Angucycline Glycosides from Mangrove-Derived Streptomyces diastaticus subsp. SCSIO GJ056
Abstract
:1. Introduction
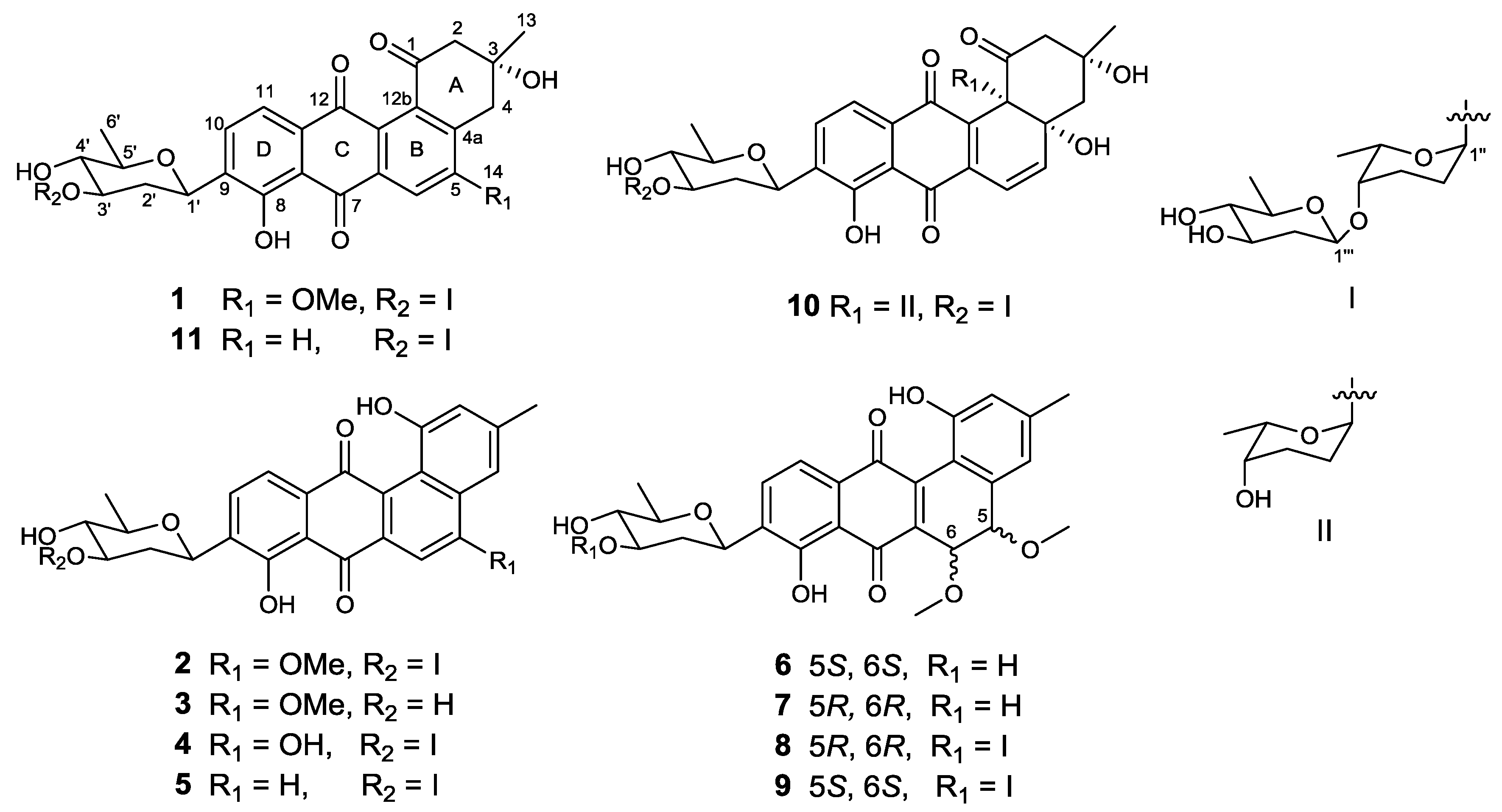
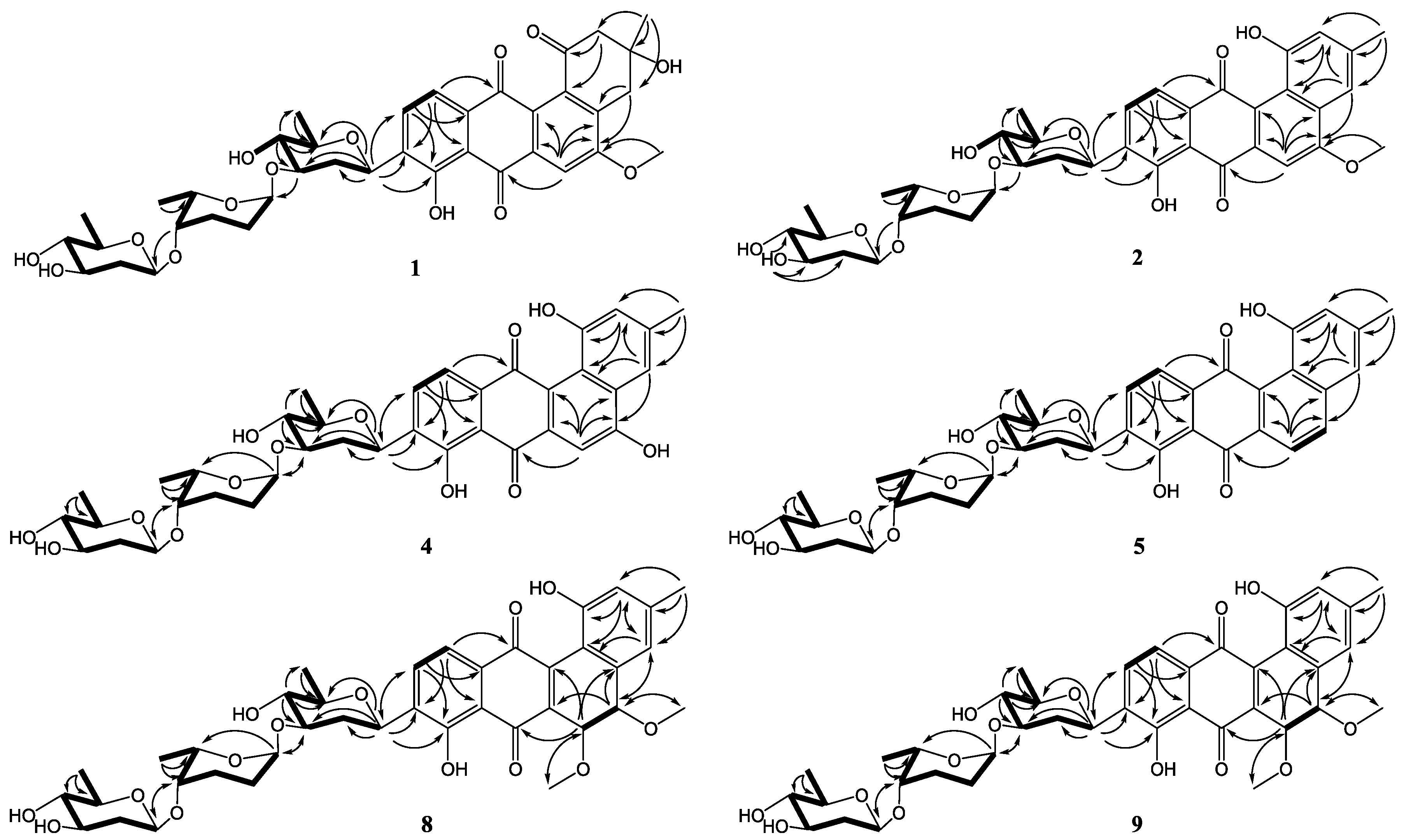
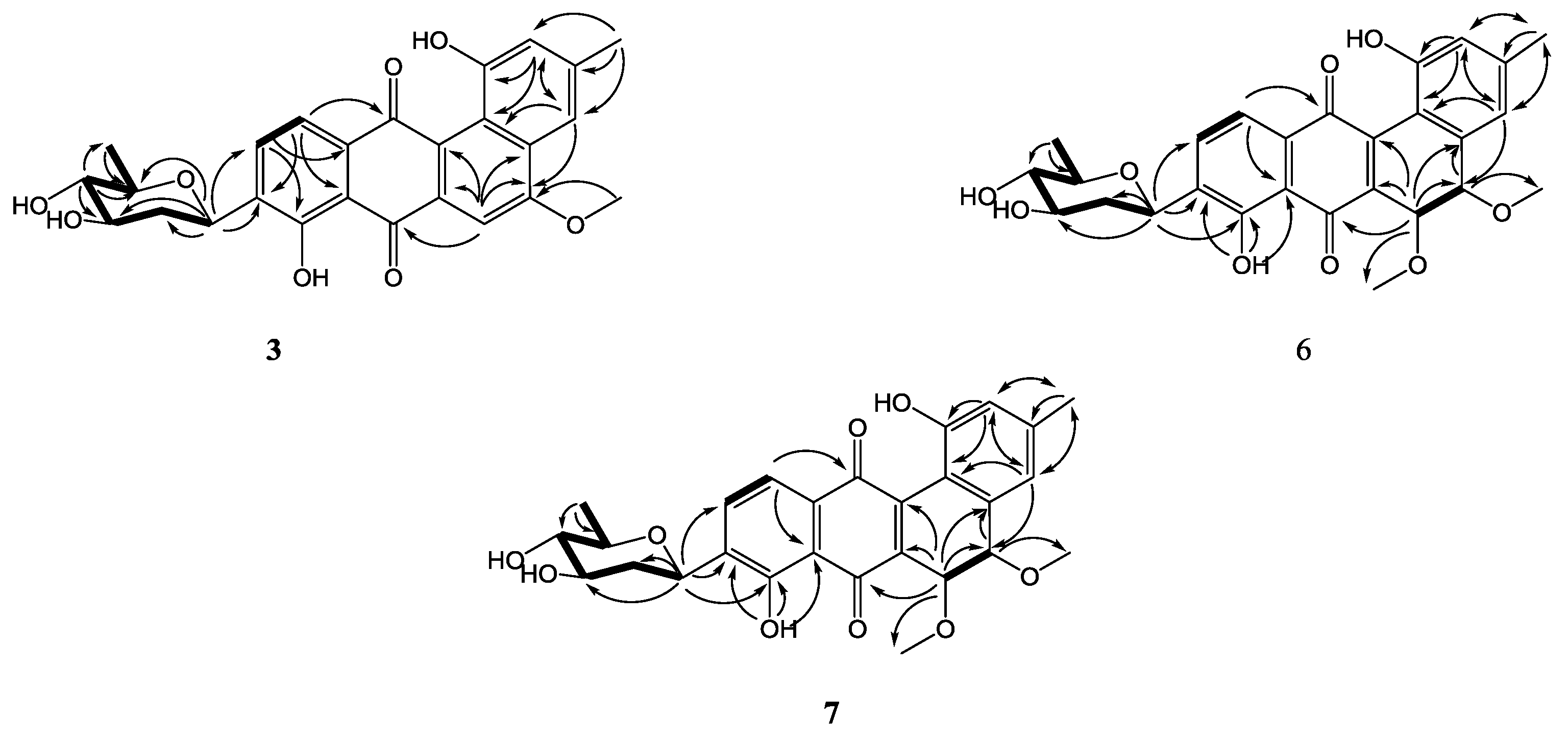
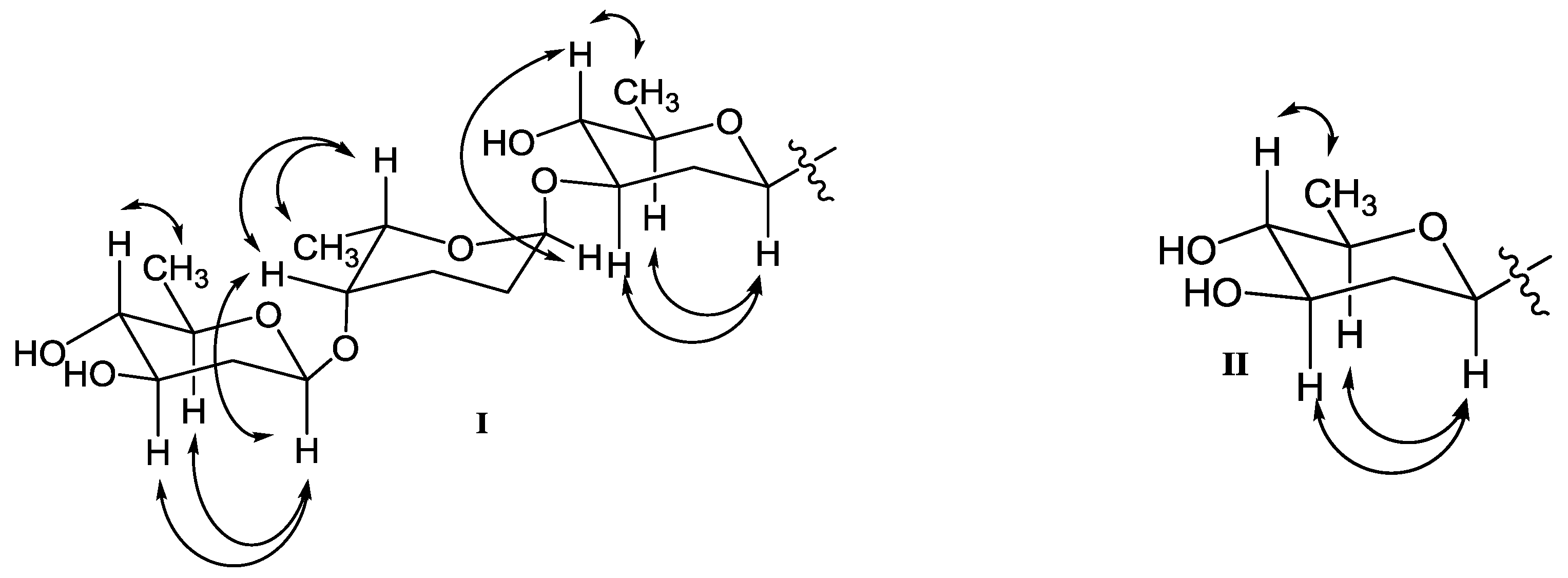
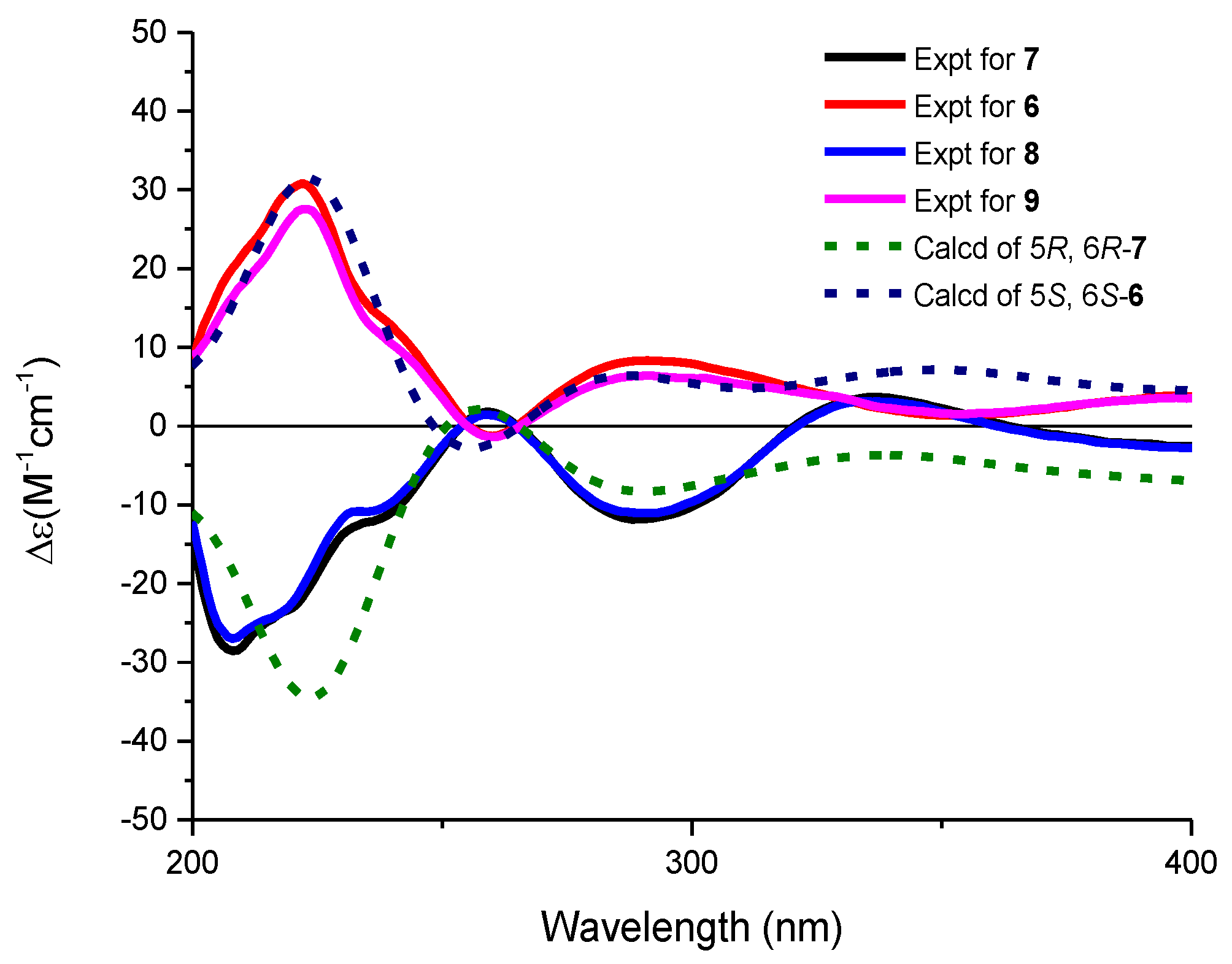
2. Results and Discussion
3. Experimental Section
3.1. General Experimental Procedures
3.2. Bacterial Materials
3.3. Fermentation, Extraction, and Isolation of the Compounds
3.4. Spectral Data
3.5. Electronic Circular Dichroism (ECD) Calculation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rohr, J.; Thiericke, R. Angucycline group antibiotics. Nat. Prod. Rep. 1992, 9, 103–137. [Google Scholar] [CrossRef] [PubMed]
- Kharel, M.K.; Pahari, P.; Shepherd, M.D.; Tibrewal, N.; Nybo, S.E.; Shaaban, K.A.; Rohr, J. Angucyclines: Biosynthesis, mode-of-action, new natural products, and synthesis. Nat. Prod. Rep. 2012, 29, 264–325. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Huang, H.; Chen, Q.; Luo, M.; Sun, A.; Song, Y.; Ma, J.; Ju, J. Identification of the grincamycin gene cluster unveils divergent roles for GcnQ in different hosts, tailoring the L-rhodinose moiety. Org. Lett. 2013, 15, 3254–3257. [Google Scholar] [CrossRef] [PubMed]
- Hertweck, C.; Luzhetskyy, A.; Rebets, Y.; Bechthold, A. Type II polyketide synthases: Gaining a deeper insight into enzymatic teamwork. Nat. Prod. Rep. 2007, 24, 162–190. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Liu, G.; Li, J.; Huang, H.; Zhang, X.; Zhang, H.; Ju, J. Cytotoxic and antibacterial angucycline- and prodigiosin-analogues from the deep-sea derived Streptomyces sp. SCSIO 11594. Mar. Drugs 2015, 13, 1304–1316. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Yu, J.; Ling, H.; Song, Y.; Yuan, J.; Ju, J.; Tao, Y.; Huang, H. Grincamycins I–K, cytotoxic angucycline glycosides derived from marine-derived actinomycete Streptomyces lusitanus SCSIO LR32. Planta Med. 2018, 84, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Duan, Y.; Cui, Z.; Wang, Z.; Li, Z.; Zhang, Y.; Ju, J.; Huang, H. Cytotoxic rearranged angucycline glycosides from deep sea-derived Streptomyces lusitanus SCSIO LR32. J. Antibiot. 2017, 70, 819–822. [Google Scholar] [CrossRef] [PubMed]
- Rohr, J.; Zeeck, A. Metabolic products of microorganisms. 240. Urdamycins, new angucycline antibiotics from Streptomyces fradiae. II. Structural studies of urdamycins B to F. J. Antibiot. 1987, 40, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Lu, X.; Ke, A.; Zheng, Z.; Lin, J.; Hao, W.; Zhu, J.; Fan, Y.; Ding, Y.; Jiang, Q.; et al. Three novel members of angucycline group from Streptomyces sp. N05WA963. J. Antibiot. 2011, 64, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Drautz, H.; Zähner, H.; Rohr, J.; Zeeck, A. Metabolic products of microorganisms. 234 Urdamycins, new angucycline antibiotics from Streptomyces fradiae. J. Antibiot. 1986, 39, 1657–1669. [Google Scholar] [CrossRef] [PubMed]
- Zeeck, A.; Rohr, J.; Sheldrick, G.M.; Jones, P.G.; Paulus, E.F. Structure of a new antibiotic and cytotoxic indicator substance, urdamycin A. J. Chem. Res. 1986, 104–105. [Google Scholar]
