyces sp. Org. Lett. 2009, 11, 3862–3865. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Movassaghi, M. Concise total synthesis and stereochemical revision of (+)-naseseazines A and B: Regioselective arylative dimerization of diketopiperazine alkaloids. J. Am. Chem. Soc. 2011, 133, 14940–14943. [Google Scholar] [CrossRef] [PubMed]
- Buedenbender, L.; Grkovic, T.; Duffy, S.; Kurtböke, D.I.; Avery, V.M.; Carroll, A.R. Naseseazine C, a new anti-plasmodial dimeric diketopiperazine from a marine sediment derived Streptomyces sp. Tetrahedron Lett. 2016, 57, 5893–5895. [Google Scholar] [CrossRef]
- Xiong, Z.Q.; Liu, Q.X.; Pan, Z.L.; Zhao, N.; Feng, Z.X.; Wang, Y. Diversity and bioprospecting of culturable actinomycetes from marine sediment of the Yellow Sea, China. Arch. Microbiol. 2015, 197, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Sun, C.; Zheng, M.; Harmer, J.R.; Yu, M.; Zhang, Y.; Peng, H.; Zhu, D.; Deng, Z.; Chen, S.L.; et al. Efficient biosynthesis of heterodimeric C3-aryl pyrroloindoline alkaloids. Nat. Commun. 2018, 9, 4428. [Google Scholar] [CrossRef]
- Varoglu, M.; Corbett, T.H.; Valeriote, F.A.; Crews, P. Asperazine, a selective cytotoxic alkaloid from a sponge-derived culture of Aspergillus niger. J. Org. Chem. 1997, 62, 7078–7079. [Google Scholar] [CrossRef]
- Varoglu, M.; Crews, P. Biosynthetically diverse compounds from a saltwater culture of sponge-derived Aspergillus niger. J. Nat. Prod. 2000, 63, 41–43. [Google Scholar] [CrossRef]
- Xu, B.; Zou, K.; Cheng, F. Alkaloids from Penicillium oxalicum, a fungus residing in Acrida cinerea. Adv. Mater. Res. 2014, 881–883, 442–445. [Google Scholar] [CrossRef]
- Govek, S.P.; Overman, L.E. Total synthesis of (+)-asperazine. Tetrahedron 2007, 63, 8499–8513. [Google Scholar] [CrossRef]
- Ding, G.; Jiang, L.; Guo, L.; Chen, X.; Zhang, H.; Che, Y. Pestalazines and pestalamides, bioactive metabolites from the plant pathogenic fungus Pestalotiopsis theae. J. Nat. Prod. 2008, 71, 1861–1865. [Google Scholar] [CrossRef] [PubMed]
- Zulqarnain; Iqbal, Z.; Cox, R.; Anwar, J.; Ahmad, N.; Khan, K.; Iqbal, M.; Manzoor, N.; Nhattak, S.U. Antifungal activity of compounds isolated from Aspergillus niger and their molecular docking studies with tomatinase. Nat. Prod. Res. 2018, 24, 1–5. [Google Scholar] [CrossRef]
- Xiao, J.; Zhang, Q.; Gao, Y.Q.; Shi, X.W.; Gao, J.M. Antifungal and antibacterial metabolites from an endophytic Aspergillus sp. associated with Melia azedarach. Nat. Prod. Res. 2014, 28, 1388–1392. [Google Scholar] [CrossRef]
- Cai, S.; Kong, X.; Wang, W.; Zhou, H.; Zhu, T.; Li, D.; Gu, Q. Aspergilazine A, a diketopiperazine dimer with a rare N-1 to C-6 linkage, from a marine-derived fungus Aspergillus taichungensis. Tetrahedron Lett. 2012, 53, 2615–2617. [Google Scholar] [CrossRef]
- Yun, K.; Khong, T.T.; Leutou, A.S.; Kim, G.D.; Hong, J.; Lee, C.H.; Son, B.W. Cristazine, a new cytotoxic dioxopiperazine alkaloid from the mudflat-sediment-derived fungus Chaetomium cristatum. Chem. Pharm. Bull. 2016, 64, 59–62. [Google Scholar] [CrossRef]
- Geiger, W.B. Chetomin an antibiotic substance from Chaetomium cochliodes. Arch. Biochem. 1949, 21, 125–131. [Google Scholar] [PubMed]
