Abstract
Marine fungi are a promising source of novel bioactive natural products with diverse structure. In our search for new bioactive natural products from marine fungi, three new phenone derivatives, asperphenone A–C (1–3), have been isolated from the ethyl acetate extract of the fermentation broth of the mangrove-derived fungus, Aspergillus sp. YHZ-1. The chemical structures of these natural products were elucidated on the basis of mass spectrometry, one- and two-dimensional NMR spectroscopic analysis and asperphenone A and B were confirmed by single-crystal X-ray crystallography. Compounds 1 and 2 exhibited weak antibacterial activity against four Gram-positive bacteria, Staphylococcus aureus CMCC(B) 26003, Streptococcus pyogenes ATCC19615, Bacillus subtilis CICC 10283 and Micrococcus luteus, with the MIC values higher than 32.0 µM.
1. Introduction
Marine filamentous fungi are a rich source of antimicrobial compounds, anti-inflammatory, anticancer and antiviral agents [1,2,3,4,5]. Among all the marine-sourced fungi, the species in the genus Aspergillus (Trichocomaceae) are an important source for novel pharmacological metabolites such as polyketides, alkaloids and terpenoids [6,7,8,9]. The mangrove plants inhabit the intertidal zones in the tropics and subtropics, providing a very unique habitat for animals and microbes. Thus, the mangrove-associated fungi, with the majority coming from endophytic species, have attracted much attention to discover structurally diverse and bioactive secondary metabolites. In the past several years, our research group has explored many novel secondary metabolites from mangrove-derived endophytic fungi, such as sesquiterpenoids diaporols A-I, diaporine with regulation activity of macrophage differentiation and other polyketides [10,11,12]. As part of our ongoing search for novel biologically active natural products from microbe from special environment [13,14], we chemically investigated an endophytic fungus, Aspergillus sp. YHZ-1, from mangrove plant from Hainan Island, China. Fractionation of the ethyl acetate extract of its liquid fermentation broth led to the discovery of three new phenone derivatives, which we have named asperphenone A–C (1–3). Compounds 1 and 2 were tested and displayed weak antibacterial activity against four Gram-positive bacteria. Herein, we report the isolation and structure elucidation of these new compounds and their antibacterial activity.
2. Results
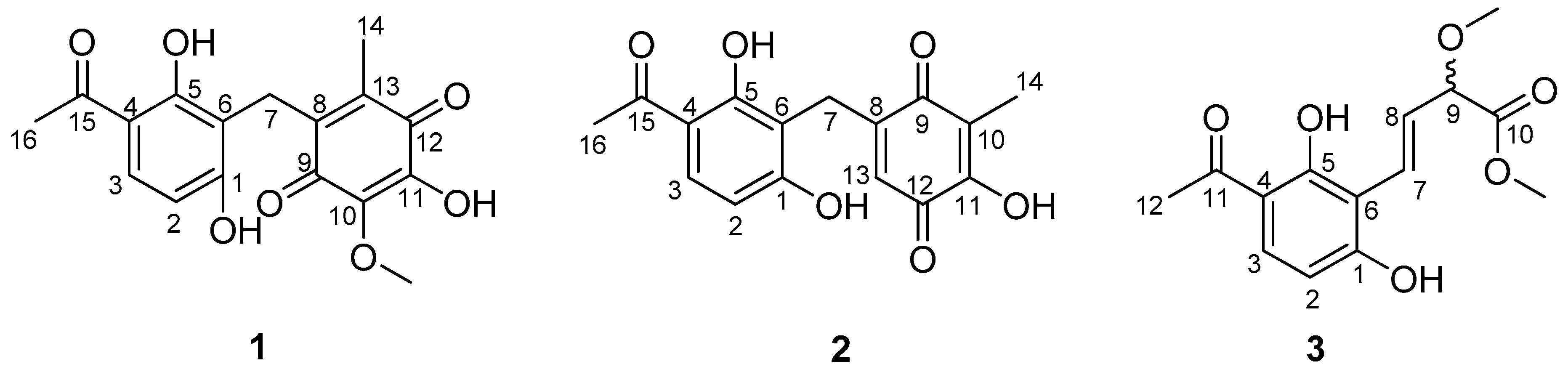
A large-scale fermentation broth (50 L) of Aspergillus sp. YHZ-1 was collected and extracted with ethyl acetate three times to produce a crude extract. The subsequent fractionation by repeated column chromatography over silica gel, octadecylsilyl silica gel (ODS), Sephadex LH-20 and semi-preparative reversed-phase high performance liquid chromatography (HPLC) yielded three new phenone derivatives, asperphenone A–C (1–3) (Figure 1).
