Abstract
Chemical investigations on the fermentation extract obtained from an ascidian-derived Streptomyces sp. (USC-16018) yielded a new ansamycin polyketide, herbimycin G (1), as well as a known macrocyclic polyketide, elaiophylin (2), and four known diketopiperazines (3–6). The structures of the compounds were elucidated based on 1D/2D NMR and MS data. The absolute configuration of 1 was established by comparison of experimental and predicted electronic circular dichroism (ECD) data. Antiplasmodial activities were tested for the natural products against chloroquine sensitive (3D7) and chloroquine resistant (Dd2) Plasmodium falciparum strains; the two polyketides (1–2) demonstrated an inhibition of >75% against both parasite strains and while 2 was highly cytotoxic, herbimycin G (1) showed no cytotoxicity and good predicted water solubility.
1. Introduction
Malaria is a vector-borne disease caused by protozoan parasites of the genus Plasmodium, which results in more than 400,000 deaths annually [1]. The life cycle of the malaria parasite involves two hosts. The disease is contracted when an Anopheles mosquito feeds on human blood and simultaneously injects sporozoites into the bloodstream. The sporozoites are then carried to the liver, where they reproduce asexually to give rise to thousands of merozoites that spread through the body via the bloodstream [2,3]. During this intraerythrocytic cycle, the clinical manifestations of the disease appear, and the human host experiences strong fevers and chills [3,4]. Within the red blood cells, merozoites develop into ring forms and then trophozoites, which mature to form multinucleated schizonts that eventually burst and generate new merozoites. A small proportion of trophozoites produce male and female gametocytes. The mature gametocytes are transmitted back to a mosquito as it takes a blood meal. Within the gut of the mosquito, they undergo gametogenesis and fertilization, then develop into oocysts, which asexually produce new sporozoites. These migrate to the mosquito’s salivary glands and the life cycle of the parasite begins again once the mosquito feeds on a new human host [3]. The drug resistance against the currently available antimalarials is a major concern [5] and, therefore, new drugs with novel targets are urgently needed.
Macrolide polyketides have been considered as possible candidates for antimalarials [6]. For instance, the macrolide compound azithromycin exhibits antiplasmodial activity by targeting the relict plastid in the Plasmodium parasite and inhibiting protein synthesis in this organelle [7]. The ansamycin polyketide, geldanamycin, inhibits heat shock proteins, which are highly expressed in the early blood stage of the malaria parasite and thus inhibits the growth of the parasite [8]. Actinomycetes are a class of bacteria known to produce diverse macrolide polyketides. In order to further characterise new antiplasmodial natural products belonging to this chemical class, we targeted microbial isolates from underexplored ecological niches and 120 actinomycetes were isolated from Australian ascidian species [9].
A new approach utilising two-dimensional heteronuclear single quantum coherence—total correlation spectroscopy nuclear magnetic resonance (HSQC-TOCSY NMR) profiles and biological activity data [10] allowed us to target antiplasmodial polyketide-producing strains based on identified regions of interest in the NMR spectrum that corresponded to correlations between upfield methyl proton resonances (δH 0.50–2.00) and carbon resonances between δC 60–80 ppm, which were associated with polyketide-type compounds. Based on this approach, Streptomyces sp. (USC-16018) derived from the colonial ascidian Symplegma rubra was selected because its crude extract HSQC-TOCSY NMR profile indicated the presence of several polyketide resonances (Supplementary Material, Figure S1). Furthermore, the extracts exhibited a 97% inhibition of Plasmodium falciparum at 0.2 μg/mL. The Streptomyces sp. was fermented on a larger scale and the purification of the ethyl acetate (EtOAc) extract derived from this culture resulted in the isolation of two antiplasmodial polyketides, herbimycin G (1), a new geldanamycin analogue, and the known antibiotic elaiophylin (2), as well as four known diketopiperazines (3–6) (Figure 1).
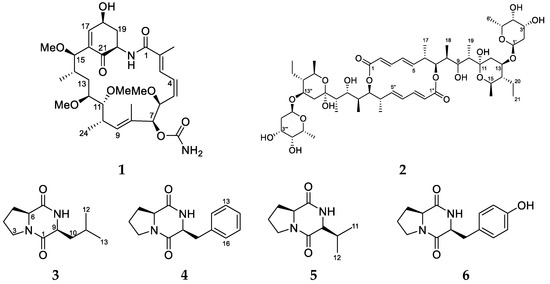
Figure 1.
The polyketides and peptides isolated from the ascidian-associated Streptomyces sp. (USC-16018): herbimycin G (1); elaiophylin (2); l-Pro-l-Leu (3); l-Pro-l-Phe (4); l-Pro-l-Val (5); l-Pro-l-Tyr (6).
2. Results and Discussion
Fermentations of Streptomyces sp. (USC-16018) were scaled up to 55 culture plates for two solid media, oatmeal agar (OMA) and glucose-starch-tryptone media (GST), which yielded 1.6 g and 0.8 g of extract, respectively, after the extraction with EtOAc. The extracts were purified by repeated reversed-phase C18 high-performance liquid chromatography (HPLC) and the analysis of fractions by 1H NMR led to the identification of two polyketides (1 and 2) and four diketopiperazine compounds (3–6).