- Waksman, S.A.; Bugie, E. Chaetomin, a new antibiotic substance produced by Chaetomium cochliodes. J. Bact. 1944, 48, 527–530. [Google Scholar]
- Brewer, D.; Duncan, J.M.; Jerram, W.A.; Leach, C.K.; Safe, S.; Taylor, A.; Vining, L.C.; Archibald, R.M.; Stevenson, R.G.; Mirocha, C.J.; et al. Ovine ill-thrift in Nova Scotia. 5. The production and toxicology of chetomin, a metabolite of chaetomium spp. Can. J. Microbiol. 1972, 18, 1129–1137. [Google Scholar] [CrossRef] [PubMed]
- Brewer, D.; McInnes, A.G.; Smith, D.G.; Taylor, A.; Walter, J.A.; Loosli, H.R.; Kis, Z.L. Sporidesmins. Part 16. The structure of chetomin, a toxic metabolite of Chaetomium cochliodes, by nitrogen-15 and carbon-13 nuclear magnetic resonance spectroscopy. J. Chem. Soc. Perkin Trans. 1 1987, 1248–1251. [Google Scholar] [CrossRef]
- McInnes, A.G.; Taylor, A.; Walter, J.A. The structure of chetomin. J. Am. Chem. Soc. 1976, 98, 6741. [Google Scholar] [CrossRef]
- Safe, S.; Taylor, A. Sporidesmins. Part XIII. Ovine Ill-thrift in Nova Scotia. Part III. The characterisation of chetomin a toxic metabolite of Chaetomium cochliodes and Chaetomium globosum. J. Chem. Soc. Perkin Trans. 1 1972, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, T.; Kadota, S.; Nakamura, K.; Nishi, A.; Taga, T.; Kaji, T.; Osaki, K.; Tubaki, K. Dethio-tetra(methylthio)chetomin, a new antimicrobial metabolite of Chaetomium globosum KINZE ex FR. Structure and partial synthesis from chetomin. Chem. Pharm. Bull. 1982, 30, 3846–3848. [Google Scholar] [CrossRef][Green Version]
- Brewer, D.; Calder, F.W.; MacIntyre, T.M.; Taylor, A. Ovine ill-thrift in Nova Scotia: I. The possible regulation of the rumen flora in sheep by the fungal flora of permanent pasture. J. Agric. Sci. 1971, 76, 465–477. [Google Scholar] [CrossRef]
- Brewer, D.; Hannah, D.E.; Taylor, A. The biological properties of 3,6-epidithiadiketopiperazines: Inhibition of growth of Bacills subtilis by gliotoxins, sporidesmins, and chetomin. Can. J. Microbiol. 1966, 12, 1187–1195. [Google Scholar] [CrossRef]
- Brewer, D.; Hannah, D.E.; Rahman, R.; Taylor, A. The growth of Bacillus subtilis in media containing chetomin, sporidesmin, and gliotoxin. Can. J. Microbiol. 1967, 13, 1451–1460. [Google Scholar] [CrossRef]
- Jen, W.C.; Jones, G.A. Effects of chetomin on growth and acidic fermentation products of rumen bacteria. Can. J. Microbiol. 1983, 29, 1399–1404. [Google Scholar] [CrossRef] [PubMed]
- Cook, K.M.; Hilton, S.T.; Mecinović, J.; Motherwell, W.B.; Figg, W.D.; Schofield, C.J. Epidithiodiketopiperazines block the interaction between hypoxia-inducible factor-1α (HIF-1α) and p300 by a zinc ejection mechanism. J. Biol. Chem. 2009, 284, 26831–26838. [Google Scholar] [CrossRef] [PubMed]
- Kessler, J.; Hahnel, A.; Wichmann, H.; Rot, S.; Kappler, M.; Bache, M.; Vordemark, D. HIF-1α inhibition by siRNA or chetomin in human malignant glioma cells: Effects on hypoxic radioresistance and monitoring via CA9 expression. BMC Cancer 2010, 10, 605. [Google Scholar] [CrossRef]
- Horiuchi, A.; Hayashi, T.; Kikuchi, N.; Hayashi, A.; Fuseya, C.; Shiozawa, T.; Konishi, I. Hypoxia upregulates ovarian cancer invasiveness via the binding of HIF-1alpha to a hypoxia-induced, methylation-free hypoxia response element of S100A4 gene. Int. J. Cancer 2012, 131, 1755–1767. [Google Scholar] [CrossRef] [PubMed]