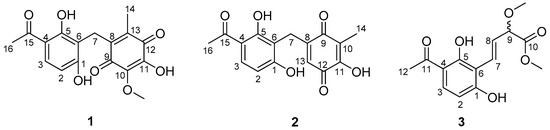
Figure 1.
The structures of asperphenone A–C (1–3) isolated from Aspergillus sp. YHZ-1.
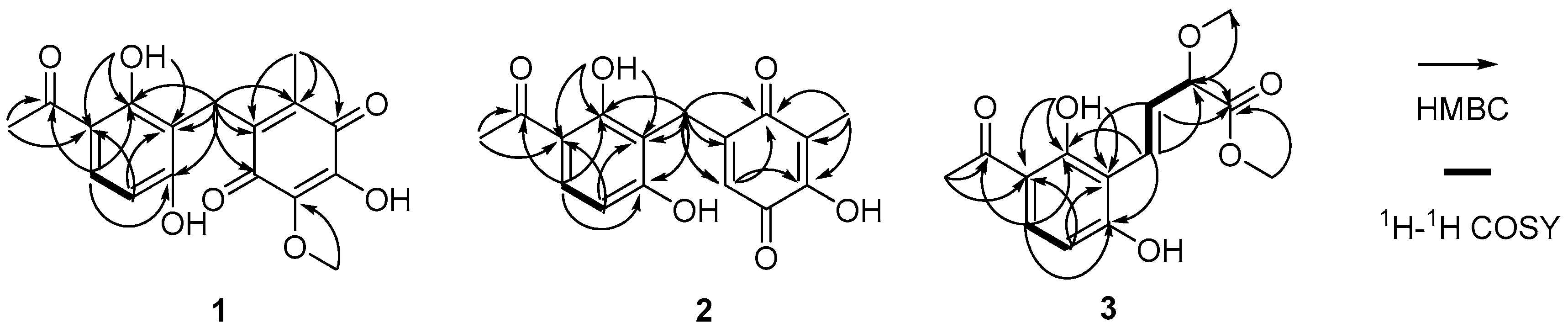
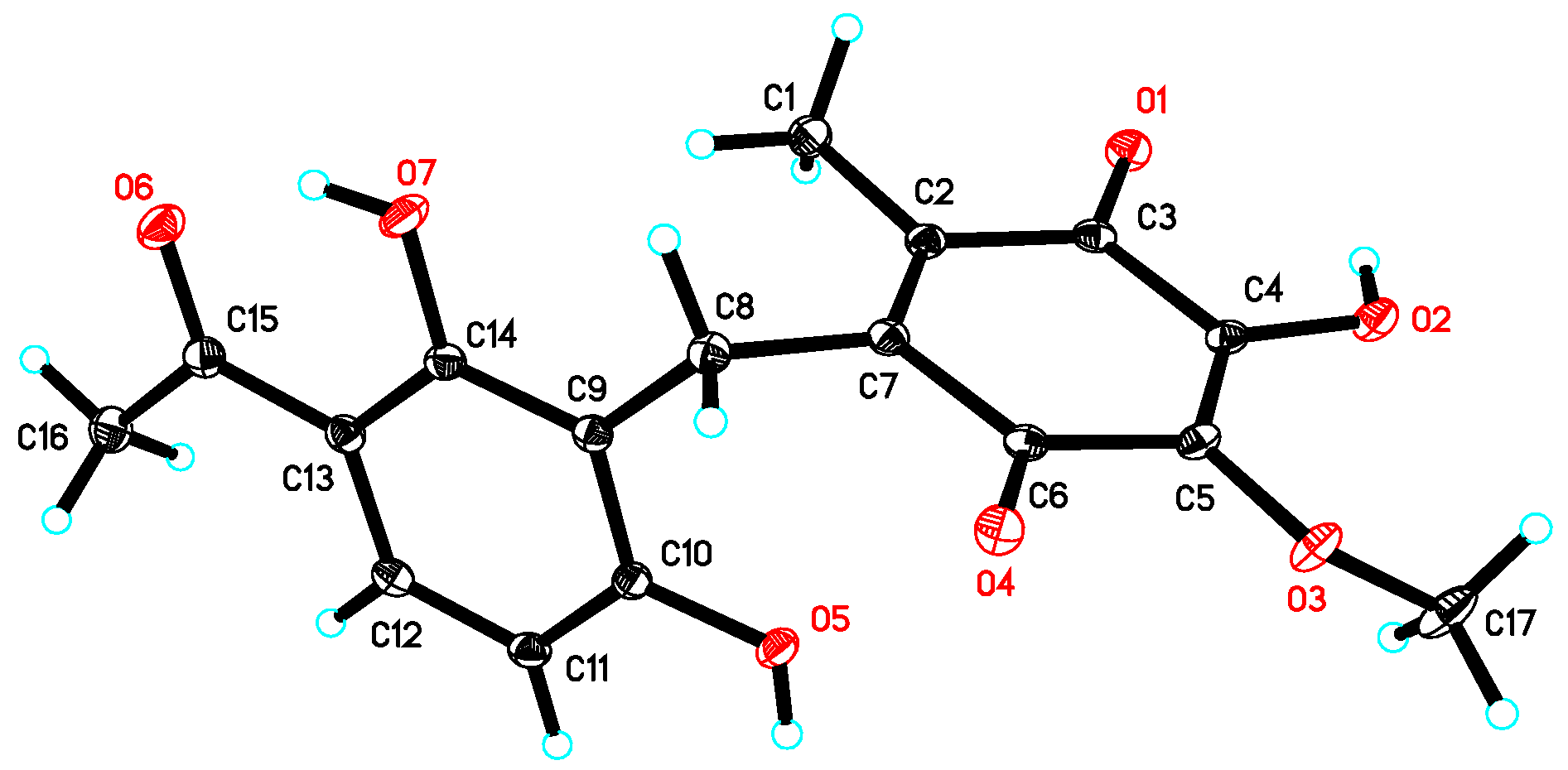
Asperphenone A (1) was isolated as a reddish-brown needle crystal. The molecular formula of 1 was determined to be C17H16O7 on the basis of the high-resolution (HR) ESI-MS (m/z 355.2716 [M + Na]+, calcd. 355.2706) along with the 1H and 13C NMR data (Table 1), indicating ten degrees of unsaturation. The 1H, 13C and HSQC NMR spectra (in supplementary materials) in DMSO-d6 revealed signals of two methyls (δC/δH 11.6/1.73, C-14/H3-14; δC/δH 26.6/2.52, C-16/H3-16), one oxygenated methyl (δC/δH 60.3/3.78, 10-OMe), one methylene (δC/δH 19.4/3.70, C-7/H2-7), one ketone carbon (δC 203.8, C-15), two α,β-unsaturated carbonyl carbons (δC 182.7, C-9; δC 185.1, C-12), three phenolic hydroxyl protons (δH 10.25, 1-OH; δH 13.21, 5-OH; δH 10.85, 11-OH) and ten aromatic/olefinic carbons, whose chemical shift values indicated the presence of one 1,2,3,4-tetrasubstituted benzene ring (δC 162.7, C-1; δC/δH 107.8/6.42 (d, J = 8.8 Hz), C-2/H-2; δC/δH 131.8/7.67 (d, J = 8.8 Hz), C-3/H-3; δC 112.8, C-4; δC 162.4, C-5; δC 112.0, C-6) and four quaternary carbons (δC 