Herbimycin G (1) was isolated as a dark brown solid with a UV maximum at 245 nm. A sodiated molecule ion at m/z 601.3111 [M + Na]+ obtained from high resolution-electrospray ionisation-quadrupole time of flight-mass spectrometry (HR-ESI-QTOF-MS) together with 1H and 13C NMR data suggested that 1 had a molecular formula of C30H46N2O9. The 1H NMR spectrum of 1 in DMSO-d6 had resonances that could be assigned to an amide proton at δH 8.02 (d, J = 9.4 Hz); five olefinic proton resonances at δH 7.13 (dd, J = 12.2 Hz, 1.2 Hz), δH 6.65 (ddd, J = 3.3 Hz, 1.9 Hz, 1.4 Hz), δH 6.48 (ddd, J = 12.2 Hz, 10.9 Hz, 1.4 Hz), δH 5.65 (dd, J = 10.9 Hz, 7.9 Hz), and δH 5.20 (d, J = 7.7 Hz); and an oxygenated methine at δH 4.68 (dddd, J = 10.6 Hz, 5.1 Hz, 4.7 Hz, 2.5 Hz) that correlated to a hydroxy resonance at δH 5.56 (d, J = 5.1 Hz) in the correlation spectroscopy (COSY) spectrum. A further five oxygenated methines were observed at δH 4.56 (dd, J = 7.9 Hz, 1.4 Hz), δH 5.16 (s), δH 3.15 (m), δH 3.33 (m), and δH 4.15 (d, J = 1.4 Hz), and four methoxy-resonances occurred between δH 3.13–3.35 ppm. The 1H NMR spectrum contained an additional three methines at δH 4.55, δH 2.45, δH 1.51; two methylene pairs δH 1.55 and δH 1.41, as well as δH 1.96 and δH 2.26, and four methyl signals δH 1.84 (d, J = 1.2 Hz), δH 1.62 (s), δH 0.94 (d, J = 6.7 Hz), and δH 0.69 (d, J = 6.7 Hz). The 13C and heteronuclear single quantum correlation (HSQC) NMR data showed 30 carbon resonances (Table 1). This data suggested that 1 was a close analogue to previously reported herbimycins, containing an identical macrolactam ring to herbimycins A, D, E, and 17,19-dimethylthioherbimycin A (Supplementary Material, Figure S2) [11,12,13]. The structural differences arose through different substitution of the benzoquinone moiety between C-15 and the amide proton. In herbimycin G (1), the loss of an olefinic methine was observed while a methylene (δH 1.96, δH 2.26/δC 39.2) and methine (δH 4.55/δC 53.2) were gained. The COSY and HSQC analyses revealed an isolated spin system—1-hydroxy-3-amino-propyl—which included the exchangeable amide proton (δH 8.02). Total correlation spectroscopy (TOCSY) correlations from the olefinic proton resonating at δH 6.65/δC 149.9 to all protons of this spin system implied that the olefinic proton was part of the system. Analysis of the heteronuclear multiple-bond correlation (HMBC) spectrum confirmed that the olefinic carbon (C-17) was alpha to the hydroxyl. The deshielded carbon resonance of C-17 indicated another electron withdrawing group beta to the double bond proton and a strong HMBC correlation from H-17 to a carbon at δC 197.1 determined it to be a carbonyl group. H-15 (δH 4.15) showed an additional two unassigned HMBC correlations to the olefinic carbon C-17 as well as a quaternary carbon at δH 134.0, linking the spin systems to the macrolactam ring at C-15. As one of the methylene protons (H-19b) also showed a correlation to the carbonyl resonance, it was concluded that this fragment consisted of a 4-hydroxyl-cyclohexenone system. The final structure was consistent with the sodium adduct ion m/z 601.3111 [M + Na]+ as well as the degrees of hydrogen deficiency calculated from the molecular formula.

Table 1.
The NMR data for herbimycin G (1) (800 MHz in DMSO-d6, δ in ppm).
Previously reported geldanamycins and herbimycins contained a 2,6-disubstituted-p-benzoquinone ring or a hydroquinone ring bonded to the macrolactam system, whereas herbimycin G (1), contains a 2,6-disubstituted 4-hydroxyl-cyclohexenone ring and, therefore, has an additional two stereogenic centres. Consequently, a ROESY spectrum was obtained in order to elucidate the relative configuration of the nine stereogenic centres in 1. ROESY correlations between H-6 (δH 4.57) and H-7 (δH 5.16) showed that the two protons were cis to each other, indicating that the O-methyl at C-6 and the carbamate group attached to C-7 were on the β-face of the molecule. Based on the ROESY exchange (antiphase to the ROESY correlations) correlations, the exchangeable carbamate NH2 protons could be assigned as δH 6.30 and δH 6.66. A weak long-range correlation between H-7 and the methyl protons H-24 at δH 0.94 established the relative configuration of the two-spin system relative to C-7. The methyl protons at δH 0.94 also correlated to the methoxy protons at δH 3.35, and H-13a (δH 1.41), H-14 (δH 1.51), and H-15 (δH 4.15) were assigned to the α-face of the molecule. A weak ROESY correlation from H-15 to H-18 (δH 4.68) allowed for the determination of the configuration of the 4-hydroxyl-cyclohexenone system relative to the macrolactam ring.