- Viziteu, E.; Grandmougin, C.; Goldschmidt, H.; Seckinger, A.; Hose, D.; Klein, B.; Moreaux, J. Chetomin, targeting HIF-1α/p300 complex, exhibits antitumour activity in multiple myeloma. Br. J. Cancer 2016, 114, 519–523. [Google Scholar] [CrossRef] [PubMed]
- Staab, A.; Loeffler, J.; Said, H.M.; Diehlmann, D.; Katzer, A.; Beyer, M.; Fleischer, M.; Schwab, F.; Baier, K.; Einsele, H.; et al. Effects of HIF-1 inhibition by chetomin on hypoxia-related transcription and radiosensitivity in HT 1080 human fibrosarcoma cells. BMC Cancer 2007, 7, 213. [Google Scholar] [CrossRef] [PubMed]
- Indelicato, M.; Pucci, B.; Schito, L.; Reali, V.; Aventaggiato, M.; Mazzarino, M.C.; Stivala, F.; Fini, M.; Russo, M.A.; Tafani, M. Role of hypoxya and autophagy in MDA-MB-231 invasiveness. J. Cell Physiol. 2010, 223, 359–368. [Google Scholar] [PubMed]
- Dewangan, J.; Srivastava, S.; Mishra, S.; Pandey, P.K.; Divakar, A.; Rath, S.K. Chetomin induces apoptosis in human triple-negative breast cancer cells by promoting calcium overload and mitochondrial dysfunction. Biochem. Biophys. Res. Commun. 2018, 495, 1915–1921. [Google Scholar] [CrossRef] [PubMed]
- Hiraki, M.; Hwang, S.Y.; Cao, S.; Ramadhar, T.R.; Byun, S.; Yoon, K.W.; Lee, J.H.; Chu, K.; Gurkar, A.U.; Kolev, V.; et al. Small-molecule reactivation of mutant p53 to wild-type-like p53 through the p53-Hsp40 regulatory axis. Chem. Biol. 2015, 22, 1206–1216. [Google Scholar] [CrossRef] [PubMed]
- Yano, K.; Horinaka, M.; Yoshida, T.; Yasuda, T.; Taniguchi, H.; Goda, A.E.; Wakada, M.; Yoshikawa, S.; Nakamura, T.; Kawauchi, A.; et al. Chetomin induces degradation of XIAP and enhances TRAIL sensitivity in urogenital cancer cells. Int. J. Oncol. 2011, 38, 365–374. [Google Scholar]
- Takahashi, M.; Takemoto, Y.; Shimazu, T.; Kawasaki, H.; Tachibana, M.; Shinkai, Y.; Takagi, M.; Shin-ya, K.; Igarashi, Y.; Ito, A.; et al. Inhibition of histone H3K9 methyltransferases by gliotoxin and related epipolythiodioxopiperazines. J. Antibiot. 2012, 65, 263–265. [Google Scholar] [CrossRef]
- Hwang, I.I.; Watson, I.R.; Der, S.D.; Ohh, M. Loss of VHL confers hypoxia-inducible factor (HIF)-dependent resistance to vesicular stomatitis virus: Role of HIF in antiviral response. J. Virol. 2006, 80, 10712–10723. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, H.; Sumino, M.; Okuyama, E.; Ishibashi, M. Immunomodulatory constituents from an Ascomycete, Chaetomium seminudum. J. Nat. Prod. 2004, 67, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Herath, K.B.; Jayasuriya, H.; Ondeyka, J.G.; Polishook, J.D.; Bills, G.F.; Dombrowski, A.W.; Cabello, A.; Vicario, P.P.; Zweerink, H.; Guan, Z.; et al. Isolation and structures of novel fungal metabolites as chemokine receptor (CCR2) antagonists. J. Antibiot. 2005, 58, 686–694. [Google Scholar] [CrossRef] [PubMed]