- Pérez, M.; Schleissner, C.; Rodríguez, P.; Zúñiga, P.; Benedit, G.; Sánchez-Sancho, F.; de la Calle, F. PM070747, a new cytotoxic angucyclinone from the marine-derived Saccharopolyspora taberi PEM-06-F23-019B. J. Antibiot. 2009, 62, 167–169. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.T.; Carney, J.R.; Gould, S.J. Cloning and heterologous expression of the entire gene clusters for PD 116740 from Streptomyces strain WP 4669 and tetrangulol and tetrangomycin from Streptomyces rimosus NRRL 3016. J. Bacteriol. 1997, 179, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Ohmori, K.; Suzuki, K. Stereochemical relay via axially chiral styrenes: Asymmetric synthesis of the antibiotic TAN-1085. Angew. Chem. Int. Ed. Engl. 2009, 48, 5633–5637. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Tanaka, Y.; Ohmori, K.; Suzuki, K. Synthesis and stereochemical assignment of angucycline antibiotic, PD-116740. Chem. Lett. 2008, 37, 470–471. [Google Scholar] [CrossRef]
- Ohmori, K.; Mori, K.; Ishikawa, Y.; Tsuruta, H.; Kuwahara, S.; Harada, N.; Suzuki, K. Concise total synthesis and structure assignment of TAN-1085. Angew. Chem. Int. Ed. Engl. 2004, 43, 3167–3171. [Google Scholar] [CrossRef] [PubMed]
- Hauser, F.M.; Dorsch, W.A.; Mal, D. Total synthesis of (±)-O-methyl PD 116740. Org. Lett. 2002, 4, 2237–2239. [Google Scholar] [CrossRef] [PubMed]
| Position | 1 a | 2 b | 3 c | |||
|---|---|---|---|---|---|---|
| δC | δH | δC | δH | δC | δH | |
| 1 | 198.4 | 155.3 | 155.2 | |||
| 2 | 53.2 | 2.90, dd (14, 1.1); 3.0, m | 118.7 | 6.99, s | 118.8 | 6.98, s |