Further strong correlations between H-18 and H-19b (δH 2.26) to H-20 (δH 4.55), indicated their position to be on the α-face. The hydroxyl group, H-19a (δH 1.96), and the amide proton (δH 8.02) faced in the β-direction. No COSY correlation was observed between H-17 and H-18, indicating that the two protons were at an angle close to 90° and the small long-range couplings (J = 1.8 Hz) between H-17 and H-19b suggested a “W” arrangement. The ROESY correlations between the methyl resonating at δH 0.69 and δH 1.96 indicated that the macrolactam folded over the 4-hydroxyl-cyclohexenone. The two-dimensional structure was therefore established and redrawn in Maestro 2016 (Maestro Technologies, Inc., Trenton, NJ, USA) to perform a Monte-Carlo Multiple Minimisation (MCMM) search using the OPLS2005 force field (Schrödinger, New York, NY, USA) to generate the energy minimised conformation of 1 (Figure 2a). The previously observed ROESY correlations were compared to the energy-minimised 3D structure of 1, confirming its relative configuration. The absolute configuration of 1 was determined by comparison of its experimental and predicted electronic circular dichroism (ECD) spectra, calculated using time-dependent density functional theory (TDDFT). A conformational search using MacroModel (Version 10.8, Schrödinger, LLC, New York, NY, USA) was performed on the energy minimised structure of 1 using the force field parameters as above with extended torsional sampling options. All structures within a 10.0 kJ/mol energy window were saved, generating 13 conformers. Geometry optimisation and Gibbs free energy calculations using Gaussian 16 (Revision A.03, Gaussian, Inc., Wallingford, CT, USA) at the B3LYP/31G(d) energy level was then performed on each of the conformers. Following this, the ECD spectra of each of the geometry optimised conformers was calculated using Gaussian 16 at the same theoretical level. A Boltzmann-weighted ECD spectrum was then calculated using SpecDis (Version 1.7, Würzburg, Germany) The experimental ECD spectrum of 1 showed good agreement with the calculated ECD of (6S,7S,10S,11R,12S,14S,15R,18S,20R)-1 (Figure 2b).
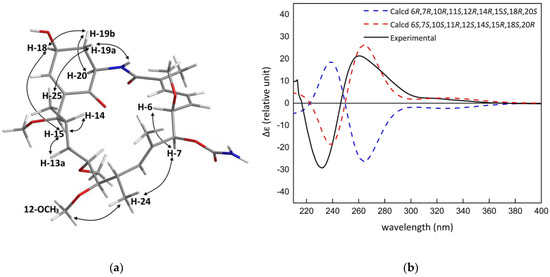
Figure 2.
(a) The key ROESY correlations of 1; (b) the comparison of the experimental electronic circular dichroism (ECD) spectrum of 1 with those calculated at the B3LYP/6-31G(d) level.
The configuration of the macrolactam ring is consistent with herbimycin analogues previously reported in the literature, but herbimycin G (1) is the first example of an ansamycin compound that contains a 2,6-disubstituted 4-hydroxyl-cyclohexenone ring. A total of eleven herbimycin analogues, including herbimycin G, have now been reported from the Streptomyces sp. strains (Supplementary Material, Figure S2) [11,12,13,14,15,16]. Benzoquinone ansamycins have been identified as potent inhibitors of the Hsp90, an important cancer target and, more recently, a novel target for antimalaria drugs. Geldanamycin had reached clinical trials as an anticancer drug, however, the compound had low water solubility and exhibited high liver toxicity. As a result, a number of synthetically modified geldanamycin analogues were made and among those tanespimycin (17-N-allylamino-17-demethoxygeldanamycin, 17-AAG) advanced to phase three in anticancer clinical trials for multiple myeloma [17]. In vitro studies have found that geldanamycin equally inhibits the growth of chloroquine-sensitive and chloroquine-resistant Plasmodium falciparum strains (IC50 20 nM) [18]. The 17-AAG analogue was reported to be effective against Plasmodium yoelii in mouse models when injected intra-peritoneal in doses of 7.2 mg/kg. However, while a single dose of chloroquine was sufficient to fully clear the animals from the parasites, a second dose of 17-AAG was needed [8]. In the same study, 17-AAG-treated mice that were reinfected with P. yoelii showed low percentages of parasitemia, which completely cleared after 9 days; the chloroquine-treated mice took a week longer to recover from the parasites [8]. Such reports suggest that ansamycin analogues may be good candidates for antiplasmodial drugs. Due to the low water solubility and high liver toxicity of geldanamycin, it is of great interest to find new water soluble and non-cytotoxic drug candidates to treat malaria infections.