- Welch, T.R.; Williams, R.M. Studies on the biosynthesis of chetomin: Enantiospecific synthesis of a putative, late-stage biosynthetic intermediate. Tetrahedron 2013, 69, 770–773. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jo, M.J.; Patil, M.P.; Jung, H.I.; Seo, Y.B.; Lim, H.K.; Son, B.W.; Kim, G.D. Cristazine, a novel dioxopiperazine alkaloid, induces apoptosis via the death receptor pathway in A431 cells. Drug Dev. Res. 2019, 80, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Liu, X.; Guo, H.; Ren, B.; Chen, C.; Piggott, A.M.; Yu, K.; Gao, H.; Wang, Q.; Liu, M.; et al. Brevianamides with antitubercular potential from a marine-derived isolate of Aspergillus versicolor. Org. Lett. 2012, 14, 4770–4773. [Google Scholar] [CrossRef] [PubMed]
- Anjaneyulu, M.; Gopal, N.; Rao, A. A novel dimeric dipeptide from the Indian-Ocean starfish Pentaceraster regulus. J. Chem. Res. Synop. 1996, 50. [Google Scholar] [CrossRef]
- Giessen, T.B.; Marahiel, M.A. Rational and combinatorial tailoring of bioactive cyclic dipeptides. Front. Microbiol. 2015, 6, 785. [Google Scholar] [CrossRef]
- Koglin, A.; Walsh, C.T. Structural insights into nonribosomal peptide enzymatic assembly lines. Nat. Prod. Rep. 2009, 26, 987–1000. [Google Scholar] [CrossRef]
- Yin, W.B.; Grundmann, A.; Cheng, J.; Li, S.M. Acetylaszonalenin biosynthesis in Neosartorya fischeri. Identification of the biosynthetic gene cluster by genomic mining and functional proof of the genes by biochemical investigation. J. Biol. Chem. 2009, 284, 100–109. [Google Scholar] [CrossRef]
- Maiya, S.; Grundmann, A.; Li, S.M.; Turner, G. The fumitremorgin gene cluster of Aspergillus fumigatus: Identification of a gene encoding brevianamide F synthetase. ChemBioChem 2006, 7, 1062–1069. [Google Scholar] [CrossRef] [PubMed]
- Schenke, D.; Böttcher, C.; Lee, J.; Scheel, D. Verticillin A is likely not produced by Verticillium sp. J. Antibiot. 2011, 64, 523–524. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.; Rosa, F.; Ribeiro, T.; Hernandez-Bautista, R.; Bonaldo, M.; Gonçalves Silva, N.; Eiríksson, F.; Thorsteinsdóttir, M.; Ussar, S.; Urbatzka, R. Identification of cyanobacterial strains with potential for the treatment of obesity-related co-morbidities by bioactivity, toxicity evaluation and metabolite profiling. Mar. Drugs 2019, 17, 280. [Google Scholar] [CrossRef] [PubMed]
- Overy, D.P.; Berrue, F.; Correa, H.; Hanif, N.; Hay, K.; Lanteigne, M.; Mquilian, K.; Duffy, S.; Boland, P.; Jagannathan, R.; et al. Sea foam as a source of fungal inoculum for the isolation of biologically active natural products. Mycology 2014, 5, 130–144. [Google Scholar] [CrossRef] [PubMed]
- Davidson, B.S. New dimensions in natural products research: Cultured marine microorganisms. Curr. Opin. Biotechnol. 1995, 6, 284–291. [Google Scholar] [CrossRef]
- Kobayashi, J.; Ishibashi, M. Bioactive metabolites of symbiotic marine microorganisms. Chem. Rev. 1993, 93, 1753–1769. [Google Scholar] [CrossRef]
- Schoch, C.L.; Crous, P.W.; Groenewald, J.Z.; Boehm, E.W.; Burgess, T.I.; de Gruyter, J.; de Hoog, G.S.; Dixon, L.J.; Grube, M.; Gueidan, C.; et al. A class-wide phylogenetic assessment of Dothideomycetes. Stud. Mycol. 2009, 64, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Overy, D.P.; Bayman, P.; Kerr, R.G.; Bills, G.F. An assessment of natural product discovery from marine (sensu strictu) and marine-derived fungi. Mycology 2014, 5, 145–167. [Google Scholar] [CrossRef]
