Effect of Marine Microalga Chlorella pyrenoidosa Ethanol Extract on Lipid Metabolism and Gut Microbiota Composition in High-Fat Diet-Fed Rats
Abstract
1. Introduction
2. Results
2.1. Compound Detection Using UPLC-Q-TOF-MS/MS
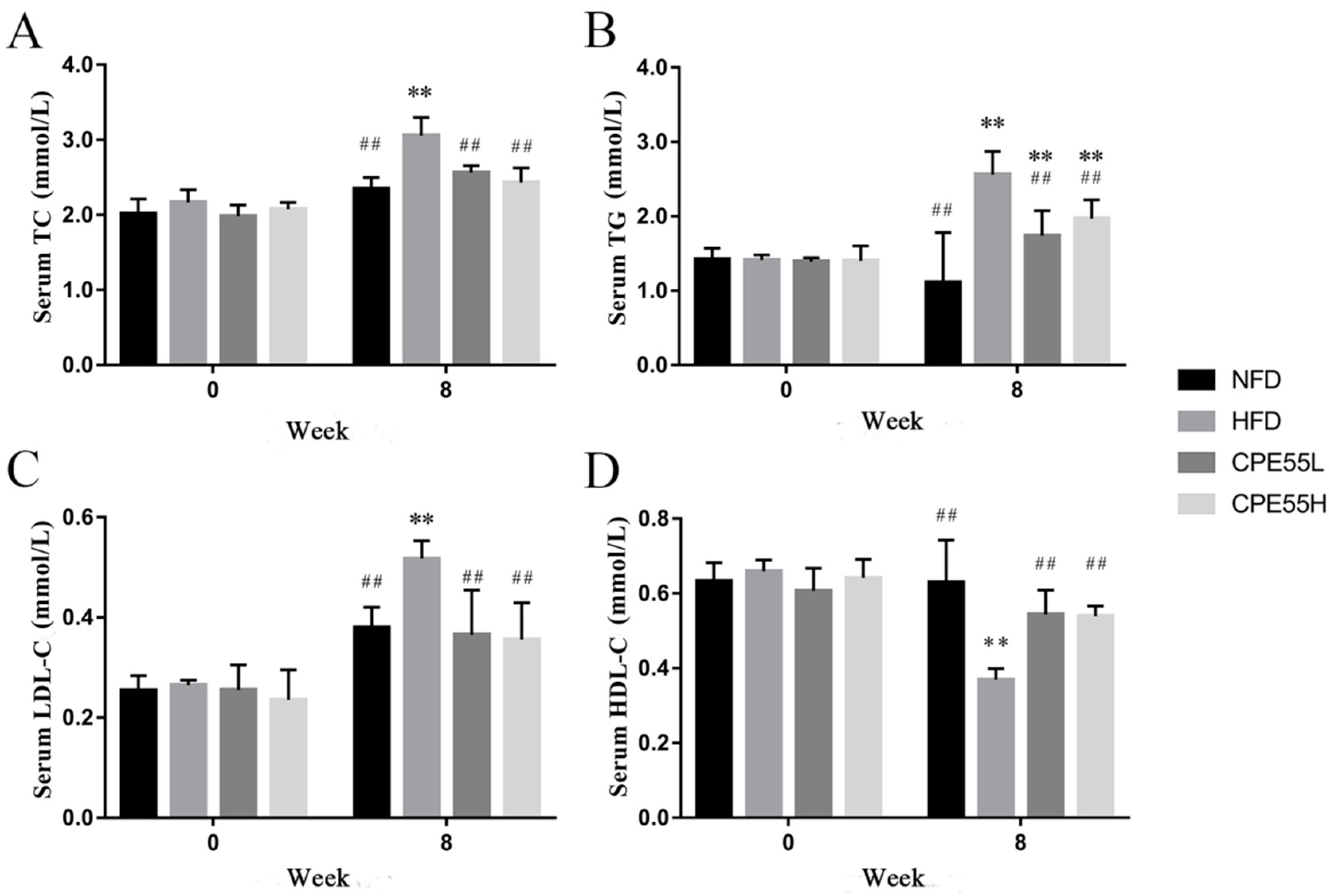
2.2. Effect of CPE55 on Body Weight and Serum Lipids Parameters of Hyperlipidemic Rats
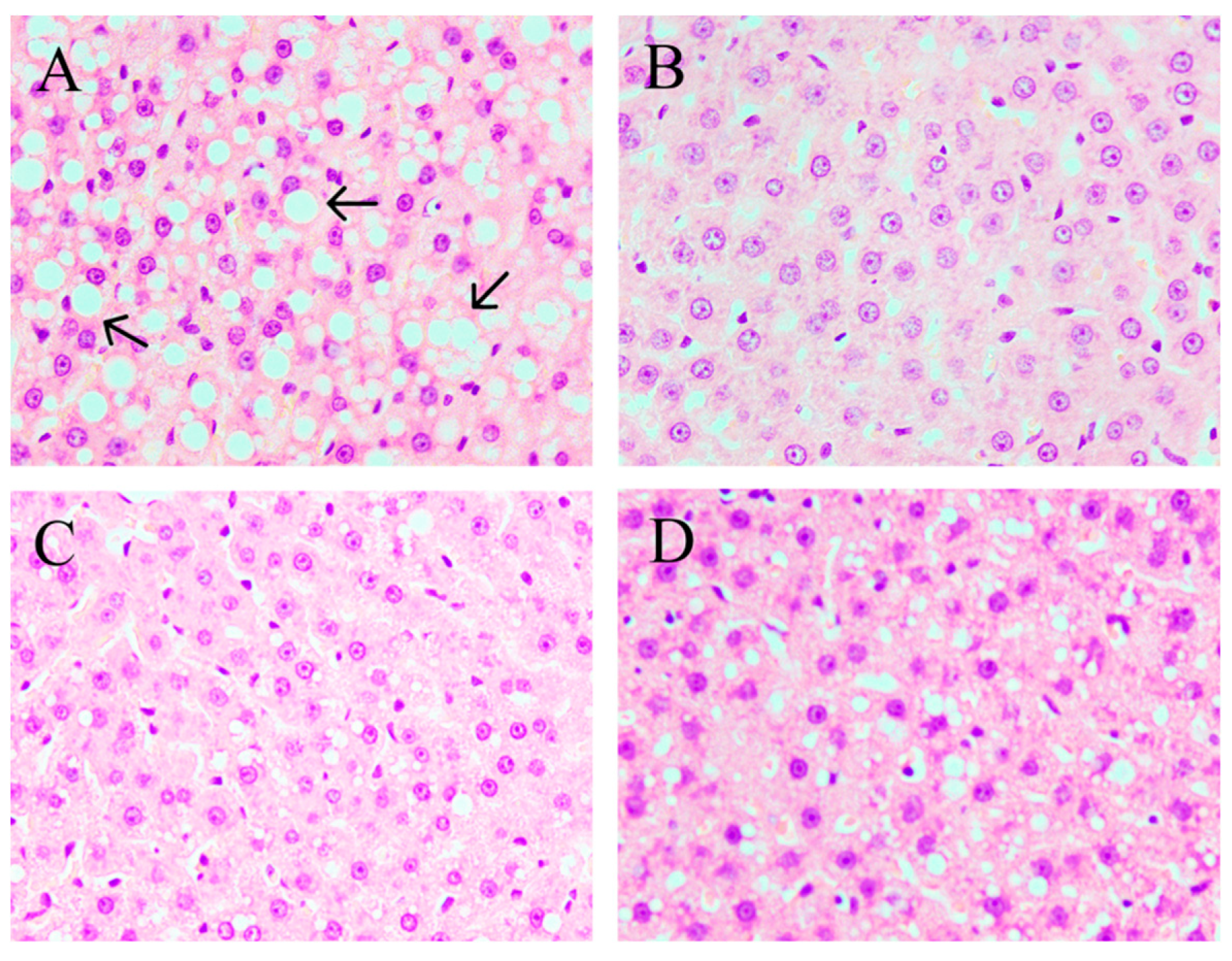
2.3. CPE55 Attenuates HFD-Induced Lipid Steatosis
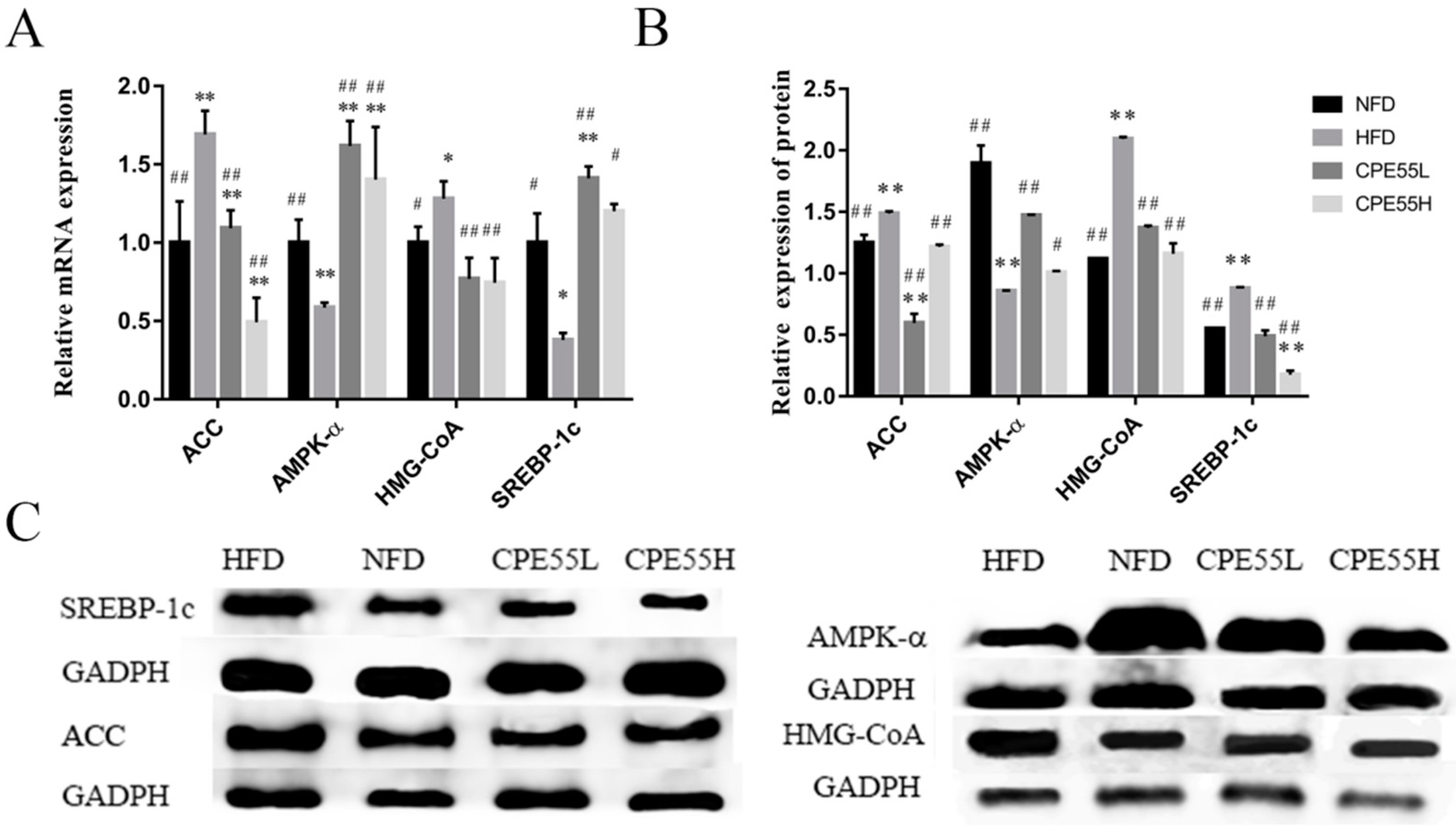
2.4. Effect of CPE55 on Liver Genes and Protein Expressions
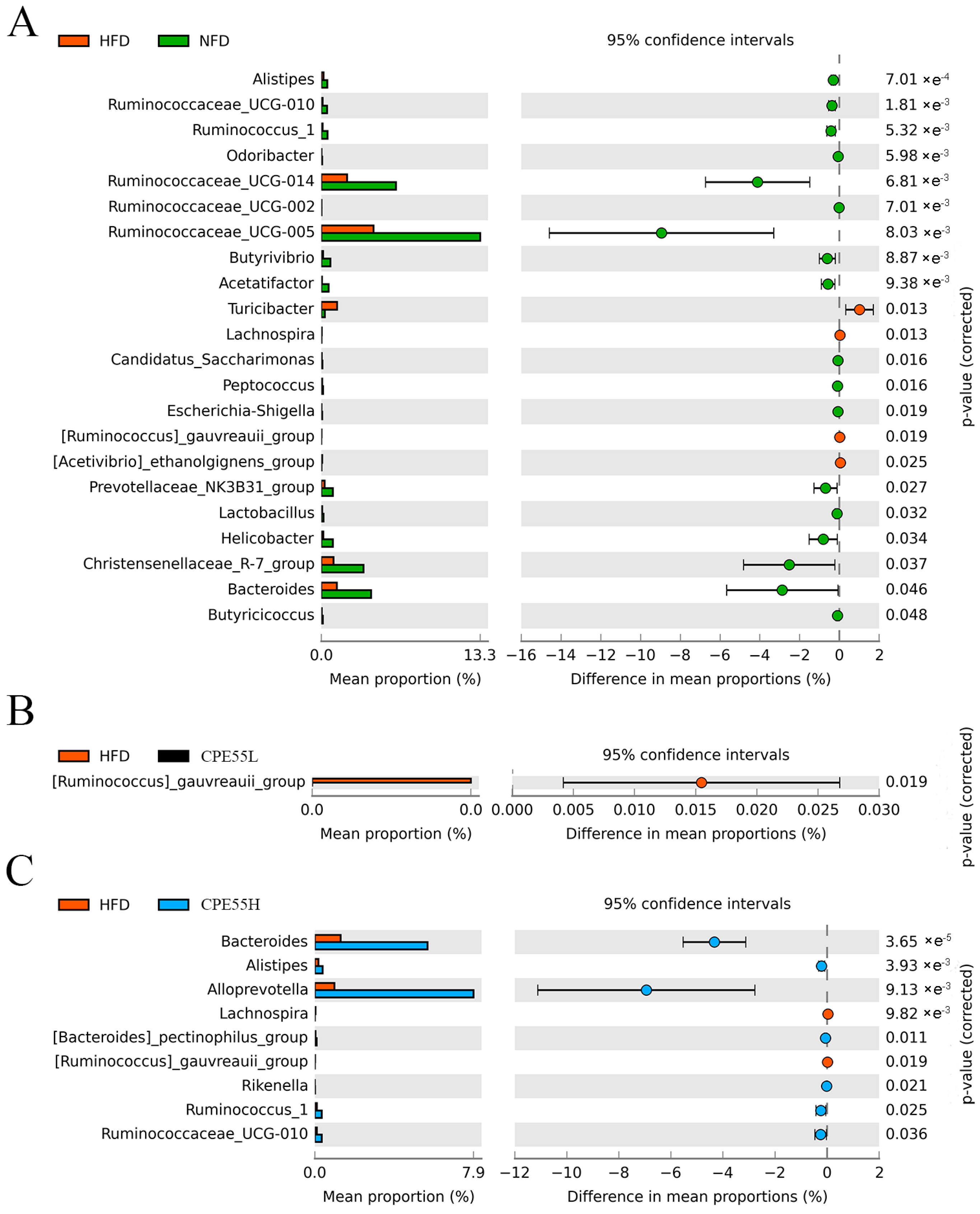
2.5. CPE55 Regulates Caecum Microbiota Composition on Hyperlipidemic Rats
2.6. Effect of CPE55 on Total Bile Acid in Faecal
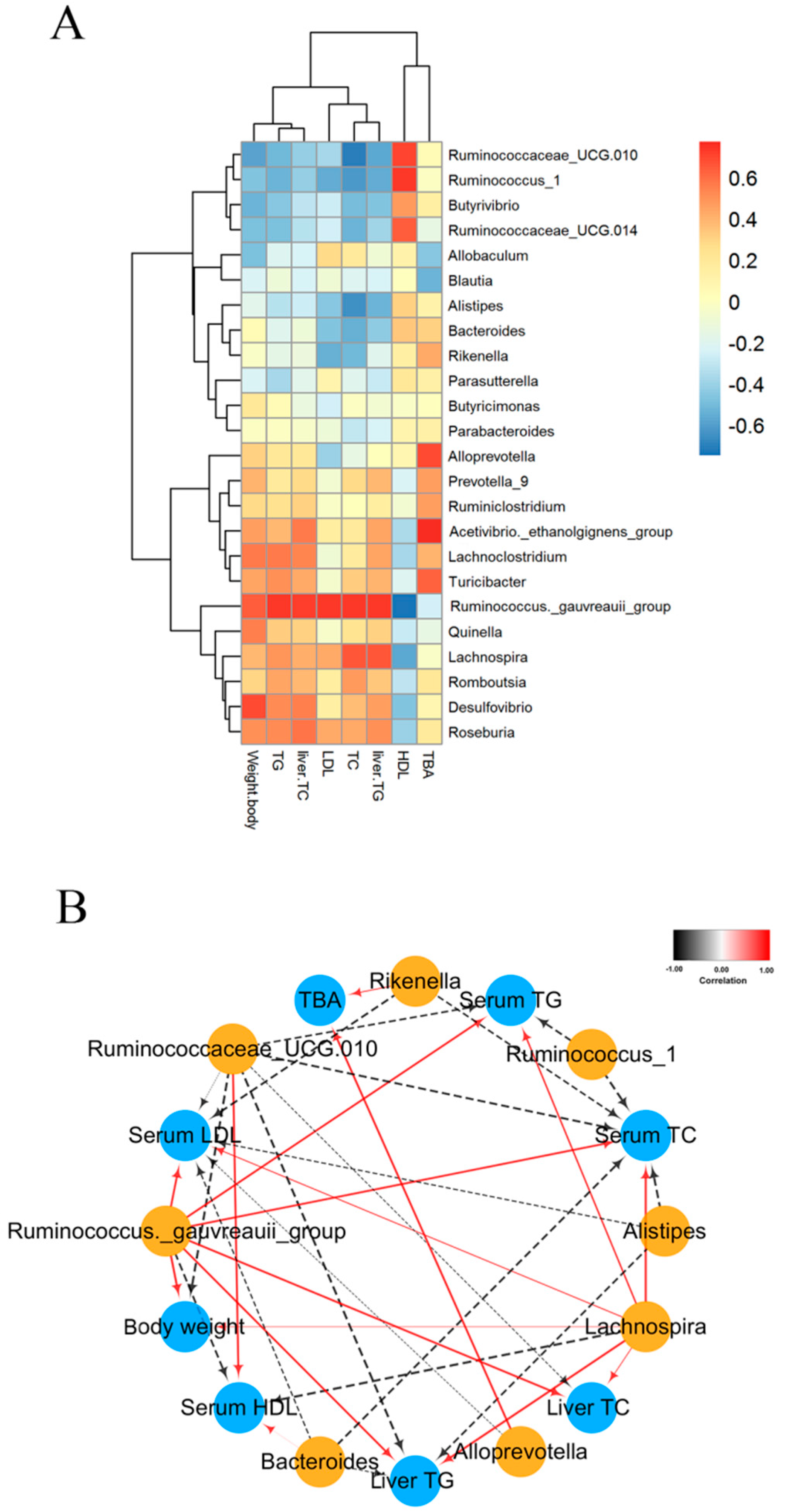
2.7. Correlation between Biological Indicators and Cecal Microbiota
3. Discussion
4. Materials and methods
4.1. Preparation of C. pyrenoidosa Extract and UPLC-QTOF-MS/MS Analysis
4.2. Animals
4.3. Sample Collection
4.4. Serum and Liver Biochemical Index Analysis
4.5. Hepati Histopathology Analysis
4.6. RT-qPCR Analysis
4.7. Western Blot Analysis
4.8. Extraction of DNA from Cecal Samples and 16S rRNA Sequencing
4.9. Bioinformatics Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sarti, C.; Gallagher, J. The metabolic syndrome: Prevalence, CHD risk, and treatment. J. Diabetes Complicat. 2006, 20, 121–132. [Google Scholar] [CrossRef]
- Tds, A.; Aubertinleheudre, M.; Carvalho, L.P.; Máximo, R.O.; Corona, L.P.; Trp, B.; Nunes, D.P.; Jlf, S.; Yao, D.; Lebrão, M.L. Dynapenic obesity as an associated factor to lipid and glucose metabolism disorders and metabolic syndrome in older adults—Findings from SABE study. Clin. Nutr. 2017, 37, 1360–1366. [Google Scholar]
