The Anemonia viridis Venom: Coupling Biochemical Purification and RNA-Seq for Translational Research
Abstract
1. Introduction
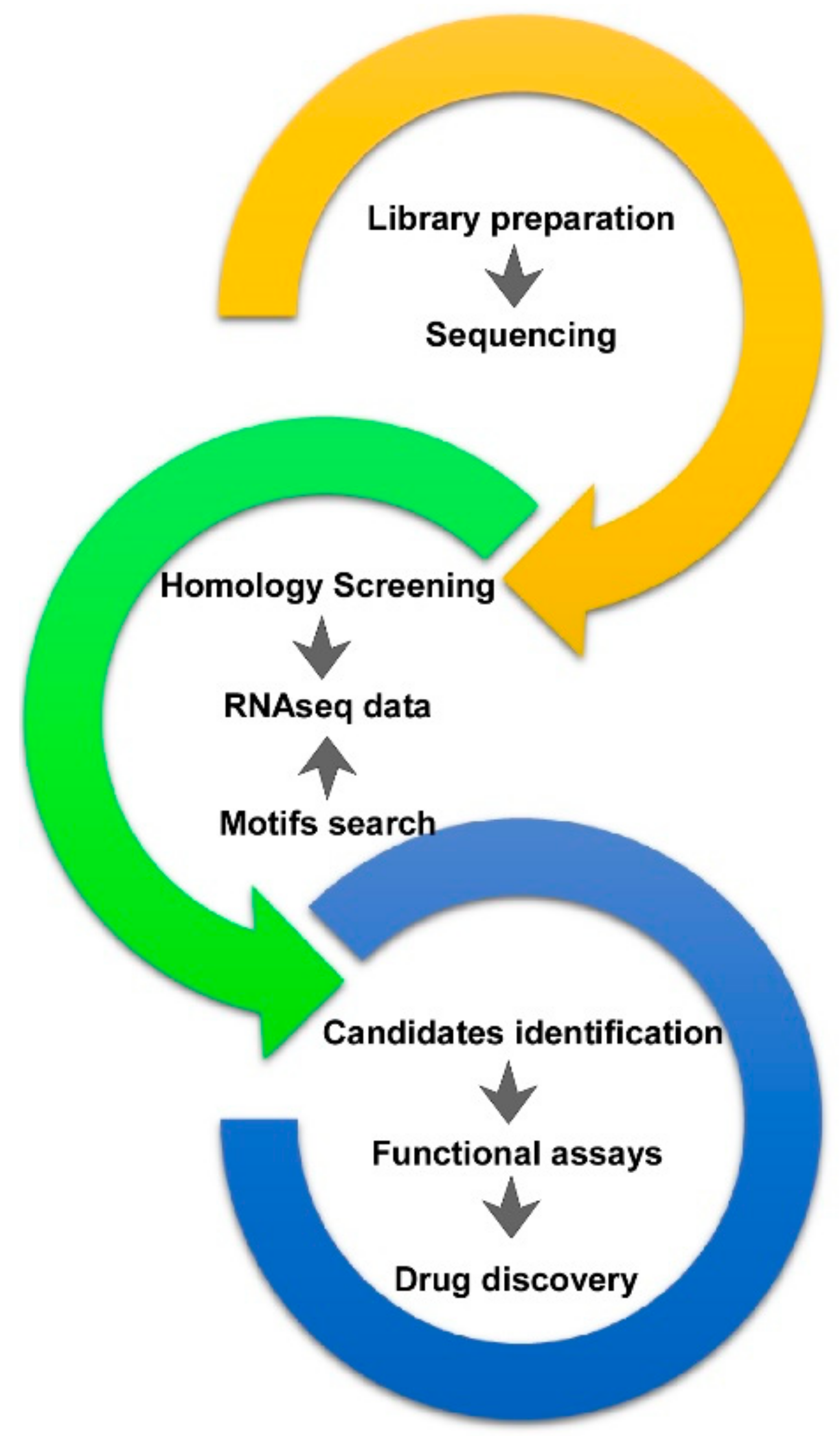
2. A. viridis RNA Datasets and Data Mining
3. The Multifaceted Molecular Arsenal of Sea Anemone A. viridis