136.0, C-8; δC 138.8, C-10; δC 143.3, C-11; δC 144.5, C-13). The tetrasubstituted benzene ring was determined as a 1,3-dihydroxy-4-acetophenone moiety, which was deduced by analysis of the 1H-1H COSY correlation from H-2 to H-3 and the HMBC correlations from H-2 to C-4 and C-6, from H-3 to C-1, C-5 and C-15, from H3-16 to C-4 and C-15 and from 5-OH to C-4, C-5 and C-6 (Figure 2). HMBC correlations from H2-7 to C-1, C-5 and C-6 permitted the linkage of C-7 to C-6 of the acetophenone moiety. The presence of a p-benzoquinone moiety could be deduced from the chemical shifts of C8-C13 in 1, which was partially confirmed by the observation of HMBC correlations from H3-14 to C-8, C-12 and C-13 and from H2-7 to C-8, C-9 and C13. Meanwhile, the connectivity of the p-benzoquinone and acetophenone moieties via the methylene C-7 was established by the relevant HMBC correlations. However, the positions of the hydroxyl group and methoxyl group on the p-benzoquinone moiety cannot be determined as their 2D correlations cannot be observed. Finally, a high-quality single crystal of 1 was obtained and the full structure of 1 was determined by the X-ray crystallographic analysis in a Cu Kα radiation in low temperature (Figure 3), which also confirmed the proposed structure of 1.

