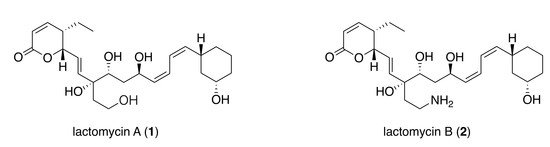
Lactomycins A–C, Dephosphorylated Phoslactomycin Derivatives that Inhibit Cathepsin B, from the Marine-derived Streptomyces sp. ACT232
Abstract
:1. Introduction
2. Results
2.1. Isolation
2.2. Structure Elucidation
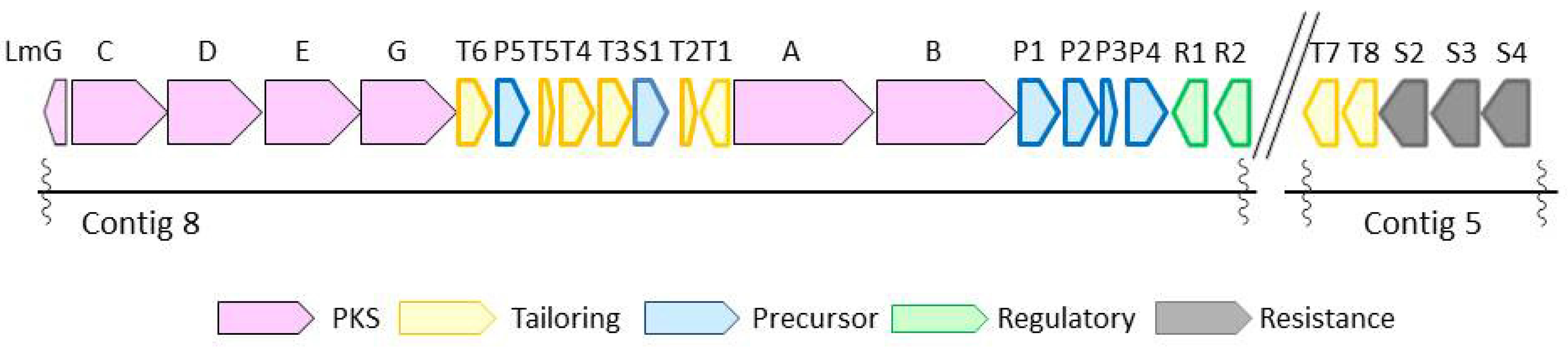
2.3. Biosynthetic Gene Cluster
2.4. Biological Activity
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Collection and Identification of the Microorganism
4.3. Fermentation, Extraction, and Isolation
4.4. Identification of the Biosynthetic Gene Cluster
4.5. Cathepsin B Inhibitory Assay
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Munro, M.H.G.; Blunt, J.W.; Dumdei, E.J.; Hickford, S.J.H.; Lill, R.E.; Li, S.; Battershill, C.N.; Duckworth, A.R. The discovery and development of marine compounds with pharmaceutical potential. J. Biotechnol. 1999, 70, 15–25. [Google Scholar] [CrossRef]
- Gerwick, W.H.; Moore, B.S. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Irie, R.; Takda, K.; Ise, Y.; Ohtsuka, S.; Okada, S.; Gustafson, K.; Matsunaga, S. Structure revision of poecillastrin C and the absolute configuration of the β-hydroxyaspartic acid residue. Org. Lett. 2017, 19, 5395–5397. [Google Scholar] [CrossRef] [PubMed]
- Gondi, C.S.; Rao, J.S. Cathepsin B as a cancer target. Expert Opin. Ther. Targets 2013, 17, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Takada, K.; Nogi, Y.; Okada, S.; Matsunaga, S. Lower homologues of ahpatinin, aspartic protease inhibitores, from a marine Streptomyces sp. J. Nat. Prod. 2014, 77, 1749–1752. [Google Scholar] [CrossRef] [PubMed]
- Shibata, T.; Kurihara, S.; Oikawa, T.; Ohkawa, N.; Shimazaki, N.; Sasagawa, K.; Kobayashi, T.; Kohama, T.; Asai, F.; Shiraishi, A.; Sugimura, Y. Preparation of leustroducsin H and the structure-activity relationship of its derivatives. J. Antibiot. 1995, 48, 1518–1520. [Google Scholar] [CrossRef] [PubMed]
- Trost, B.M.; Knopf, J.D.; Brindle, C.S. Synthetic strategies employed for the construction of fostriecin and related natural products. Chem. Rev. 2016, 116, 15035–15088. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, C.R.; Trager, W.F. Absolute configuration of acenocoumarin. J. Med. Chem. 1979, 22, 1122–1124. [Google Scholar] [CrossRef] [PubMed]
- Fushimi, S.; Nishikawa, S.; Shimazu, A.; Seto, H. Studies on new phosphate ester antifungal antibiotics phoslactomycins. I. Taxonomy, fermentation, purification and biological activities. J Antibiot. 1989, 42, 1019–1025. [Google Scholar] [CrossRef] [PubMed]