- Kong, X.; Gao, Y.; Geng, X.; Xie, G.; Hao, S.; Li, Y.; Zhang, Z. Effect of lipid lowering tablet on blood lipid in hyperlipidemia model rats. Saudi J. Biol. Sci. 2018, 25, 715–718. [Google Scholar] [CrossRef] [PubMed]
- Gera, T.; Sachdev, H.P.; Nestel, P. Effect of iron supplementation on physical performance in children and adolescents: Systematic review of randomized controlled trials. Indian Pediatr. 2007, 44, 15–24. [Google Scholar] [PubMed]
- Sugimoto, T.; Sato, M.; Dehle, F.C.; Brnabic, A.J.M.; Weston, A.; Burge, R. Lifestyle-related metabolic disorders, osteoporosis, and fracture risk in Asia: A systematic review. Value Health Reg. Issues 2016, 9, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Licata, A.; Giammanco, A.; Minissale, M.G.; Pagano, S.; Petta, S.; Averna, M. Liver and statins: A critical appraisal of the evidence. Curr. Med. Chem. 2018. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Wu, Y.J.; Yang, C.F.; Liu, B.; Huang, Y.F. Hypotensive, hypoglycemic and hypolipidemic effects of bioactive compounds from microalgae and marine microorganisms. Int. J. Food Sci. Technol. 2015, 50, 1705–1717. [Google Scholar] [CrossRef]
- Chen, Y.X.; Liu, X.Y.; Wu, L.X.; Tong, A.J.; Zhao, L.N.; Liu, B.; Zhao, C. Physicochemical characterization of polysaccharides from Chlorella pyrenoidosa and its anti-ageing effects in Drosophila melanogaster. Carbohydr. Polym. 2018, 185, 120–126. [Google Scholar] [CrossRef]
- Safi, C.; Zebib, B.; Merah, O.; Pontalier, P.Y.; Vaca-Garcia, C. Morphology, composition, production, processing and applications of Chlorella vulgaris: A review. Renew. Sustain. Energy Rev. 2014, 35, 265–278. [Google Scholar] [CrossRef]
- Renju, G.L.; Muraleedhara, K.G.; Bandugula, V.R. Effect of lycopene isolated from Chlorella marina on proliferation and apoptosis in human prostate cancer cell line PC-3. Tumour Biol. 2014, 35, 10747–10758. [Google Scholar] [CrossRef]