Table 1.
1H and 13C NMR data for compounds 1–3.
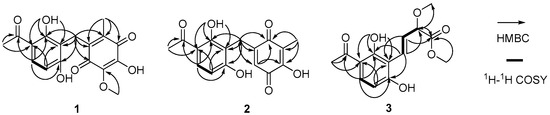
Figure 2.
The key 2D NMR correlations of asperphenone A–C (1–3).
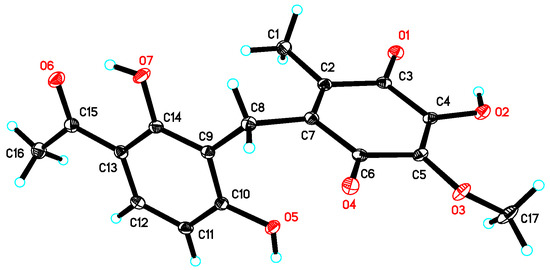
Figure 3.
X-ray crystal structure of 1.
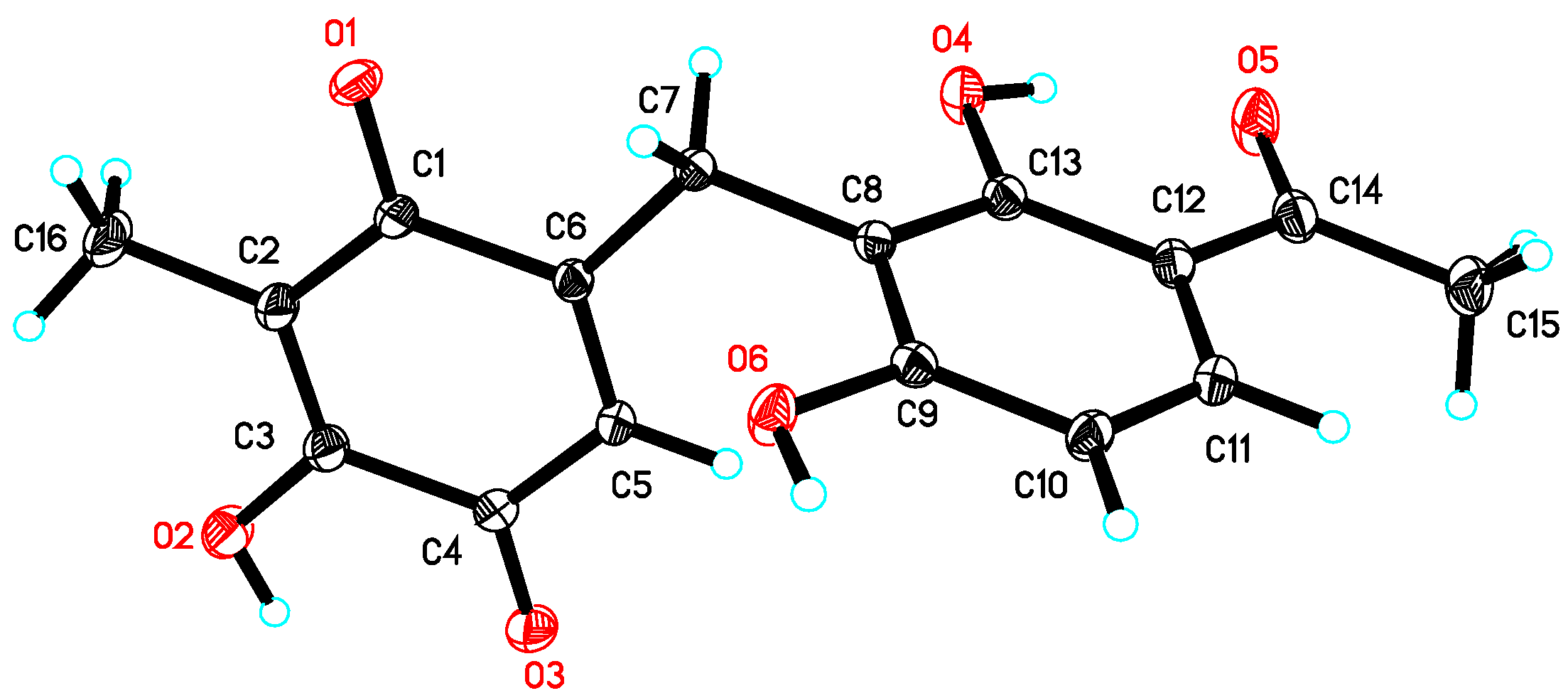
Asperphenone B (2) was obtained as a yellow needle crystal, with the molecular formula C16H14O6 (ten degrees of unsaturation) as derived from the HRESIMS data (m/z 325. 1326 [M + Na]+, calcd. 325.0683). The 13C NMR spectrum showed sixteen carbon signals for two methyls (C-14 and C-16), one methylene (C-7), one ketone carbon (C-15), two α,β-unsaturated carbonyl carbons (C-9 and C-12) and ten aromatic/olefinic carbons. Comparison of the MS, 1H and 13C NMR data of 2 with those of 1 revealed the presence of the same 1,3-dihydroxy-4-acetophenone moiety as 1 and a different substituted p-benzoquinone moiety. Further HMBC correlations (Figure 2) from H3-14 (δH 1.82) to C-9 (δC 187.2), C-10 (δC 116.9) and C-11(δC 153.8), from H-13 (δH 5.76) to C-7 (δC 22.0), C-8 (δC 148.5), C-9 and C-11 and from H2-7 (δH 3.61) to C-8, C-9 and C-13 (δC 127.1) along with the unsaturation requirement of 2 confirmed the substituted pattern of the p-benzoquinone moiety. Finally, the connection between the two moieties mentioned above was through C-7 by interpretation of the relevant HMBC correlations from H2-7. Detailed NMR analysis allowed the assignment of 2 as shown in Figure 1, which was also confirmed by a low-temperature single-crystal X-ray diffraction experiment (Figure 4).
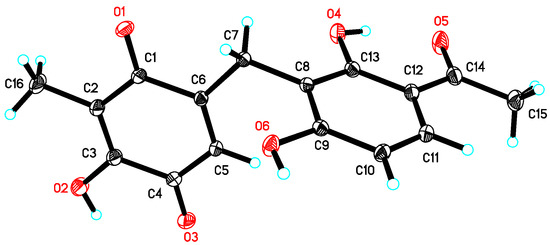
Figure 4.
X-ray crystal structure of 2.