- Hoffmeister, D.; Keller, N.P. Natural products of filamentous fungi: Enzymes, genes, and their regulation. Nat. Prod. Rep. 2007, 24, 393–416. [Google Scholar] [CrossRef]
- Amend, A.; Burgaud, G.; Cunliffe, M.; Edgcomb, V.P.; Ettinger, C.L.; Gutiérrez, M.H.; Heitman, J.; Hom, E.F.Y.; Ianiri, G.; Jones, A.C.; et al. Fungi in the marine environment: Open questions and unsolved problems. MBio 2019, 10. [Google Scholar] [CrossRef]
- Canu, N.; Belin, P.; Thai, R.; Correia, I.; Lequin, O.; Seguin, J.; Moutiez, M.; Gondry, M. Incorporation of non-canonical amino acids into 2,5-diketopiperazines by cyclodipeptide synthases. Angew. Chem. Int. Ed. Engl. 2018, 57, 3118–3122. [Google Scholar] [CrossRef]
- Rinke, C.; Schwientek, P.; Sczyrba, A.; Ivanova, N.N.; Anderson, I.J.; Cheng, J.F.; Darling, A.; Malfatti, S.; Swan, B.K.; Gies, E.A.; et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 2013, 499, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.T. Insights into the chemical logic and enzymatic machinery of NRPS assembly lines. Nat. Prod. Rep. 2016, 33, 127–135. [Google Scholar] [CrossRef]
- Spiteller, P. Chemical ecology of fungi. Nat. Prod. Rep. 2015, 32, 971–993. [Google Scholar] [CrossRef]
- Degrassi, G.; Aguilar, C.; Bosco, M.; Zahariev, S.; Pongor, S.; Venturi, V. Plant growth-promoting Pseudomonas putida WCS358 produces and secretes four cyclic dipeptides: Cross-talk with quorum sensing bacterial sensors. Curr. Microbiol. 2002, 45, 250–254. [Google Scholar] [CrossRef]
- Holden, M.T.; Ram Chhabra, S.; de Nys, R.; Stead, P.; Bainton, N.J.; Hill, P.J.; Manefield, M.; Kumar, N.; Labatte, M.; England, D.; et al. Quorum-sensing cross talk: Isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other Gram-negative bacteria. Mol. Microbiol. 1999, 33, 1254–1266. [Google Scholar] [CrossRef]
- Park, D.K.; Lee, K.E.; Baek, C.H.; Kim, I.H.; Kwon, J.H.; Lee, W.K.; Lee, K.H.; Kim, B.S.; Choi, S.H.; Kim, K.S. Cyclo(Phe-Pro) modulates the expression of ompU in Vibrio spp. J. Bacteriol. 2006, 188, 2214–2221. [Google Scholar] [CrossRef]
- Gardiner, D.M.; Cozijnsen, A.J.; Wilson, L.M.; Pedras, M.S.; Howlett, B.J. The sirodesmin biosynthetic gene cluster of the plant pathogenic fungus Leptosphaeria maculans. Mol. Microbiol. 2004, 53, 1307–1318. [Google Scholar] [CrossRef]
- Li, J.; Wang, W.; Xu, S.X.; Magarvey, N.A.; McCormick, J.K. Lactobacillus reuteri-produced cyclic dipeptides quench agr-mediated expression of toxic shock syndrome toxin-1 in staphylococci. Proc. Natl. Acad. Sci. USA 2011, 108, 3360–3365. [Google Scholar] [CrossRef]
- Ortiz-Castro, R.; Diaz-Perez, C.; Martinez-Trujillo, M.; Del Rio, R.E.; Campos-Garcia, J.; Lopez-Bucio, J. Trans kingdom signaling based on bacterial cyclodipeptides with auxin activity in plants. Proc. Natl. Acad. Sci. USA 2011, 108, 7253–7258. [Google Scholar] [CrossRef]
- Orr, J.G.; Leel, V.; Cameron, G.A.; Marek, C.J.; Haughton, E.L.; Erick, L.J.; Trim, J.E.; Hawksworth, G.M.; Halestrap, A.P.; Wright, M.C. Mechanism of action of the antifibrogenic compound gliotoxin in rat liver cells. Hepatology 2004, 40, 232–242. [Google Scholar] [CrossRef]
- Debatin, K.M.; Poncet, D.; Kromer, G. Chemotherapy: Targeting the mitochondrial death pathway. Oncogene 2002, 21, 8786–8803. [Google Scholar] [CrossRef]