In herbimycin G (1), the benzoquinone was reduced to a 4-hydroxyl-cyclohexenone and, thus, potentially has better water solubility. Therefore, we assessed the cLogP and other Lipinski parameters of all known herbimycins, as well as several related ansamycin compounds (Figure 3; Supplementary Material, Table S1). Compared to geldanamycin, most herbimycin analogues, except for herbimycin D, F, and TAN-420E, exhibited similar or lower cLogP values. Herbimycin G (1) and TAN-420B had the lowest cLogP—0.68 and 0.63, respectively—indicating better water solubility. The molecular weights of all assessed herbimycin and geldanamycin compounds were non-compliant with the Lipinski rule of five that uses four physiochemical properties (molecular weight < 500 Da, partition coefficient cLogP < 5, hydrogen bond donors < 5, and hydrogen bond acceptors < 10) to assess drug-likeness [19]. Macrocycles frequently violate the rule of five but, nonetheless, have been successful as clinical leads (such as rifamycin) or candidates (such as erythromycin) and have made their way to the market [20]. This is due to their high binding affinity, stability, and specificity for a molecular target [20].
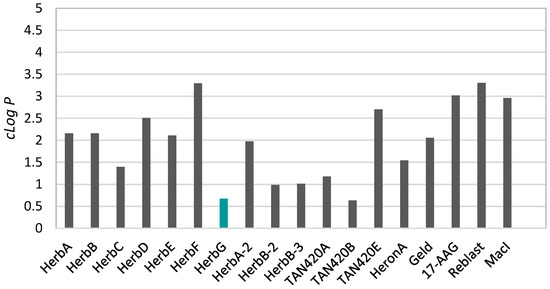
Figure 3.
The analysis of the cLogP values of herbimycins and related ansamycin compounds (note: structures and keys for abbreviations for the presented compounds are given in the Supplementary Material Table S1 and Figure S2).
In addition to the new herbimycin analogue, EtOAc extracts from the Streptomyces sp. (USC-16018) grown on the GST media yielded the known polyketide, elaiophylin (2), and four diketopiperazines (3–6). The obtained spectroscopic NMR data for 3–6, were compared to the literature and Cyclo-l-Pro-l-Leu (3), Cyclo-l-Pro-l-Phe (4), Cyclo-l-Pro-l-Val (5), and Cyclo-l-Pro-l-Tyr (6) were identified [21,22]. Additionally, a positive low resolution–electrospray ionisation–mass spectrometry (LR-ESI-MS) protonated molecular ion at m/z 1024.65 was observed and the molecular formula for 2 was established to be C54H88O18. Based on the 1H, COSY, HSQC, and HMBC spectra, a partial structure was elucidated containing 44 protons, 27 carbons, and 10 oxygen atoms. This only accounted for half the number of carbons and hydrogens, suggesting that 2 was a symmetric dimer. The comparison of these NMR data with the carbon data documented in the literature identified 2 as the known antibiotic compound elaiophylin [23]. Several Streptomyces species are known to produce elaiophylin and geldanamycin compounds. Recently, the complete genome mining of four geldanamycin producing Streptomyces sp. strains identified up to 56 putative biosynthetic gene clusters and found highly similar geldanamycin and elaiophylin gene clusters in all the strains [24]. Geldanamycin and herbimycin have the same ansamycin carbon skeleton. It is therefore assumed that both biosynthetic pathways exhibit very similar polyketide synthase (PKS) activity. In fact, Rasher and co-workers [25] proposed that both compounds are “assembled from the 3-amino-5-hydroxybenzoic acid (AHBA) starter unit by successive condensation of two-carbon building blocks in seven chain-elongation steps that use one malonyl, four methylmalonyl, and two methoxymalonyl extender units” and structural differences are introduced through enzymatic modifications post PKS assembly [25]. Such findings yet again highlight the biosynthetic potential of the Streptomyces species. It is very likely that the Streptomyces sp. (USC-16018), under different laboratory conditions, will be able to produce a wide range of additional secondary metabolites.
With the aim to discover marine actinomycete-derived antiplasmodial compounds, 1–6 were screened for activity against the drug-sensitive 3D7 strain and the chloroquine- and pyrimethamine-resistant Dd2 strain of P. falciparum (Table 2). Elaiophylin (2) inhibited both the 3D7 and Dd2 parasites with similar potency, IC50 = 778 nM and 598 nM, respectively. Antiplasmodial activity in the same range (IC50 214 nM) against the K1 strain was previously reported in the literature [26]. Herbimycin G showed no cytotoxicity and some degree of parasite growth inhibition, corresponding to a 77% inhibition against 3D7 at 40 μM. The inhibition curve did not reach the full inhibition plateau and, therefore, an IC50 value could not be determined (Figure 4). The remaining compounds were devoid of antiplasmodial or cytotoxic activity, while the activity of the reference compounds was in the expected range (Table 2).

Table 2.
The antiplasmodial activity and cytotoxicity of compounds 1–6 and the six reference compounds.