- Fushimi, S.; Furihata, K.; Seto, H. Studies of new phosphate ester antifungal antibiotics phoslactomycins. II. Structure elucidation of phoslactomycins A to F. J. Antibiot. (Tokyo) 1989, 42, 1026–1036. [Google Scholar] [CrossRef] [PubMed]
- Mizuhara, N.; Usuki, Y.; Ogita, M.; Fujita, K.I.; Kuroda, M.; Doe, M.; Iio, H.; Tanaka, T. Identification of phoslactomycin E as a metabolite inducing hyphal morphological abnormalities in Aspergillus fumigatus IFO 5840. J. Antibiot. (Tokyo) 2007, 60, 762–765. [Google Scholar] [CrossRef] [PubMed]
- Fotso, S.; Graupner, P.; Xiong, Q.; Hahn, D.; Avila-Adame, C.; Davis, G. Phoslactomycins from Streptomyces sp. MLA1839 and their biological activities. J. Nat. Prod. 2013, 76, 1509–1513. [Google Scholar] [CrossRef] [PubMed]
- Kohama, T.; Nakamura, T.; Kinoshita, T.; Kaneko, I.; Shiraishi, A. Novel microbial metabolites of the phoslactomycins family induce production of colony-stimulating factors by bone marrow stromal cells. II. Isolation, physico-chemical properties and structure determination. J. Antibiot. (Tokyo) 1993, 46, 1512–1519. [Google Scholar] [CrossRef] [PubMed]
- Kohama, T.; Maeda, H.; Sakai, J.I.; Shiraishi, A.; Yamashita, K. Leustroducsin B, a New Cytokine Inducer Derived from an Actinomycetes, Induces Thrombocytosis in Mice. J. Antibiot. (Tokyo) 1996, 49, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Simizu, S.; Teruya, T.; Nogawa, T.; Aono, H.; Ueki, M.; Uramoto, M.; Kobayashi, Y.; Osada, H. Deamino-hydroxy-phoslactomycin B, a biosynthetic precursor of phoslactomycin, induces myeloid differentiation in HL-60 cells. Biochem. Biophys. Res. Commun. 2009, 383, 406–410. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. AntiSMASH 3.0―A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef] [PubMed]
- Palaniappan, N.; Kim, B.S.; Sekiyama, Y.; Osada, H.; Reynolds, K.A. Enhancement and selective production of phoslactomycin B, a protein phosphatase IIa inhibitor, through identification and engineering of the corresponding biosynthetic gene cluster. J. Biol. Chem. 2003, 278, 35552–35557. [Google Scholar] [CrossRef] [PubMed]
- Alhamadsheh, M.M.; Palaniappan, N.; DasChouduri, S.; Reynolds, K.A. Modular polyketide synthases and cis double bond formation: Establishment of activated cis-3-cyclohexylpropenoic acid as the diketide intermediate in phoslactomycin biosynthesis. J. Am. Chem. Soc. 2007, 129, 1910–1911. [Google Scholar] [CrossRef] [PubMed]