- Sohtome, Y.; Sodeoka, M. Development of chaetocin and S-adenosylmethionine analogues as tools for studying protein methylation. Chem. Rec. 2018, 18, 1660–1671. [Google Scholar] [CrossRef]
- Fujishiro, S.; Dodo, K.; Iwasa, E.; Teng, Y.; Sohtome, Y.; Hamashima, Y.; Ito, A.; Yoshida, M.; Sodeoka, M. Epidithiodiketopiperazine as a pharmacophore for protein lysine methyltransferase G9a inhibitors: Reducing cytotoxicity by structural simplification. Bioorg. Med. Chem. Lett. 2013, 23, 733–736. [Google Scholar] [CrossRef]
- Jiang, D.S.; Fang, Z.; Zhu, X.H.; Wei, X. The promising therapeutic agents for heart diseases: Histone Methyltransferase inhibitors. Int. J. Cardiol. 2017, 239, 6. [Google Scholar] [CrossRef]
- Ciarkowski, J. CNDO/2 quantum-mechanical calculations of the conformational flexibility of the diketopiperazine skeleton. Biopolymers 1984, 23, 397–407. [Google Scholar] [CrossRef]
- Liskamp, R.M.J.; Rijkers, D.T.S.; Kruijtzer, J.A.W.; Kemmink, J. Peptides and proteins as a continuing exciting source of inspiration for peptidomimetics. ChemBioChem 2011, 12, 1626–1653. [Google Scholar] [CrossRef]
- Ray, J.G.; Ly, J.T.; Savin, D.A. Peptide-based lipid mimetics with tunable core properties via thiol–alkyne chemistry. Polym. Chem. 2011, 2, 1536–1541. [Google Scholar] [CrossRef]
- Hadden, M.K.; Blagg, B.S.J. Dimeric approaches to anti-cancer chemotherapeutics. Anticancer Agents Med. Chem. 2008, 8, 807–816. [Google Scholar] [CrossRef]
- Jervis, P.J.; Moulis, M.; Jukes, J.-P.; Ghadbane, H.; Cox, L.R.; Cerundolo, V.; Besra, G.S. Towards multivalent CD1d ligands: Synthesis and biological activity of homodimeric α-galactosyl ceramide analogues. Carbohydr. Res. 2012, 356, 152–162. [Google Scholar] [CrossRef]
- Lian, G.; Yu, B. Naturally occurring dimers from chemical perspective. Chem. Biodivers. 2010, 7, 2660–2691. [Google Scholar] [CrossRef]
- MacDonald, S.J.; Watson, K.G.; Cameron, R.; Chalmers, D.K.; Demaine, D.A.; Fenton, R.J.; Gower, D.; Hamblin, J.N.; Hamilton, S.; Hart, G.J.; et al. Potent and long-acting dimeric inhibitors of influenza virus neuraminidase are effective at a once-weekly dosing regimen. Antimicrob. Agents Chemother. 2004, 48, 4542–4549. [Google Scholar] [CrossRef]
- Muñoz-Torrero, D.; Camps, P. Dimeric and hybrid anti-Alzheimer drug candidates. Curr. Med. Chem. 2006, 13, 399–422. [Google Scholar] [CrossRef]
- Ni, F.; Kota, S.; Takahashi, V.; Strosberg, A.D.; Snyder, J.K. Potent inhibitors of hepatitis C core dimerization as new leads for anti-hepatitis C agents. Bioorg. Med. Chem. 2011, 21, 2198–2202. [Google Scholar] [CrossRef]
- Raspaglio, G.; Ferlini, C.; Mozzetti, S.; Prislei, S.; Gallo, D.; Das, N.; Scambia, G. Thiocolchicine dimers: A novel class of topoisomerase-I inhibitors. Biochem. Pharm. 2005, 69, 113–121. [Google Scholar] [CrossRef]
- Gomes, N.G.M.; Dasari, R.; Chandra, S.; Kiss, R.; Kornienko, A. Marine invertebrate metabolites with anticancer activities: Solutions to the “supply problem”. Mar. Drugs 2016, 14, 98. [Google Scholar] [CrossRef]
- Kim, J.; Ashenhurst, J.A.; Movassaghi, M. Total synthesis of (+)-11,11’-dideoxyverticillin A. Science 2009, 324, 238–241. [Google Scholar] [CrossRef]