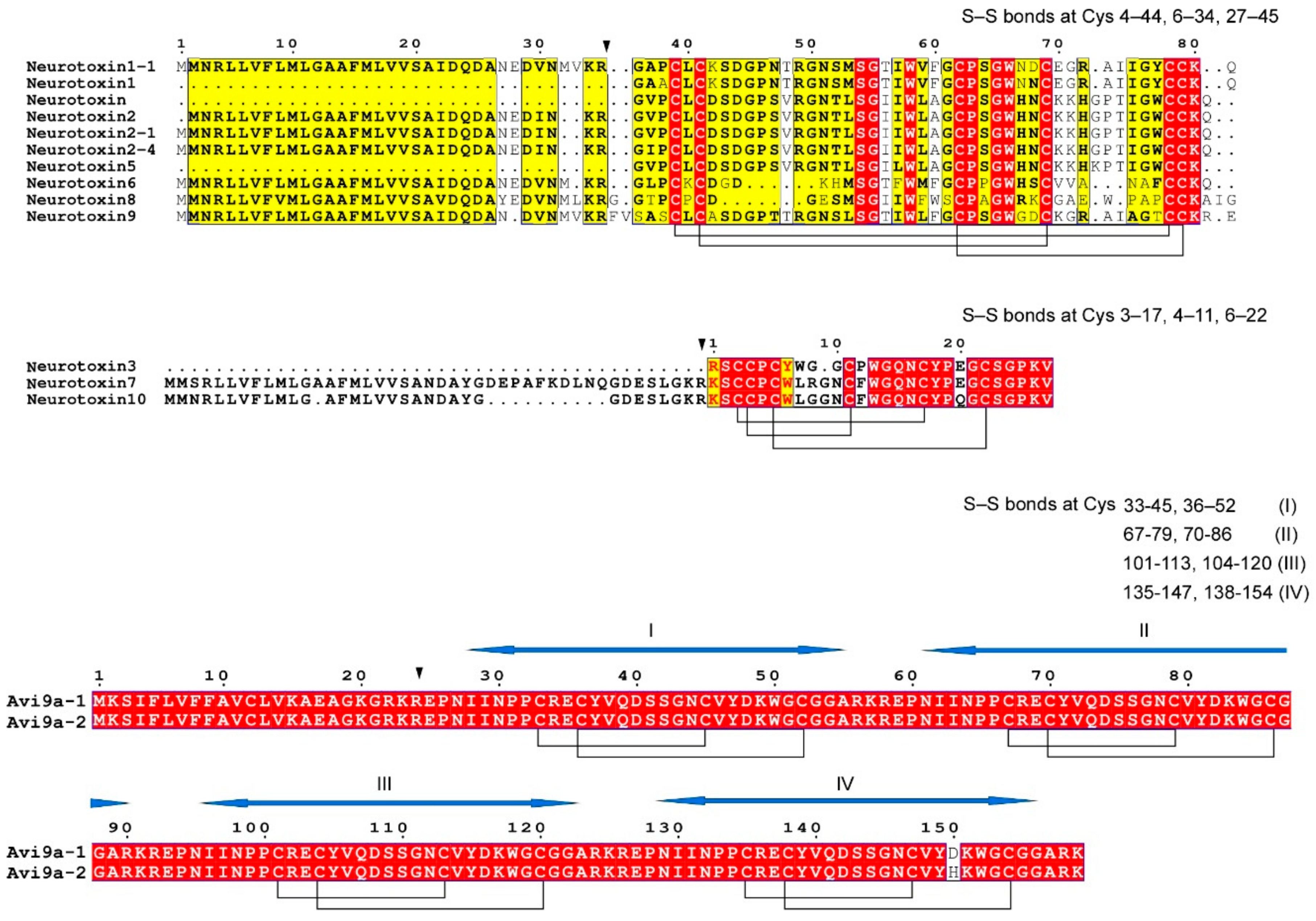
3.1. Sodium Channel Peptide Toxins
- (1)
- neurotoxin 1 (δ-actitoxin-Avd1a; ATX Ia);
- (2)
- neurotoxin 2 (δ-actitoxin-Avd1c; ATX II);
- (3)
- neurotoxin 5 (δ-actitoxin-Avd1d; ATX-V).