- Ghatge, M.S.; Reynolds, K.A. The plmS2-Encoded Cytochrome P450 Monooxygenase Mediates Hydroxylation of Phoslactomycin B in Streptomyces sp. Strain HK803. J. Bacteriol. 2005, 187, 7970–7976. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.L.; Zhao, J.; Liu, W.; Gao, J.F.; Tao, L.M.; Pan, H.X.; Tang, G.L. Identification of phoslactomycin biosynthetic gene clusters from Streptomyces platensis SAM-0654 and characterization of PnR1 and PnR2 as positive transcriptional regulators. Gene 2012, 509, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Ghatge, M.S.; Palaniappan, N.; Alhamadsheh, M.M.; DiBari, J.; Reynolds, K.A. Application of a newly identified and characterized 18-O-acyltransferase in chemoenzymatic synthesis of selected natural and nonnatural bioactive derivatives of phoslactomycins. Appl. Environ. Microbiol. 2009, 75, 3469–3476. [Google Scholar] [CrossRef] [PubMed]
- Lecaille, F.; Kaleta, J.; Bpoemme, D. Human and parasitic papain-like cysteine proteases: Their role in physiology and pathology and recent developments in inhibitor design. Chem. Rev. 2002, 102, 4459–4488. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, T.; Seto, H.; Beppu, T. Leptomycins A and B, new antifungal antibiotics. I. Taxonomy of the producing strain and their fermentation, purification and characterization. J. Antibiot. (Tokyo) 1983, 36, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, T.; Seto, H.; Beppu, T. Leptomycins A and B, new antifungal antibiotics II Structure elucidation. J. Antibiot. (Tokyo) 1983, 36, 646–650. [Google Scholar] [PubMed]
- Hiwasa, T.; Fujita-Yoshigaki, J.; Shirouzu, M.; Koide, H.; Sawada, T.; Sakiyama, S.; Yokoyama, S. c-Ha-Ras mutants with point mutations in Gln-Val-Val region have reduced inhibitory activity toward cathepsin B. Cancer Lett. 1993, 69, 161–165. [Google Scholar] [CrossRef]

| Position | 1 (DMSO-d6) | 2 (CD3OD) | 3 (CD3OD) | |||||
|---|---|---|---|---|---|---|---|---|
| δC, Type | δH (J in Hz) | HMBC | δC, Type | δH (J in Hz) | HMBC | δC, Type | δH (J in Hz) | |
| 1 | 164.0, C | 166.4, C | 166.4, C | |||||
| 2 | 120.5, CH | 5.96, d (9.7) | 1, 4 | 121.1, CH | 6.03, d (9.8) | 1, 4 | 120.8, CH | 6.02, d (9.8) |
| 3 | 151.5, CH | 7.03, dd (4.7, 9.7) | 1, 4, 5 | 152.8, CH | 7.10, dd (5.0, 9.8) | 1, 5 | 152.6, CH | 7.10, dd (4.7, 9.8) |
| 4 | 39.1, CH | 2.48, m | 40.6, CH | 2.56, m | 40.5, CH | 2.53, m | ||
| 5 | 80.7, CH | 5.02, dd (6.5, 4.2) | 1, 3, 6, 7, 22 | 82.4, CH | 5.10, t (4.7) | 82.2, CH | 5.06 dd (4.3, 6.7) | |