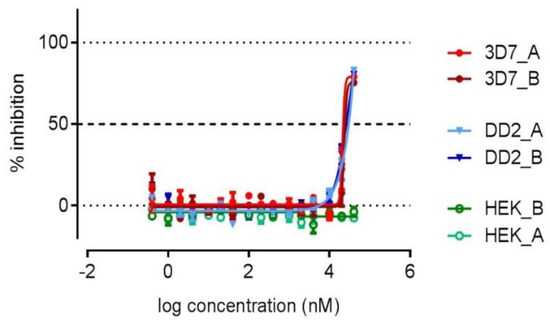
Figure 4.
The dose-response curve for 1 against 3D7, Dd2, and HEK-293 in duplicates.
These results showed that marine actinomycetes were a valuable source for the discovery of new secondary metabolites. Several herbimycin, geldanamycin, and other ansamycin compounds have previously been reported from terrestrial and marine Streptomyces species. To our knowledge, the new compound, herbimycin G (1), is the first example of such a compound containing a 2,6-disubstituted 4-hydroxyl-cyclohexenone ring. Herbimycin G exhibited weak antiplasmodial activity with no cytotoxicity. We investigated the physiochemical properties of the reported herbimycins and related analogues and found that 1 had a lower cLogP compared to the other herbimycin analogues, potentially related to the different ring structure, suggesting a good bioavailability. As herbimycin G did not show cytotoxicity against HEK293, it appears promising to test this compound for other biological activity, such as antitumour activity, in the future.
3. Materials and Methods
3.1. General Experimental Section
The NMR spectra were recorded at 25 °C on a Bruker Avance III HDX 800 MHz spectrometer with a triple (TCI) resonance 5 mm cryoprobe (Bruker, Billerica, MA, USA). The 1H and 13C chemical shifts were referenced to the DMSO-d6 solvent peak at δH 2.50 and δC 39.5, respectively. Standard parameters were used for 2D NMR spectra, including gCOSY, gHSQC, gHMBC, and ROESY. The HR-ESI-QTOF-MS data were recorded on a 6530 Accurate Mass Q-TOF (Agilent Technologies, Santa Clara, CA, USA) workstation using positive electrospray ionisation, with the mobile phase ACN/H2O containing 0.1% formic acid. A LaChrom Merck Hitachi L7100 series pump (Tokyo, Japan) equipped with a Hitachi L7450 PDA detector (Tokyo, Japan) was used for HPLC separations. Prior to HPLC separation, the microbial crude extract was adsorbed onto Alltech Davisil 35–75 μm 150 Å C18 (Columbia, MD, USA). All separations were performed with a Thermo Hypersil-Keystone Betasil 5μm Å C18 HPLC column (21.2 mm × 150 mm) (Waltham, MA, USA). The optical rotations were measured on a JASCO P-1020 polarimeter (Easton, MD, USA). The UV spectra were obtained on a Shimadzu UV-1800 UV/vis spectrophotometer (Kyoto, Japan). The CD spectra were recorded on a JASCO J-715 spectropolarimeter (Easton, MD, USA). All solvents were Scharlau HPLC grade (Barcelona, Spain), except the EtOAc used for extractions was of the Honeywell HPLC grade (Morris Plains, NJ, USA), and for MS, Fisher Scientific LC/MS grade acetonitrile (Hampton, NH, USA) was used. The H2O was Millipore Milli-Q PF filtered (Sartorius, Göttingen, Germany).
3.2. Biological Material
Streptomyces sp. (USC-16018) was isolated from the ascidian Symplegma rubra collected on July 15, 2015, Hastings Point, NSW, Australia [9]. The ascidian specimen was identified by Merrick Ekins (Queensland Museum, Brisbane, Australia). The isolate was cultured using selective media and identified via 16S rRNA sequencing. The 16S sequence was deposited at GenBank (accession number MF773774).
3.3. Fermentation, Extraction, and Isolation
Streptomyces sp. (USC-16018) was grown on 55 OMA (20 g oatmeal, 20 g bacteriological agar, 3 g yeast extract, 1 L deionised H2O) and 55 GST (5 g glucose, 2 g starch, 3 g beef extract, 5 g yeast extract, 5 g tryptone, 4 g CaCO3, 4 g NaCl, 1 g NaSO4, 0.5 g KCl, 2 g MgCl, 0.5 g KH2PO4, 20 g bacteriological agar, 1 L deionised H2O) agar plates for 2 weeks at 28 °C. The plates were then exhaustively extracted by shaking in ethyl acetate. The extracts were dried under a reduced vacuum to yield 1.6 g of dark brown gum for OMA cultures and 0.8 g of light red gum for GST cultures. Each of the extracts was dissolved in a small amount of MeOH and C18 silica gel was added in a 1:2 ratio. The solvent was evaporated and the extract adsorbed on the C18 was transferred to a refillable HPLC guard column (10 mm × 20 mm) which was attached to a C18 Betasil HPLC column. At a flow rate of 9 mL/min, the isocratic HPLC conditions of the 10% aqueous MeOH were initially employed for 10 min, followed by a linear gradient over 40 min from 10% to 100% MeOH, finishing with another 10 min isocratic conditions at 100% MeOH. Sixty one-minute-fractions were collected and analysed by 1H NMR spectroscopy. Of the OMA extract, fractions 31–35 were combined and further purified by eluting with a gradient from 25% to 60% aqueous MeOH over 40 min to yield 0.9 mg of compound 1. Fraction 46 and 26 of the GST extract HPLC separation yielded 0.6 mg and 0.7 mg of compound 2 and 3, respectively. Fractions 20–25 were combined and repurified to yield 1.2 mg of compound 4 and 2.1 mg of compound 5. A total of 0.9 mg of compound 2 were also detected in the OMA extract.