Asperphenone C (3) was isolated as a colorless needle crystal that analyzed for the molecular formula C14H16O6 (seven degrees of unsaturation) by HRESIMS (m/z 303. 1215 [M + Na]+, calcd. 303.0839) in combination with 13C NMR data (Table 1). The 13C and DEPT135 NMR spectra of 3 displayed signals for fourteen carbons, including one methyl carbon (δC 26.3, C-12), two oxygenated methyl carbons (δC 57.0, 9-OMe; δC 52.0, 10-OMe), one oxygenated methine (δC 83.3, C-9), one ketone carbon (δC 204.3, C-11), one carboxylic carbon (δC 171.7, C-10) and eight aromatic/olefinic carbons. In this molecule, the same moiety, 1,3-dihydroxy-4-acetophenone as found in 1, could be deduced through analysis of the 1H-1H COSY correlation from H-2 (δH 6.55, d, J = 8.6 Hz) to H-3 (δH 7.71, d, J = 8.6 Hz) and HMBC correlations from H-2 to C-4 (δC 113.8) and C-6 (δC 111.4), from H-3 to C-1 (δC 163.3), C-5 (δC 164.4) and C-11, from 5-OH (δH 13.73) to C-4, C-5 and C-6 and from H3-12 (δH 2.56) to C-4 and C-11. 1H-1H COSY correlations from H-7 (δH 7.07, dd, J = 16.2, 0.8 Hz) to H-8 (δH 6.78, dd, J = 16.2, 7.0 Hz) and from H-8 to H-9 (δH 4.42, dd, J = 7.0, 0.8 Hz) and HMBC cross-peaks from H-8 to the carboxylic carbon C-10 established the fragment C-7-C-8-C-9-C-10, which was attached to the acetophenone moiety through C-6-C-7 linkage on the basis of the HMBC correlations from H-8 to C-6 and from H-7 to C-1 and C-5. 3JC-H diagnostic correlations from the oxygenated methyl protons at δH 3.37 to C-9 and from the oxygenated methyl protons at δH 3.70 to C-10 demonstrated that the methoxyl groups should be located at C-9 and C-10, respectively. Thus, the structure of compound 3 was elucidated as shown in Figure 1. However, the absolute configuration of C-9 has not been determined.