- Boyd, E.M.; Sperry, J. Total synthesis of (–)-aspergilazine A. Org. Lett. 2014, 16, 5056–5059. [Google Scholar] [CrossRef]
- Liang, K.; Deng, X.; Tong, X.; Li, D.; Ding, M.; Zhou, A.; Xia, C. Copper-mediated dimerization to access 3a,3a’-bispyrrolidinoindoline: Diastereoselective synthesis of (+)-WIN 64821 and (–)-ditryptophenaline. Org. Lett. 2015, 17, 206–209. [Google Scholar] [CrossRef]
- Loach, R.P.; Fenton, O.S.; Movassaghi, M. Concise total synthesis of (+)-asperazine, (+)-pestalazine A, and (+)-iso-pestalazine A. Structure revision of (+)-pestalazine A. J. Am. Chem. Soc. 2016, 138, 1057–1064. [Google Scholar] [CrossRef]
- Tadano, S.; Mukaeda, Y.; Ishikawa, H. Bio-inspired dimerization reaction of tryptophan derivatives in aqueous acidic media: Three-step syntheses of (+)-WIN 64821, (–)-ditryptophenaline, and (+)-naseseazine B. Angew. Chem. Int. Ed. 2013, 52, 7990–7994. [Google Scholar] [CrossRef]
- Kim, J.; Movassaghi, M. Biogenetically-inspired total synthesis of epidithiodiketopiperazines and related alkaloids. Acc. Chem. Res. 2015, 48, 1159–1171. [Google Scholar] [CrossRef]
- Boyer, N.; Morrison, K.C.; Kim, J.; Hergenrother, P.J.; Movassaghi, M. Synthesis and anticancer activity of epipolythiodiketopiperazine alkaloids. Chem. Sci. 2013, 4, 1646–1657. [Google Scholar] [CrossRef]
- Iwasa, E.; Hamashima, Y.; Fujishiro, S.; Higuchi, E.; Ito, A.; Yoshida, M.; Sodeoka, M. Total synthesis of (+)-chaetocin and its analogues: Their histone methyltransferase G9a inhibitory activity. J. Am. Chem. Soc. 2010, 132, 4078–4079. [Google Scholar] [CrossRef]
- Teng, Y.; Iuchi, K.; Iwasa, E.; Fujishiro, S.; Hamashima, Y.; Dodo, K.; Sodeoka, M. Unnatural enantiomer of chaetocin shows strong apoptosis-inducing activity through caspase-8/caspase-3 activation. Bioorg. Med. Chem. Lett. 2010, 20, 5085–5088. [Google Scholar] [CrossRef]
- Wada, M.; Suzuki, H.; Kato, M.; Oikawa, H.; Tsubouchi, A.; Oguri, H. Stereodivergent synthesis of bispyrrolidinoindoline alkaloidal scaffolds and generation of a lead candidate with stereospecific antiproliferative activity. ChemBioChem 2019, 20, 1273–1281. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomes, N.G.M.; Pereira, R.B.; Andrade, P.B.; Valentão, P. Double the Chemistry, Double the Fun: Structural Diversity and Biological Activity of Marine-Derived Diketopiperazine Dimers. Mar. Drugs 2019, 17, 551. https://doi.org/10.3390/md17100551
Gomes NGM, Pereira RB, Andrade PB, Valentão P. Double the Chemistry, Double the Fun: Structural Diversity and Biological Activity of Marine-Derived Diketopiperazine Dimers. Marine Drugs. 2019; 17(10):551. https://doi.org/10.3390/md17100551
Chicago/Turabian StyleGomes, Nelson G. M., Renato B. Pereira, Paula B. Andrade, and Patrícia Valentão. 2019. "Double the Chemistry, Double the Fun: Structural Diversity and Biological Activity of Marine-Derived Diketopiperazine Dimers" Marine Drugs 17, no. 10: 551. https://doi.org/10.3390/md17100551
APA StyleGomes, N. G. M., Pereira, R. B., Andrade, P. B., & Valentão, P. (2019). Double the Chemistry, Double the Fun: Structural Diversity and Biological Activity of Marine-Derived Diketopiperazine Dimers. Marine Drugs, 17(10), 551. https://doi.org/10.3390/md17100551