Herbimycin G (1): C30H46N2O9; brown solid; [α]D + 8.6 (c 0.007 g/100 mL, MeOH), UV (MeOH) λmax (log ε) 245 (1.93) nm; 1H NMR (800 MHz, DMSO) and 13C NMR (200 MHz, DMSO) data, Table 1; (+)-HR-ESIMS m/z 601.3111 [M + Na]+ (calculated for C30H46N2NaO9, 601.3096); all 1D and 2D NMR spectra in Supplementary Material, Figures S3–S8.
Elaiophylin (2): C54H88O18; brown solid; 1H NMR (800 MHz, DMSO) and 13C NMR (200 MHz, DMSO) data, Supplementary Material, Table S2; (+)-LR-ESIMS m/z 1024.6 [M + H]+ (calculated for C54H88O18, 1024.60).
Cyclo-l-Pro-l-Leu (3): C11H18N2O2; yellow solid; UV (MeOH) λmax (log ε) 277 (0.71) nm, 329 (−0.01) nm; 1H NMR (800 MHz, DMSO) and 13C NMR (200 MHz, DMSO) data, Supplementary Material, Table S3.
Cyclo-l-Pro-l-Phe (4): C14H16N2O2; yellow solid; UV (MeOH λmax (log ε) 216 (1.58) nm; 1H NMR (800 MHz, DMSO) and 13C NMR (200 MHz, DMSO) data, Supplementary Material, Table S3.
Cyclo-l-Pro-l-Val (5): C10H14N2O2; yellow solid; UV (MeOH λmax (log ε) 215 (1.72) nm; 1H NMR (800 MHz, DMSO) and 13C NMR (200 MHz, DMSO) data, Supplementary Material, Table S3.
Cyclo-l-Pro-l-Tyr (6): C14H16N2O3; yellow solid; UV (MeOH λmax (log ε) 215 (1.72) nm; 1H NMR (800 MHz, DMSO) and 13C NMR (200 MHz, DMSO) data, Supplementary Material, Table S3.
3.4. Molecular Modelling Calculations
The generation of conformers was performed using the Schrödinger MacroModel 2016 (Version 10.8, Schrödinger, LLC, New York, NY, USA) using the protocol reported by Willoughby and co-workers [27]. First-principle calculations based on the density functional theory (DFT) were carried out to optimize the atomic structures at the B3LYP/6-31G(d) level with the Gaussian 16 (Revision A.03, Gaussian, Inc., Wallingford, CT, USA) suite of programs [28]. The electronic transition and rational strength were calculated using the time-dependent DFT (TDDFT) method at the same theoretical level. The solvent effect in the MeOH solution was considered during all calculations using the Polarizable Continuum Model (PCM) [29]. The Boltzmann-weighted UV and ECD were calculated using the freely available software SpecDis (Version 1.7, Würzburg, Germany) [30] and GaussSum (Version 3.0, Cambridge, UK) [31] using a sigma/gamma value of 0.25 eV. The Lipinski parameters were calculated in ChemBio3D Ultra 14.0 (PerkinElmer, Waltham, MA, USA).
3.5. Biological Activity Testing
P. falciparum parasites (3D7 and Dd2 strains) were grown in RPMI 1640 supplemented with 25 mM HEPES, 5% AB human male serum, 2.5 mg/mL Albumax II, and 0.37 mM hypoxanthine. The parasites were subjected to two rounds of sorbitol synchronization before undergoing compound treatment. The ring stage parasites were exposed to the experimental compounds in 384-wells imaging microplates (PerkinElmer CellCarrier, Waltham, MA, USA), as previously described in Reference [32]. The plates were incubated for 72h at 37 °C, 90% N2, 5% CO2, 5% O2, then the parasites were stained with 2-(4-amidinophenyl)-1H–indole-6-carboxamidine (DAPI), and imaged using an Opera QEHS micro-plate confocal imaging system (PerkinElmer, Waltham, MA, USA). The images were analysed as previously described in Reference [32].
Human Embryonic Kidney cells (HEK293) were maintained in the Dulbecco’s Modified Eagle Medium (DMEM) medium supplemented with 10% FBS. The HEK293 cells were exposed to the compounds in TC-treated 384-wells plates (Falcon, Durham, NC, USA). The Plates were incubated for 72 h at 37 °C, 5% CO2, then the media was removed from the wells and replaced with an equal volume of 44 µM resazurin. After an additional 5–6 h incubation at standard conditions, the total fluorescence (excitation/emission: 530 nm/595 nm) was measured using the Envision plate reader (PerkinElmer, Waltham, MA, USA).