In the primary screen for antibacterial compounds, compounds 1 and 2 were tested against four Gram-positive bacteria, Staphylococcus aureus CMCC(B) 26003, Streptococcus pyogenes ATCC19615, Bacillus subtilis CICC 10283 and Micrococcus luteus. As summarized in Table 2, Both of these two compounds showed weak activity against the tested bacteria, Staphylococcus aureus CMCC(B) 26003, Streptococcus pyogenes ATCC19615, Bacillus subtilis CICC 10283 and Micrococcus luteus, with the MIC values higher than 32.0 µM.

Table 2.
Antibacterial activities of compounds 1 and 2 (MIC, μM).
3. Materials and Methods
3.1. General Experimental Procedures
The HR-ESI-MS data were obtained on an Agilent 6210 time of flight LC-MS instrument (Agilent Technologies Inc., Palo Alto, CA, USA). NMR experiments were conducted on a Bruker DPX-400 NMR spectrometer (400 MHz for 1H NMR and 100 MHz for 13C NMR) or Bruker DRX-600 spectrometer (600 MHz for 1H NMR and 150 MHz for 13C NMR) (Bruker Corporation, Karlsruhe, Germany). The chemical shifts were given in δ (ppm) and referenced to the solvent signal (DMSO-d6, δH 2.50, δC 39.5; acetone-d6, δH 2.05, δC 29.8). Column chromatography (CC) was accomplished on silica gel (200–300 mesh, Qingdao Marine Chemical Inc., Qingdao, China), ODS (40–70 µm, Merck Company, Darmstadt, Germany) and Sephadex LH-20 (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). Semi-preparative reverse-phase (RP) HPLC was performed on a Hitachi HPLC system with a L-7110 pump, a L-7420 UV/vis detector and an Hypersil RP-C18 column (5 µm, 250 × 10.0 mm, Thermo Fisher Scientific, Waltham, MA, USA). Thin-layer chromatography (TLC) was conducted on silica gel GF254 (10–20 µm, Qingdao Marine Chemical Inc., Qingdao, China). All chemicals used were of HPLC grade or analytical grade.
3.2. Strain Isolation and Cultivation
The fungus Aspergillus sp. YHZ-1 was isolated by one of the authors (Y.-Q.Z.) from an unidentified mangrove plant from Hainan Island, China, in October 2015. The isolate was identified as Aspergillus sp. by its morphological characteristics and the voucher specimen (IFB-YHZ-1) was deposited in the Institute of Functional Biomolecules, State Key Laboratory of Pharmaceutical Biotechnology, Nanjing University. The strain was cultivated on MEA agar plate (consisting of 20 g/L malt extract, 20 g/L sucrose, 1 g/L peptone, 20 g/L agar and deionized water) at 30 °C for 5 days. Then small agar plugs with mycelia were inoculated into five 1 L-Erlenmeyer flasks, each containing 400 mL MEA liquid medium, which were cultivated at 28 °C with 108 rpm/min. After 3 days of fermentation, 10 mL of the seed cultures were inoculated into 200 flasks with 250 mL ME liquid medium and fermented on a rotary shaker with 120 rpm/min at 28 °C for 14 days.
3.3. Extraction and Isolation
The entire filtrate of the fermented broth (50 L) was harvested and extracted three times with an equivalent volume of ethyl acetate at room temperature. The organic solvent was evaporated in vacuo to yield 25 g of crude extract. Then the crude extract was fractionated by silica gel (200–300 mesh) CC eluting with a gradient of CH2Cl2-MeOH mixtures (v/v, 100:0, 100:1, 100:2, 100:4, 100:8, 100:16, 100:32, 0:100) to give 8 fractions (Fr.1–8). Fr.3 (CH2Cl2-MeOH, v/v, 100:2) was subsequently subjected to ODS CC with MeOH-H2O (v/v, 30:70, 40:60, 50:50, 60:40, 70:30, 100:0) to afford 6 sub fractions (Fr.3.1–Fr.3.6). Finally, compounds 1 (12.5 mg), 2 (11.2 mg), 3 (6.3 mg) were purified from Fr.3.4 and Fr.3.5 by semi-preparative reverse-phase HPLC (2 mL/min, detector UV λmax 254 nm, CH3CN-H2O 55:45).
3.4. Crystal Data of 1
The single crystal X-ray diffraction data of compound 1 was collected on a Bruker APEX-II diffractometer at 130 K with Cu Kα radiation (λ = 1.54178 Å). The structure was solved using the program SHELXS-97 and refined by full-matrix least-squares on F2. Crystal data of compound 1 have been deposited with the Cambridge Crystallographic Data Centre (deposition No. CCDC 1584390), which can be obtained free of charge via www.ccdc.cam.ac.uk/data_request/cif.