- Jia, Q.; Sang, M.K. Effects of the molecular weight and protein and sulfate content of Chlorella ellipsoidea polysaccharides on their immunomodulatory activity. Int. J. Biol. Macromol. 2017, 107, 70–77. [Google Scholar]
- Da, S.F.V.; Conzferreira, M.E.; Lima, L.M.; Frasés, S.; De, S.W.; Sant’Anna, C. Green production of microalgae-based silver chloride nanoparticles with antimicrobial activity against pathogenic bacteria. Enzym. Microb. Technol. 2016, 97, 114–121. [Google Scholar]
- Sibi, G.; Rabina, S. Inhibition of pro-inflammatory mediators and cytokines by Chlorella vulgaris extracts. Pharmacogn. Res. 2016, 8, 118–122. [Google Scholar] [CrossRef]
- Bae, M.J.; Shin, H.S.; Chai, O.H.; Han, J.G.; Shon, D.H. Inhibitory effect of unicellular green algae (Chlorella vulgaris) water extract on allergic immune response. J. Sci. Food Agric. 2013, 93, 3133–3136. [Google Scholar] [CrossRef] [PubMed]
- An, B.K.; Kim, K.E.; Jeon, J.Y.; Lee, K.W. Effect of dried Chlorella vulgaris and Chlorella growth factor on growth performance, meat qualities and humoral immune responses in broiler chickens. Springerplus 2016, 5, 718. [Google Scholar] [CrossRef] [PubMed]
- Fallah, A.A.; Sarmast, E.; Habibian, S.D.; Engardeh, J.; Mahmoodnia, L.; Khaledifar, A.; Jafari, T. Effect of Chlorella supplementation on cardiovascular risk factors: A meta-analysis of randomized controlled trials. Clin. Nutr. 2018, 37, 1892–1901. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, N.; Konishi, F.; Kumamoto, S.; Maruyama, I.; Ando, Y.; Yanagita, T. Beneficial effects of Chlorella on glucose and lipid metabolism in obese rodents on a high-fat diet. Obes. Res. Clin. Pract. 2013, 7, E95–E105. [Google Scholar] [CrossRef] [PubMed]