| 3 | 71.8 | 140.8 | 140.9 | |||
| 4 | 37.5 | 3.18, m; 2.80, d (18.1) | 113.0 | 7.58, s | 113.2 | 7.58, m |
| 4a | 137.9 | 126.0 | 126.0 | |||
| 5 | 160.9 | 160.3 | 160.5 | |||
| 6 | 108.2 | 7.65, s | 99.8 | 7.54, s | 100.0 | 7.54, s |
| 6a | 135.1 | 135.8 | 136.1 | |||
| 7 | 187.9 | 187.5 | 187.6 | |||
| 7a | 114.8 | 113.9 | 114.1 | |||
| 8 | 158.0 | 156.8 | 157.0 | |||
| 9 | 136.6 | 136.8 | 137.3 | |||
| 10 | 133.9 | 7.81, d (7.8) | 133.4 | 7.82, d (7.6) | 133.3 | 7.80, m |
| 11 | 119.5 | 7.59, d (7.8) | 119.9 | 7.68, d (7.6) | 120.0 | 7.67, m |
| 11a | 133.8 | 133.7 | 133.8 | |||
| 12 | 182.3 | 185.6 | 186.0 | |||
| 12a | 128.4 | 129.8 | 129.9 | |||
| 12b | 137.4 | 120.4 | 120.5 | |||
| 13 | 30.1 | 1.42, s | 21.2 | 2.42, s | 21.2 | 2.41, s |
| 14 | 56.5 | 3.99, s | 56.6 | 4.14, s | 56.6 | 4.13, s |
| 1′ | 71.1 | 4.82, d (10.6) | 70.4 | 4.8, d (13.7) | 70.8 | 4.78, d (10.9) |
| 2′ | 37.6 | 1.41, m | 39.7 | 1.35, m | 40.0 | 1.31, dd (11.4) |
| 2.42, dd (12.3, 4.2) | 2.01, m | 2.28, d (10.2) | ||||
| 3′ | 81.6 | 3.67, ddd (11.4, 8.4, 5.1) | 74.6 | 3.69, ddd (11.1, 8.9, 4.9) | 71.7 | 3.55, t (11.8) |
| 4′ | 76.0 | 3.13, m | 74.5 | 3.06, td (8.9, 5.4) | 77.1 | 2.90, t (8.8) |
| 5′ | 76.1 | 3.45, m | 76.2 | 3.47, q (6.0) | 76.3 | 3.38, m |
| 6′ | 18.4 | 1.37, d (6.1) | 18.5 | 1.30, d (6.1) | 18.5 | 1.28, d (6.1) |
| 1″ | 97.4 | 4.96, s | 91.9 | 4.90, d (2.3) | ||
| 2″ | 25.1 | 1.47, m | 24.1 | 1.29, m | ||
| 2.06, m | 1.83, m | |||||
| 3″ | 24.5 | 1.9, m | 24.1 | 1.76, m; 1.95, m | ||
| 4″ | 76.2 | 3.49, m | 75.4 | 3.44, m | ||