Crystal data for 1: molecular formula C17H16O7, Mr = 332.30, monoclinic crystals, a = 13.2620 (3) Å, b = 16.2202 (4) Å, c = 7.0447 (2) Å, α = 90.00°, β = 97.7510° (10), γ = 90.00°, Z = 4, μ = 0.977 mm−1, F (000) = 696 and T = 130 K; Crystal dimensions: 0.15 × 0.1 × 0.05 mm3, Volume = 1501.56 (7) Å3, 10,794 reflections measured, 2783 independent reflections (Rint = 0.0472), the final R indices [I > 2σ(I)] R1 = 0.0469, wR2 = 0.1286, R indices (all data) R1 = 0.0500, wR2 = 0.1330. The goodness of fit on F2 was 1.026.
3.5. Crystal Data of 2
The single crystal data of compound 2 was collected on a Bruker APEX-II diffractometer at 130 K with Cu Kα radiation (λ = 1.54178 Å). The structure was solved using the program SHELXS-97 and refined by full-matrix least-squares on F2. Crystal data of compound 2 have been deposited with the Cambridge Crystallographic Data Centre (deposition No. CCDC 1584387), which can be obtained free of charge via www.ccdc.cam.ac.uk/data_request/cif.
Crystal data for 2: molecular formula C16H14O6, Mr = 302.27, monoclinic crystals, a = 11.4611 (4) Å, b = 4.9487 (2) Å, c = 13.6864 (5) Å, α = 90.00°, β = 111.933° (2), γ = 90.00°, Z = 2, μ = 0.909 mm−1, F (000) = 316 and T = 130 K; Crystal dimensions: 0.22 × 0.2 × 0.15 mm3, Volume = 720.07 (5) Å3, 4697 reflections measured, 1843 independent reflections (Rint = 0.0350), the final R indices [I > 2σ(I)] R1 = 0.0385, wR2 = 0.1115, R indices (all data) R1 = 0.0389, wR2 = 0.1119. The goodness of fit on F2 was 1.095.
3.6. Antibacterial Activity Assay
The in vitro antibacterial activity of compounds 1 and 2 were evaluated against four bacteria including Staphylococcus aureus CMCC(B) 26003, Streptococcus pyogenes ATCC19615, Bacillus subtilis CICC 10283 and Micrococcus luteus in accordance with previously reported methods [14,15]. In the assays, the medium used in the test was Müller-Hinton (MH) broth. All test compounds and the positive control ampicillin were dissolved in dimethyl sulfoxide (DMSO). The minimum inhibitory concentration (MIC) values were determined in the 96-well plates (triplicate) and determined as the lowest sample concentration exhibiting no bacterial growth.
4. Conclusions
Three new phenone derivatives, asperphenone A–C (1–3), were isolated from the ethyl acetate extract of the fermentation broth of the mangrove-derived fungus Aspergillus sp. YHZ-1. The chemical structures of these compounds were elucidated on the basis of HR-ESI-MS, 1D and 2D NMR spectroscopic analysis, as well as X-ray crystallographic data. Both of the tested compounds 1 and 2 displayed weak antibacterial activity against four Gram-positive bacteria, Staphylococcus aureus CMCC(B) 26003, Streptococcus pyogenes ATCC19615, Bacillus subtilis CICC 10283 and Micrococcus luteus, indicating that the mangrove-associated fungi are still a rich source for discovering diverse new bioactive natural products which could be used as lead compounds in drug development.
Supplementary Materials
The 1D and 2D NMR spectra for compounds 1–3 are available online at http://www.mdpi.com/1660-3397/16/2/45/s1.
Acknowledgments
This work was financially supported by the National Natural Science Fund of China (Nos. 81673333, 81522042, 21572100, 81421091, 81500059, 41406083, 21672101 and 21661140001), SBK2015022152 and Jiangsu Provincial Key Medical Discipline (laboratory) (No. ZDXKA2016020).