- Craig, P.M.; Moyes, C.D.; Lemoine, C. Sensing and responding to energetic stress: Evolution of the AMPK network. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2017, 224, 156–159. [Google Scholar] [CrossRef]
- Raghow, R.; Yellaturu, C.; Deng, X.; Park, E.A.; Elam, M.B. SREBPs: The crossroads of physiological and pathological lipid homeostasis. Trends Endocrinol. Metab. 2008, 19, 65–73. [Google Scholar] [CrossRef]
- Silambarasan, T.; Manivannan, J.; Raja, B.; Chatterjee, S. Prevention of cardiac dysfunction, kidney fibrosis and lipid metabolic alterations in l-NAME hypertensive rats by sinapic acid-Role of HMG-CoA reductase. Eur. J. Pharmacol. 2016, 777, 113–123. [Google Scholar] [CrossRef]
- Mckenney, J.M. Something important is missing in the ACC/AHA cholesterol treatment guidelines. J. Am. Pharm. Assoc. 2015, 55, 324–329. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Bibiloni, R.; Knauf, C.; Waget, A.; Neyrinck, A.M.; Delzenne, N.M.; Burcelin, R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 2008, 57, 1470–1481. [Google Scholar] [CrossRef] [PubMed]
- Norris, G.H.; Jiang, C.; Ryan, J.; Porter, C.M.; Blesso, C.N. Milk sphingomyelin improves lipid metabolism and alters gut microbiota in high fat diet-fed mice. J. Nutr. Biochem. 2016, 30, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, K.S.; Maier, T.V.; Walker, A.; Heinzmann, S.S.; Forcisi, S.; Martinez, I.; Walter, J.; Schmittkopplin, P. Challenges of metabolomics in human gut microbiota research. Int. J. Med. Microbiol. 2016, 306, 266–279. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Yang, C.; Liu, B.; Lin, L.; Sarker, S.D.; Nahar, L.; Yu, H.; Cao, H.; Xiao, J. Bioactive compounds from marine macroalgae and their hypoglycemic benefits. Trends Food Sci. Technol. 2018, 72, 1–12. [Google Scholar] [CrossRef]