The raw data were normalized using the in-plate positive and negative controls to obtain the normalized % inhibition data, which was then used to calculate the IC50 values through a 4-parameter logistic curve fitting in the GraphPad Prism (Version 6.0, La Jolla, CA, USA). The experiments were carried out in two independent biological replicates, each consisting of two technical replicates.
Supplementary Materials
The following are available online at http://www.mdpi.com/1660-3397/16/6/189/s1, Figure S1: HSQC-TOCSY NMR spectrum; Figure S2: Herbimycin analogues and related ansamycins; Figure S3–S8: 1D and 2D NMR spectra of 1; Table S1: Physiochemical parameters of herbimycins and related ansamycin compounds; Table S2–S3: NMR data for 2–5.
Author Contributions
L.B. performed the fermentation of the microbial strain, isolated and elucidated the natural products and prepared the manuscript; L.P.R. conducted theoretical calculations for the ECD spectra; L.L. and V.M.A. performed biological activity testing; D.I.K. and A.R.C. were the supervisors of the L.B.’s PhD project and provided advice on the research design and feedback to the manuscript.
Funding
This work was funded by an Australian Postgraduate Award (APA) provided by the Australian Commonwealth Government awarded to L.B. and an Australian Research Council grant No. LP120200557 awarded to V.M.A..
Acknowledgments
The authors would like to thank Tanja Grkovic for providing valuable feedback to the manuscript. They gratefully acknowledge the support of the Griffith University eResearch Services Team and the use of the High-Performance Computing Cluster “Gowonda” to calculate ECD spectra. L.B. acknowledges the provision of an Australian Postgraduate Award and Griffith University International Postgraduate Research Scholarship. The authors thank the Medicines for Malaria Venture (MMV) for ongoing support and acknowledge the Australian Red Cross Blood Bank for the provision of fresh red blood cells, without which this research could not have been performed.
Conflicts of Interest
The authors declare no conflict of interest.
References
- World Health Organization. World Malaria Report 2017. Available online: http://www.who.int/malaria/publications/world-malaria-report-2017/report/en/ (accessed on 1 May 2018).
- Tuteja, R. Malaria—An overview. FEBS J. 2007, 274, 4670–4679. [Google Scholar] [CrossRef] [PubMed]
- Cowman, A.F.; Healer, J.; Marapana, D.; Marsh, K. Malaria: Biology and disease. Cell 2016, 167, 610–624. [Google Scholar] [CrossRef] [PubMed]
- Michalakis, Y.; Renaud, F. Malaria: Evolution in vector control. Nature 2009, 462, 298–300. [Google Scholar] [CrossRef] [PubMed]
- Menard, D.; Dondorp, A. Antimalarial drug resistance: A threat to malaria elimination. Cold Spring Harbor Perspect. Med. 2017, 7, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Goodman, C.D.; Useglio, M.; Peirú, S.; Labadie, G.R.; McFadden, G.I.; Rodríguez, E.; Gramajo, H. Chemobiosynthesis of new antimalarial macrolides. Antimicrob. Agents Chemother. 2013, 57, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, A.B.S.; Sun, Q.; Nkrumah, L.J.; Dunne, M.W.; Sacchettini, J.C.; Fidock, D.A. In Vitro Efficacy, Resistance Selection, and Structural Modeling Studies Implicate the Malarial Parasite Apicoplast as the Target of Azithromycin. J. Biol. Chem. 2007, 282, 2494–2504. [Google Scholar] [CrossRef] [PubMed]
- Mout, R.; Xu, Z.-D.; Wolf, A.K.; Davisson, V.J.; Jarori, G.K. Anti-malarial activity of geldanamycin derivatives in mice infected with Plasmodium yoelii. Malar. J. 2012, 11, 54. [Google Scholar] [CrossRef] [PubMed]
- Buedenbender, L.; Carroll, A.R.; Ekins, M.; Kurtböke, D.İ. Taxonomic and metabolite diversity of actinomycetes associated with three Australian ascidians. Diversity 2017, 9, 53. [Google Scholar] [CrossRef]
- Buedenbender, L.; Habener, L.J.; Grkovic, T.; Kurtböke, D.İ.; Duffy, S.; Avery, V.M.; Carroll, A.R. HSQC–TOCSY fingerprinting for prioritization of polyketide- and peptide-producing microbial isolates. J. Nat. Prod. 2018, 81, 957–965. [Google Scholar] [CrossRef] [PubMed]