| 6 | 123.7, CH | 5.69, dd (6.5, 15.6) | 5, 8 | 127.4, CH | 5.97, ma | 5, 7, 8 | 124.5, CH | 5.87, dd (6.7, 15.7) |
| 7 | 137.4, CH | 5.84, d (15.6) | 5, 6, 8 | 136.9, CH | 5.97, ma | 5, 6, 8 | 139.4, CH | 6.00, d (15.7) |
| 8 | 76.6, C | 77.9, C | 75.6, C | |||||
| 9 | 72.6, CH | 3.53, d (10.0) | 7, 8, 10, 11, 24 | 74.7, CH | 3.72, brd (9.6) | 8 | 74.9, CH | 3.68, d (9.7) |
| 10a | 39.6, CH2 | 0.98, m | 9 | 40.5, CH2 | 1.34, m | 11 | 40.1, CH2 | 1.31, m |
| 10b | 1.58, ddd(1.4, 10.0, 12.8) | 11 | 1.75, m | 9 | 1.73, m | |||
| 11 | 63.5, CH | 4.58, ddd (1.8, 8.4, 10.0) | 9, 10, 13 | 65.5, CH | 4.80b | 65.0, CH | 4.80, brt (10) | |
| 12 | 137.6, CH | 5.40, brt (9) | 10, 14 | 135.8, CH | 5.45, brt (9) | 14 | 135.7, CH | 5.46, brt (9) |
| 13 | 121.8, CH | 6.09b | 11, 15 | 124.4, CH | 6.27, t (11) | 11 | 123.8, CH | 6.26, t (11) |
| 14 | 122.5, CH | 6.10b | 12, 16 | 123.0, CH | 6.23, t (11) | 16 | 122.9, CH | 6.23, t (11) |
| 15 | 137.8, CH | 5.25, brt (9) | 13, 17, 21 | 139.3, CH | 5.32, brt (9) | 13 | 138.7, CH | 5.31, brt (9) |
| 16 | 35.0, CH | 2.45, m | 36.4, CH | 2.55, m | 36.1, CH | 2.55, m | ||
| 17a | 42.7, CH2 | 0.98, m | 43.0, CH2 | 1.02, m | 42.8, CH2 | 1.03, m | ||
| 17b | 1.68, m | 1.82, m | 16, 18 | 1.84, m | ||||
| 18 | 68.5, CH | 3.37, m | 70.8, CH | 3.54, m | 70.3, CH | 3.53, m | ||
| 19a | 35.6, CH2 | 0.99, m | 36.1, CH2 | 1.13, m | 35.6, CH2 | 1.13, m | ||
| 19b | 1.78, m | 1.92, m | 1.92, m | |||||
| 20a | 23.9, CH2 | 1.26, m | 25.1, CH2 | 1.37, m | 24.8, CH2 | 1.38, m | ||
| 20b | 1.64, m | 1.78, m | 1.78, m | |||||
| 21a | 32.4, CH2 | 0.89, m | 33.3, CH2 | 0.99, m | 33.0, CH2 | 0.98, m | ||
| 21b | 1.44, m | 1.55, m | 1.56, m | |||||
| 22a | 21.7, CH2 | 1.32, m | 3, 4, 5, 23 | 22.8, CH2 | 1.47, m | 3, 4, 5, 23 | 22.5, CH2 | 1.46, m |
| 22b | 1.47, m | 3, 4, 5, 23 | 1.65, m | 3, 4, 23 | 1.65, m | |||
| 23 | 11.3, CH3 | 0.80, d (7.4) | 4, 22 | 11.4, CH3 | 0.96, t (7.4) | 4, 22 | 11.1, CH3 | 0.96, d (7.4) |
| 24 | 40.0, CH2 | 1.74, m | 7, 8, 9, 25 | 35.2, CH2 | 1.98, m | 7, 8, 25 | 24.8, CH3 | 1.27, s |
| 25a | 57.9, CH2 | 3.46, m | 8, 24 | 37.4, CH2 | 2.97, m | 24 | ||
| 25b | 3.53, m | 8, 24 | 3.03, m | 8, 24 | ||||
| Gene | Protein Homologues, Identity (%) | Proposed Function | |
|---|---|---|---|
| S.plantensis SAM-0654 | Streptomyces sp. HK803 | ||
| lmG | pnG, 87 | plm8, 83 | Thioesterase |
| lmC | pnC, 90 | plm4, 88 | PKS |
| lmD | pnD, 90 | plm5, 92 | PKS |