- Wolf, K.J.; Lorenz, R.G. Gut microbiota and obesity. Dig. Dis. 2012, 1, 1–8. [Google Scholar] [CrossRef]
- Sato, K.; Naito, M.; Yukitake, H.; Hirakawa, H.; Shoji, M.; Mcbride, M.J.; Rhodes, R.G.; Nakayama, K. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 276–281. [Google Scholar] [CrossRef]
- Velagapudi, V.R.; Hezaveh, R.; Reigstad, C.S.; Gopalacharyulu, P.; Yetukuri, L.; Islam, S.; Felin, J.; Perkins, R.; Borén, J.; Orešič, M.; et al. The gut microbiota modulates host energy and lipid metabolism in mice. J. Lipid Res. 2010, 51, 1101–1112. [Google Scholar] [CrossRef]
- Zhao, C.; Yang, C.F.; Chen, M.J.; Lv, X.C.; Liu, B.; Yi, L.Z.; Cornara, L.; Wei, M.C.; Yang, Y.C.; Tundis, R.; et al. Regulatory efficacy of brown seaweed Lessonia nigrescens extract on the gene expression profile and intestinal microflora in type 2 diabetic mice. Mol. Nutr. Food Res. 2018, 62, 1700730. [Google Scholar] [CrossRef]
- Shinya, S.; Kazuhito, H.; Yukari, E.; Hiroo, S. Hypocholesterolemic mechanism of Chlorella: Chlorella and its indigestible fraction enhance hepatic cholesterol catabolism through up-regulation of cholesterol 7alpha-hydroxylase in rats. Biosci. Biotechnol. Biochem. 2007, 71, 916–925. [Google Scholar]
- Dyrbuś, K.; Osadnik, T.; Desperak, P.; Desperak, A.; Gąsior, M.; Banach, M. Evaluation of dyslipidaemia and the impact of hypolipidemic therapy on prognosis in high and very high risk patients through the hyperlipidaemia therapy in tERtiary Cardiological cEnTer (TERCET) registry. Pharmacol. Res. 2017, 132, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Sidebottom, A.C.; Sillah, A.; Vock, D.M.; Miedema, M.D.; Pereira, R.; Benson, G.; Lindberg, R.; Boucher, J.L.; Knickelbine, T.; Vanwormer, J.J. Assessing the impact of the heart of New Ulm Project on cardiovascular disease risk factors: A population-based program to reduce cardiovascular disease. Prev. Med. (Baltim.) 