| 5″ | 67.5 | 4.09, m | 65.3 | 4.15, m | ||
| 6″ | 16.9 | 1.16, d (6.5) | 17.0 | 1.05, d (6.5) | ||
| 1″′ | 101.5 | 4.45, dd (9.7, 1.5) | 101.0 | 4.48, dd (9.7, 1.6) | ||
| 2″′ | 38.9 | 2.19, ddd (12.5, 4.8, 1.5) | 36.0 | 1.24, m | ||
| 1.60, td (12.1, 10.0) | 2.47, m | |||||
| 3″′ | 71.4 | 3.45, m | 70.3 | 3.33, m | ||
| 4″′ | 77.0 | 2.95, t (8.9) | 76.8 | 2.72, m | ||
| 5″′ | 71.7 | 3.16, m | 71.6 | 3.11, dq (9.0, 6.2) | ||
| 6″′ | 17.7 | 1.23, d (6.2) | 18.2 | 1.14, d (6.2) | ||
| Position | 4 a | 5 a | 6 b | |||
|---|---|---|---|---|---|---|
| δC | δH | δC | δH | δC | δH | |
| 1 | 155.4 | 155.0 | 157.1 | |||
| 2 | 119.6 | 6.97, m | 117.3 | 7.0, s | 126.0 | 6.73, s |
| 3 | 139.7 | 141.5 | 144.0 | |||
| 4 | 114.4 | 7.63, m | 119.5 | 7.37, s | 123.0 | 6.89, s |
| 4a | 121.7 | 138.6 | 139.2 | |||
| 5 | 163.6 | 135.4 | 8.22, d (8.5) | 78.1 | 4.23, d (2.8) | |
| 6 | 105.6 | 7.54, s | 121.5 | 8.18, d (8.5) | 70.2 | 4.91, d (2.8) |
| 6a | 136.5 | 133.8 | 141.6 | |||
| 7 | 188.7 | 188.0 | 189.1 | |||
| 7a | 114.2 | 114.4 | 113.7 | |||
| 8 | 156.8 | 156.7 | 157.6 | |||
| 9 | 137.5 | 136.8 | 135.9 | |||
| 10 | 133.6 | 7.8, m | 133.5 | 7.86, d (7.8) | 133.0 | 7.88, d (7.9) |
| 11 | 120.2 | 7.71, m | 118.7 | 7.67, d (7.8) | 121.4 | 7.80, d (7.9) |
| 11a | 134.3 | 134.5 | 131.5 | |||
| 12 | 184.6 | 186.8 | 189.0 | |||
| 12a | 130.4 | 134.4 | 143.0 | |||
| 12b | 122.1 | 119.9 | 112.9 | |||
| 13 | 21.1 | 2.40, s | 21.1 | 2.45, s | 21.4 | 2.34, s |
| 14 | 58.5 | 3.45, s | ||||
| 15 | 56.7 | 3.26, s | ||||
| 1′ | 70.6 | 4.78, dd (11.1, 3.0) | 70.5 | 4.83, d (11.4) | 71.4 | 4.92, d (11.0) |