| lmE | pnE, 91 | plm6, 91 | PKS |
| lmF | pnF, 92 | plm7, 93 | PKS |
| lmT6 | pnT6, 88 | plmT8, 88 | Dehydrogenase |
| lmP5 | pnP6, 94 | plmT7, 94 | Crotonyl CoA reductase |
| lmT5 | pnT5, 92 | plmT6, 96 | Ferredoxin |
| lmT4 | pnT4, 94 | plmT5, 98 | Protein kinase |
| lmT3 | pnT3, 97 | plmT4, 98 | Cytochrome P450 |
| lmS1 | pnS1, 94 | plmT3, 92 | Transporter |
| lmT2 | pnT2, 93 | plmT2, 92 | NAD dependent epimerase |
| lmT1 | pnT1, 83 | plmT1, 84 | Aminotransferase |
| lmA | pnA, 90 | plm1, 90 | PKS |
| lmB | pnB, 90 | plm2-3, 94 | PKS |
| lmP1 | pnP1, 92 | plmJK, 96 | AMP-dependent synthetase and ligase |
| lmP2 | pnP2, 94 | plmL, 96 | Short-chain-CoA dehydrogenase |
| lmP3 | pnP3, 93 | ChcA, 97 | Cyclohexenylcarbonyl-CoA reductase |
| lmP4 | pnP4, 87 | plmM, 92 | NADH:FMN oxidoreductase |
| lmR1 | pnR1, 86 | plmR1, 94 | Regulator |
| lmR2 | pnR2 a | plmR2, 92 | Regulator |
| lmT7 | pnT7, 92 | plmS2, 90 | Cytochrome P450 |
| lmT8 | pnT8, 90 | plmS3, 90 | 18-O-Acyltransferase |
| lmS2 | pnS2, 88 | plmS4, 85 | Transporter |
| lmS3 | PnS3, 91 | ND b | ABC transporter |
| lmS4 | PnS4, 88 | ND | ABC transporter |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Carandang, R.R.; Harada, Y.; Okada, S.; Yoshitake, K.; Asakawa, S.; Nogi, Y.; Matsunaga, S.; Takada, K. Lactomycins A–C, Dephosphorylated Phoslactomycin Derivatives that Inhibit Cathepsin B, from the Marine-derived Streptomyces sp. ACT232. Mar. Drugs 2018, 16, 70. https://doi.org/10.3390/md16020070
Sun Y, Carandang RR, Harada Y, Okada S, Yoshitake K, Asakawa S, Nogi Y, Matsunaga S, Takada K. Lactomycins A–C, Dephosphorylated Phoslactomycin Derivatives that Inhibit Cathepsin B, from the Marine-derived Streptomyces sp. ACT232. Marine Drugs. 2018; 16(2):70. https://doi.org/10.3390/md16020070
Chicago/Turabian StyleSun, Yi, Rogie Royce Carandang, Yuta Harada, Shigeru Okada, Kazutoshi Yoshitake, Shuichi Asakawa, Yuichi Nogi, Shigeki Matsunaga, and Kentaro Takada. 2018. "Lactomycins A–C, Dephosphorylated Phoslactomycin Derivatives that Inhibit Cathepsin B, from the Marine-derived Streptomyces sp. ACT232" Marine Drugs 16, no. 2: 70. https://doi.org/10.3390/md16020070
APA StyleSun, Y., Carandang, R. R., Harada, Y., Okada, S., Yoshitake, K., Asakawa, S., Nogi, Y., Matsunaga, S., & Takada, K. (2018). Lactomycins A–C, Dephosphorylated Phoslactomycin Derivatives that Inhibit Cathepsin B, from the Marine-derived Streptomyces sp. ACT232. Marine Drugs, 16(2), 70. https://doi.org/10.3390/md16020070