2018, 112, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Kim, J.; Cheng, J.; Ong, M.; Lao, W.G.; Jin, X.L.; Lin, Y.G.; Xiao, L.; Zhu, X.Q.; Qu, X.Q. Green tea polyphenols ameliorate non-alcoholic fatty liver disease through upregulating AMPK activation in high fat fed Zucker fatty rats. World J. Gastroenterol. 2017, 23, 3805–3814. [Google Scholar] [CrossRef] [PubMed]
- Hardie, D.G.; Ashford, M.L. AMPK: Regulating energy balance at the cellular and whole body levels. Physiology 2014, 29, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Soto-Acosta, R.; Mosso, C.; Cervantes-Salazar, M.; Puerta-Guardo, H.; Medina, F.; Favari, L.; Ludert, J.E.; Angel, R.M. Del The increase in cholesterol levels at early stages after dengue virus infection correlates with an augment in LDL particle uptake and HMG-CoA reductase activity. Virology 2013, 442, 132–147. [Google Scholar] [CrossRef]
- Hedin, C.; Cj, V.D.G.; Rogers, G.B.; Cuthbertson, L.; Mccartney, S.; Stagg, A.J.; Lindsay, J.O.; Whelan, K. Siblings of patients with Crohn’s disease exhibit a biologically relevant dysbiosis in mucosal microbial metacommunities. Gut 2016, 65, 944–953. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, Y.; Xu, J.; Xue, Z.; Zhang, M.; Pang, X.; Zhang, X.; Zhao, L. Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci. Rep. 2015, 5, 14405. [Google Scholar] [CrossRef]
- Fiorucci, S.; Distrutti, E. Bile acid-activated receptors, intestinal microbiota, and the treatment of metabolic disorders. Trends Mol. Med. 2015, 21, 702–714. [Google Scholar] [CrossRef]