| 2′ | 40.0 | 1.36, dd (22.4, 11.2) | 39.5 | 1.35, td (11.9, 9.9) | 39.5 | 1.46, dd (23.8, 11.7) |
| 2.02, dd (12.2, 5.0) | 2.01, ddd (12.2, 5.0, 1.5) | 2.54, dd (12.1, 3.9 ) | ||||
| 3′ | 74.9 | 3.69, ddd (11.2, 9.0, 4.9) | 74.9 | 3.70, ddd (11.1, 8.9, 4.8) | 73.2 | 3.85, m |
| 4′ | 74.8 | 3.05, t (9.0) | 74.7 | 3.06, t (8.9) | 78.2 | 3.21, t (8.9) |
| 5′ | 76.4 | 3.48, m | 76.3 | 3.48, overlapped | 76.0 | 3.53, dt (15.1, 6.1 ) |
| 6′ | 18.6 | 1.29, d (6.0) | 18.5 | 1.30, d (6.1) | 18.2 | 1.42, d (6.1) |
| 1″ | 92.4 | 4.89, m | 92.2 | 4.89, d (2.7) | ||
| 2″ | 24.3 | 1.27, m | 24.2 | 1.26, m | ||
| 1.84, m | 1.83, m | |||||
| 3″ | 24.3 | 1.77, m; 1.96, m | 24.2 | 1.77, m; 1.95, m | ||
| 4″ | 75.7 | 3.44, m | 75.6 | 3.44, overlapped | ||
| 5″ | 65.6 | 4.15, q (6.5) | 65.5 | 4.15, q ( 6.4) | ||
| 6″ | 17.1 | 1.05, d (6.5) | 17.0 | 1.04, d (6.4) | ||
| 1″′ | 101.3 | 4.47, d (9.6) | 101.2 | 4.47, d (9.7) | ||
| 2″′ | 36.3 | 1.23, m; 2.46, m | 36.2 | 1.24, m; 2.47, m | ||
| 3″′ | 70.6 | 3.32, m | 70.5 | 3.33, ddd (11.8, 8.7, 5.1) | ||
| 4″′ | 77.0 | 2.72, t (8.8) | 76.9 | 2.72, t (8.7) | ||
| 5″′ | 71.8 | 3.1, m | 71.7 | 3.1, m | ||
| 6″′ | 18.3 | 1.14, d (6.2) | 18.2 | 1.14, d (6.1) | ||
| Position | 7 | 8 | 9 | |||
|---|---|---|---|---|---|---|
| δC | δH | δC | δH | δC | δH | |
| 1 | 156.5 | 156.5 | 157.1 | |||
| 2 | 120.6 | 6.81, s | 120.6 | 6.81, s | 126.0 | 6.73, s |
| 3 | 145.4 | 145.4 | 144.0 | |||
| 4 | 119.0 | 7.17, s | 119.0 | 7.16, s | 123.0 | 6.89, s |
| 4a | 138.8 | 138.8 | 139.5 | |||
| 5 | 80.0 | 4.27, d (2.2) | 80.0 | 4.27, d (2.5) | 78.1 | 4.23, d (2.9) |