- Omura, S.; Iwai, Y.; Takahashi, Y.; Sadakane, N.; Nakagawa, A.; Oiwa, H.; Hasegawa, Y.; Ikai, T. Herbimycin, a new antibiotic produced by a strain of Streptomyces. J. Antibiot. 1979, 32, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Shaaban, K.A.; Wang, X.; Elshahawi, S.I.; Ponomareva, L.V.; Sunkara, M.; Copley, G.C.; Hower, J.C.; Morris, A.J.; Kharel, M.K.; Thorson, J.S. Herbimycins D–F, ansamycin analogues from Streptomyces sp. Rm-7-15. J. Nat. Prod. 2013, 76, 1619–1626. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Cui, J.; Lu, X.; Zheng, Z.; Liu, X.; Ni, S.; Wang, Y.; Wu, L. Methanethiol as a catabolite of methionine provides methylthio-group for chemical formation of 19-s-methylgeldanamycin and 17,19-dimethylthioherbimycin A. J. Antibiot. 2013, 66, 499–503. [Google Scholar] [CrossRef] [PubMed]
- Iwai, Y.; Nakagawa, A.; Sadakane, N.; Omura, S.; Oiwa, H.; Matsumoto, S.; Takahashi, M.; Kiai, T.; Ochiai, Y. Herbimycin B, a new benzoquinonoid ansamycin with anti-TMV and herbicidal activities. J. Antibiot. 1980, 33, 1114–1119. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Jiang, B.; Wu, L.; Nan, Y.; Cui, J.; Yu, L.; Wei, Y.; Li, J.; Shan, G. Two herbimycin analogs, 4,5-dihydro-(4s)-4-hydroxyherbimycin B and (15s)-15-hydroxyherbimycin B, from Streptomyces sp. CPCC 200291. J. Antibiot. 2015, 68, 476–480. [Google Scholar] [CrossRef] [PubMed]
- Raju, R.; Piggott, A.M.; Khalil, Z.; Bernhardt, P.V.; Capon, R.J. Heronamycin A: A new benzothiazine ansamycin from an Australian marine-derived Streptomyces sp. Tetrahedron Lett. 2012, 53, 1063–1065. [Google Scholar] [CrossRef]
- Dimopoulos, M.-A.; Mitsiades, C.S.; Anderson, K.C.; Richardson, P.G. Tanespimycin as antitumor therapy. Clin. Lymphoma Myeloma Leuk. 2011, 11, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Musiyenko, A.; Barik, S. The heat shock protein 90 of Plasmodium falciparum and antimalarial activity of its inhibitor, geldanamycin. Malar. J. 2003, 2, 30. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Levin, J.I. Macrocycles in Drug Discovery; Royal Society of Chemistry: Cambridge, UK, 2015; Volume 40. [Google Scholar]
- Furtado, N.A.; Pupo, M.T.; Carvalho, I.; Campo, V.L.; Duarte, M.C.T.; Bastos, J.K. Diketopiperazines produced by an Aspergillus fumigatus Brazilian strain. J. Braz. Chem. Soc. 2005, 16, 1448–1453. [Google Scholar] [CrossRef]
- Puopolo, G.; Cimmino, A.; Palmieri, M.; Giovannini, O.; Evidente, A.; Pertot, I. Lysobacter capsici az78 produces cyclo (l-pro-l-tyr), a 2,5-diketopiperazine with toxic activity against sporangia of Phytophthora infestans and Plasmopara viticola. J. Appl. Microbiol. 2014, 117, 1168–1180. [Google Scholar] [CrossRef] [PubMed]
- Hammann, P.; Kretzschmar, G. Secondary metabolites by chemical screening. 12. 13C NMR studies of elaiophylin derivatives. Magn. Reson. Chem. 1991, 29, 667–670. [Google Scholar] [CrossRef]
- Yin, M.; Jiang, M.; Ren, Z.; Dong, Y.; Lu, T. The complete genome sequence of Streptomyces autolyticus CGMCC 0516, the producer of geldanamycin, autolytimycin, reblastatin and elaiophylin. J. Biotechnol. 2017, 252, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Rascher, A.; Hu, Z.; Buchanan, G.O.; Reid, R.; Hutchinson, C.R. Insights into the biosynthesis of the benzoquinone ansamycins geldanamycin and herbimycin, obtained by gene sequencing and disruption. Appl. Environ. Microbiol. 2005, 71, 4862–4871. [Google Scholar] [CrossRef] [PubMed]
- Supong, K.; Sripreechasak, P.; Tanasupawat, S.; Danwisetkanjana, K.; Rachtawee, P.; Pittayakhajonwut, P. Investigation on antimicrobial agents of the terrestrial Streptomyces sp. BCC71188. Appl. Microbiol. Biotechnol. 2017, 101, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Willoughby, P.H.; Jansma, M.J.; Hoye, T.R. A guide to small-molecule structure assignment through computation of (1H and 13C) NMR chemical shifts. Nat. Protoc. 2014, 9, 643–660. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Tomasi, J.; Mennucci, B.; Cammi, R. Quantum mechanical continuum solvation models. Chem. Rev. 2005, 105, 2999–3094. [Google Scholar] [CrossRef] [PubMed]
- Bruhn, T.; Schaumloeffel, A.; Hemberger, Y.; Bringmann, G. Specdis: Quantifying the comparison of calculated and experimental electronic circular dichroism spectra. Chirality 2013, 25, 243–249. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Tenderholt, A.L.; Langner, K.M. Cclib: A library for package-independent computational chemistry algorithms. J. Comput. Chem. 2008, 29, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.; Avery, V.M. Development and optimization of a novel 384-well anti-malarial imaging assay validated for high-throughput screening. Am. J. Trop. Med. Hyg. 2012, 86, 84–92. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).