- Tyrrell, K.L.; Warren, Y.A.; Citron, D.M.; Goldstein, E.J.C. Re-assessment of phenotypic identifications of Alistipes species using molecular methods. Anaerobe 2011, 17, 130–134. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Z.; Li, Y.; Liao, S.; Wu, X.; Chang, Q.; Liang, B. Cholesterol induces lipoprotein lipase expression in a tree shrew (Tupaia belangeri chinensis) model of non-alcoholic fatty liver disease. Sci. Rep. 2015, 5, 15970. [Google Scholar] [CrossRef]
- Nie, Y.; Luo, F.; Lin, Q. Dietary nutrition and gut microflora: A promising target for treating diseases. Trends Food Sci. Technol. 2018, 75, 72–80. [Google Scholar] [CrossRef]
- Huang, J.; Lin, X.; Xue, B.; Luo, J.; Gao, L.; Wang, Y.; Ou, S.; Peng, X. Impact of polyphenols combined with high-fat diet on rats’ gut microbiota. J. Funct. Foods 2016, 26, 763–771. [Google Scholar] [CrossRef]
- Gibiino, G.; Lopetuso, L.R.; Scaldaferri, F.; Rizzatti, G.; Binda, C.; Gasbarrini, A. Exploring Bacteroidetes: Metabolic key points and immunological tricks of our gut commensals. Dig. Liver Dis. 2018, 50, 635–639. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Nyman, M.; Fåk, F. Modulation of gut microbiota in rats fed high-fat diets by processing whole-grain barley to barley malt. Mol. Nutr. Food Res. 2015, 59, 2066–2076. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.R.; Zhou, W.B.; Yang, X.L.; Tong, A.J.; Hong, J.L.; Guo, W.L.; Li, T.T.; Jia, R.B.; Pan, Y.Y.; Lin, J.; et al. The regulation mechanisms of soluble starch and glycerol for production of azaphilone pigments in Monascus purpureus FAFU618 as revealed by comparative proteomic and transcriptional analyses. Food Res. Int. 2018, 106, 626–635. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.L.; Pan, Y.Y.; Li, L.; Li, T.T.; Liu, B.; Lv, X.C. Ethanol extract of Ganoderma lucidum ameliorates lipid metabolic disorders and modulates the gut microbiota composition in high-fat diet fed rats. Food Funct. 2018, 9, 3419–3431. [Google Scholar] [CrossRef] [PubMed]