Author Contributions
Rui-Hua Jiao and Hui-Ming Ge conceived and designed the experiments; Yi-Qin Zhou and Zhi-Kai Guo cultured, isolated and identified the compounds; Xin-Zhao Deng performed the biological tests; Zhi-Kai Guo, Yi-Qin Zhou, Hao Han, Wen Wang and Lang Xiang analyzed the data; Zhi-Kai Guo, Hui-Ming Ge and Rui-Hua Jiao wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gomes, N.G.M.; Lefranc, F.; Kijjoa, A.; Kiss, R. Can some marine-derived fungal metabolites become actual anticancer agents? Mar. Drugs 2015, 13, 3950–3991. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.J.; Meng, W.; Cao, C.; Wang, J.; Shan, W.J.; Wang, Q.G. Antibacterial and antifungal compounds from marine fungi. Mar. Drugs 2015, 13, 3479–3513. [Google Scholar] [CrossRef] [PubMed]
- Moghadamtousi, S.Z.; Nikzad, S.; Kadir, H.A.; Abubakar, S.; Zandi, K. Potential antiviral agents from marine fungi: An overview. Mar. Drugs 2015, 13, 4520–4538. [Google Scholar] [CrossRef] [PubMed]
- Han, W.B.; Lu, Y.H.; Zhang, A.H.; Zhang, G.F.; Mei, Y.N.; Jiang, N.; Lei, X.; Song, Y.C.; Ng, S.W.; Tan, R.X. Curvulamine, a new antibacterial alkaloid incorporating two undescribed units from a Curvularia species. Org. Lett. 2014, 16, 5366–5369. [Google Scholar] [CrossRef] [PubMed]
- Han, W.B.; Zhang, A.H.; Deng, X.Z.; Lei, X.; Tan, RX. Curindolizine, an anti-inflammatory agent assembled via Michael addition of pyrrole alkaloids inside fungal cells. Org. Lett. 2016, 18, 1816–1819. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Xu, G.M.; Li, X.M.; Li, C.S.; Wang, B.G. Characterization of a newly isolated marine fungus Aspergillus dimorphicus for optimized production of the anti-tumor agent Wentilactones. Mar. Drugs 2015, 13, 7040–7054. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.J.; Shao, C.L.; Wu, L.Y.; Chen, M.; Wang, K.L.; Zhao, D.L.; Sun, X.P.; Chen, G.Y.; Wang, C.Y. Bioactive phenylalanine derivatives and cytochalasins from the soft coral-derived fungus, Aspergillus elegans. Mar. Drugs 2013, 11, 2054–2068. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2017, 34, 235–294. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.K.; Gai, C.J.; Cai, C.H.; Chen, L.L.; Liu, S.B.; Zeng, Y.B.; Yuan, J.Z.; Mei, W.L.; Dai, H.F. Metabolites with insecticidal activity from Aspergillus fumigatus JRJ111048 isolated from mangrove plant Acrostichum specioum endemic to Hainan island. Mar. Drugs 2017, 15, 381. [Google Scholar] [CrossRef] [PubMed]
- Zang, L.Y.; Wei, W.; Guo, Y.; Wang, T.; Jiao, R.H.; Ng, S.W.; Tan, R.X.; Ge, H.M. Sesquiterpenoids from the mangrove-derived endophytic fungus Diaporthe sp. J. Nat. Prod. 2012, 75, 1744–1749. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.C.; Ge, H.M.; Zang, L.Y.; Bei, Y.C.; Niu, Z.Y.; Wei, W.; Feng, X.J.; Ding, S.; Ng, S.W.; Shen, P.P.; et al. Diaporine, a novel endophyte-derived regulator of macrophage differentiation. Org. Biomol. Chem. 2014, 12, 6545–6548. [Google Scholar] [CrossRef] [PubMed]
- Wuringege; Guo, Z.K.; Wei, W.; Jiao, R.H.; Yan, T.; Zang, L.Y.; Jiang, R.; Tan, R.X.; Ge, H.M. Polyketides from the plant endophytic fungus Cladosporium sp. IFB3lp-2. J. Asian Nat. Prod. Res. 2013, 15, 928–933. [Google Scholar]
- Ma, S.Y.; Xiao, Y.S.; Zhang, B.; Shao, F.L.; Guo, Z.K.; Zhang, J.J.; Jiao, R.H.; Sun, Y.; Xu, Q.; Tan, R.X.; et al. Amycolamycins A and B, two enediyne-derived compounds from a locust-associated actinomycete. Org. Lett. 2017, 19, 6208–6211. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.S.; Zhang, B.; Zhang, M.; Guo, Z.K.; Deng, X.Z.; Shi, J.; Li, W.; Jiao, R.H.; Tan, R.X.; Ge, H.M. Rifamorpholines A-E, potential antibiotics from a locust-associated actinobacteria Amycolatopsis sp. Hca4. Org. Biomol. Chem. 2017, 15, 3909–3916. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).