| 6 | 67.4 | 4.99, d (3.1) | 67.4 | 4.99, d (3.1) | 70.2 | 4.91, d (2.9) |
| 6a | 140.2 | 140.2 | 141.6 | |||
| 7 | 189.0 | 188.9 | 189.1 | |||
| 7a | 113.6 | 113.5 | 113.7 | |||
| 8 | 157.6 | 157.7 | 157.7 | |||
| 9 | 139.4 | 139.7 | 135.9 | |||
| 10 | 133.2 | 7.90, d (7.9) | 133.2 | 7.89, d (7.9) | 133.0 | 7.87, d (7.8) |
| 11 | 121.5 | 7.80, d (7.9) | 121.5 | 7.78, d (7.9) | 121.4 | 7.79, d (7.8) |
| 11a | 131.3 | 131.3 | 131.4 | |||
| 12 | 188.7 | 188.8 | 189.0 | |||
| 12a | 143.1 | 143.2 | 143.0 | |||
| 12b | 112.5 | 112.5 | 112.9 | |||
| 13 | 21.9 | 2.36, s | 21.9 | 2.36, s | 21.4 | 2.34, s |
| 14 | 58.2 | 3.69, s | 58.2 | 3.69, s | 58.5 | 3.44, s |
| 15 | 59.3 | 3.42, s | 59.3 | 3.41, s | 56.7 | 3.26, s |
| 1′ | 71.4 | 4.94, d (11.0) | 71.3 | 4.89, dd (9.5, 1.1) | 71.4 | 4.87, d (10.4) |
| 2′ | 39.4 | 1.46, d (12.1) | 37.7 | 1.48, d (11.7) | 37.7 | 1.47, m |
| 2.53, dd (12.7, 3.5) | 2.48, m | 2.50, ddd (13.0, 4.9, 1.7 ) | ||||
| 3′ | 73.2 | 3.87, m | 82.4 | 3.72, ddd (11.6, 8.3, 5.1 ) | 82.5 | 3.71, ddd (11.5, 8.3, 5.1) |
| 4′ | 78.2 | 3.22, t (8.9) | 76.3 | 3.18, t (8.7) | 76.3 | 3.18, t (8.7) |
| 5′ | 76.1 | 3.54, dt (15.1, 6.0) | 76.3 | 3.51, td (12.2, 6.1) | 76.3 | 3.51, q (6.0) |
| 6′ | 18.2 | 1.43, d (6.1) | 18.6 | 1.44, d (6.0) | 18.6 | 1.44, d (6.0) |
| 1″ | 97.9 | 5.04, s | 98.0 | 5.03, s | ||
| 2″ | 25.3 | 1.55, d (13.9) | 25.4 | 1.54, d (13.8) | ||
| 2.15, m | 2.13, m | |||||
| 3″ | 24.7 | 1.95, m | 24.7 | 1.96, m | ||
| 2.13, m | 2.13, m | |||||
| 4″ | 76.1 | 3.55, m | 76.1 | 3.55, m | ||