- Li, T.T.; Liu, Y.Y.; Wan, X.Z.; Huang, Z.R.; Liu, B.; Zhao, C. Regulatory efficacy of the polyunsaturated fatty acids from microalgae Spirulina platensis on lipid metabolism and gut microbiota in high-fat diet rats. Int. J. Mol. Sci. 2018, 19, 3075. [Google Scholar] [CrossRef] [PubMed]
- Tuer, A.; Tokarz, D.; Prent, N.; Cisek, R.; Alami, J.; Dumont, D.J.; Bakueva, L.; Rowlands, J.; Barzda, V. Nonlinear multicontrast microscopy of hematoxylin-and-eosin-stained histological sections. J. Biomed. Opt. 2010, 15, 26018–26019. [Google Scholar] [CrossRef]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wan, X.; Li, T.; Liu, D.; Chen, Y.; Liu, Y.; Liu, B.; Zhang, H.; Zhao, C. Effect of Marine Microalga Chlorella pyrenoidosa Ethanol Extract on Lipid Metabolism and Gut Microbiota Composition in High-Fat Diet-Fed Rats. Mar. Drugs 2018, 16, 498. https://doi.org/10.3390/md16120498
Wan X, Li T, Liu D, Chen Y, Liu Y, Liu B, Zhang H, Zhao C. Effect of Marine Microalga Chlorella pyrenoidosa Ethanol Extract on Lipid Metabolism and Gut Microbiota Composition in High-Fat Diet-Fed Rats. Marine Drugs. 2018; 16(12):498. https://doi.org/10.3390/md16120498
Chicago/Turabian StyleWan, Xuzhi, Tiantian Li, Dan Liu, Yihan Chen, Yuanyuan Liu, Bin Liu, Huiying Zhang, and Chao Zhao. 2018. "Effect of Marine Microalga Chlorella pyrenoidosa Ethanol Extract on Lipid Metabolism and Gut Microbiota Composition in High-Fat Diet-Fed Rats" Marine Drugs 16, no. 12: 498. https://doi.org/10.3390/md16120498
APA StyleWan, X., Li, T., Liu, D., Chen, Y., Liu, Y., Liu, B., Zhang, H., & Zhao, C. (2018). Effect of Marine Microalga Chlorella pyrenoidosa Ethanol Extract on Lipid Metabolism and Gut Microbiota Composition in High-Fat Diet-Fed Rats. Marine Drugs, 16(12), 498. https://doi.org/10.3390/md16120498