| 5″ | 67.7 | 4.13, dd (12.9, 6.4 ) | 67.7 | 4.13, q (6.0) | ||
| 6″ | 17.2 | 1.24, d (6.5) | 17.1 | 1.23, d (6.5) | ||
| 1′′′ | 101.5 | 4.55, dd (9.5, 1.1) | 101.5 | 4.52, dd (9.6, 1.6) | ||
| 2′′′ | 39.3 | 1.71, dd (21.9, 12.1) | 39.3 | 1.71, td (12.1, 9.9) | ||
| 2.30, dd (12.5, 5.0 ) | 2.29, ddd (12.4, 4.9, 1.5) | |||||
| 3′′′ | 71.7 | 3.59, m | 71.7 | 3.59, m | ||
| 4′′′ | 77.7 | 3.11, t (8.9) | 77.7 | 3.11, t (8.9) | ||
| 5′′′ | 72.0 | 3.26, dq ( 9.1, 6.1) | 72.0 | 3.24, m | ||
| 6′′′ | 17.9 | 1.31, d (6.1) | 17.9 | 1.31, d (6.2) | ||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gui, C.; Liu, Y.; Zhou, Z.; Zhang, S.; Hu, Y.; Gu, Y.-C.; Huang, H.; Ju, J. Angucycline Glycosides from Mangrove-Derived Streptomyces diastaticus subsp. SCSIO GJ056. Mar. Drugs 2018, 16, 185. https://doi.org/10.3390/md16060185
Gui C, Liu Y, Zhou Z, Zhang S, Hu Y, Gu Y-C, Huang H, Ju J. Angucycline Glycosides from Mangrove-Derived Streptomyces diastaticus subsp. SCSIO GJ056. Marine Drugs. 2018; 16(6):185. https://doi.org/10.3390/md16060185
Chicago/Turabian StyleGui, Chun, Yena Liu, Zhenbin Zhou, Shanwen Zhang, Yunfeng Hu, Yu-Cheng Gu, Hongbo Huang, and Jianhua Ju. 2018. "Angucycline Glycosides from Mangrove-Derived Streptomyces diastaticus subsp. SCSIO GJ056" Marine Drugs 16, no. 6: 185. https://doi.org/10.3390/md16060185
APA StyleGui, C., Liu, Y., Zhou, Z., Zhang, S., Hu, Y., Gu, Y.-C., Huang, H., & Ju, J. (2018). Angucycline Glycosides from Mangrove-Derived Streptomyces diastaticus subsp. SCSIO GJ056. Marine Drugs, 16(6), 185. https://doi.org/10.3390/md16060185