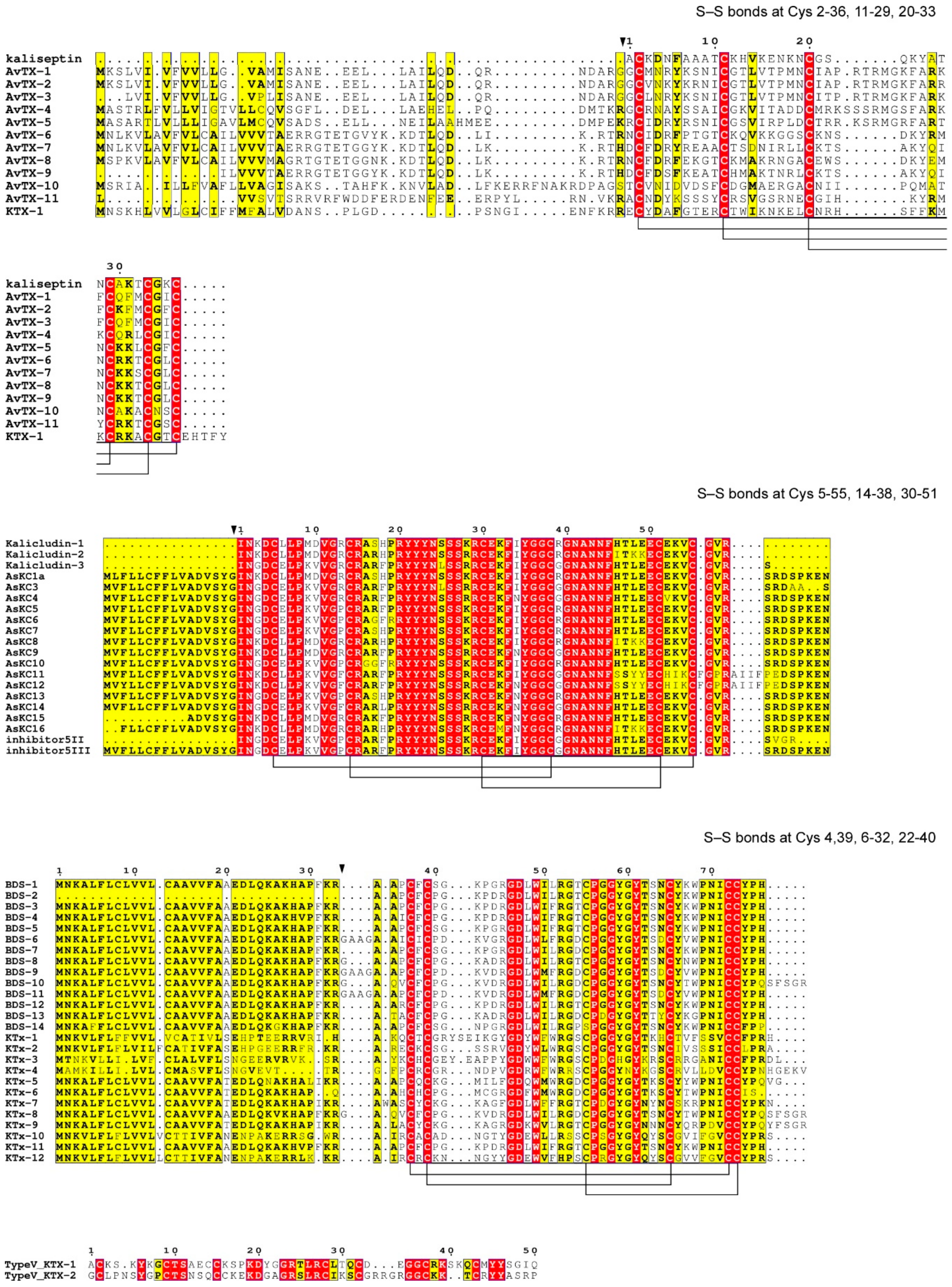
3.2. Potassium Channel Peptide Toxins
3.3. Other Candidate Toxins of A. viridis
4. From Bioprospecting to Translational Research
Author Contributions
Funding
Conflicts of Interest
References
- Molinski, T.F.; Dalisay, D.S.; Lievens, S.L.; Saludes, J.P. Drug development from marine natural products. Nat. Rev. Drug Discov. 2009, 8, 69–85. [Google Scholar] [CrossRef] [PubMed]
- Glaser, K.B.; Mayer, A.M.S. A renaissance in marine pharmacology: from preclinical curiosity to clinical reality. Biochem. Pharmacol. 2009, 78, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Rocha, J.; Peixe, L.; Gomes, N.C.M.; Calado, R. Cnidarians as a source of new marine bioactive compounds—An overview of the last decade and future steps for bioprospecting. Mar. Drugs 2011, 9, 1860–1886. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-Y.; Oliveri, P.; Gao, F.; Dornbos, S.Q.; Li, C.-W.; Bottjer, D.J.; Davidson, E.H. Precambrian animal life: probable developmental and adult cnidarian forms from Southwest China. Dev. Biol. 2002, 248, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Ruppert, E.E.; Barnes, R.D. Invertebrate Zoology, 6th ed.; Saunders College Publishing: Fort Worth, TX, USA, 1994. [Google Scholar]
- Beress, L. Biologically active compounds from coelenterates. Pure Appl. Chem. 1982, 54, 1981–1994. [Google Scholar] [CrossRef]
- Nicosia, A.; Maggio, T.; Mazzola, S.; Gianguzza, F.; Cuttitta, A.; Costa, S. Characterization of small HSPs from Anemonia viridis reveals insights into molecular evolution of alpha crystallin genes among cnidarians. PLoS ONE 2014, 9, e105908. [Google Scholar] [CrossRef] [PubMed]
- Nicosia, A.; Maggio, T.; Mazzola, S.; Cuttitta, A. Evidence of accelerated evolution and ectodermal-specific expression of presumptive BDS toxin cDNAs from Anemonia viridis. Mar. Drugs 2013, 11, 4213–4231. [Google Scholar] [CrossRef] [PubMed]
- Moran, Y.; Weinberger, H.; Sullivan, J.C.; Reitzel, A.M.; Finnerty, J.R.; Gurevitz, M. Concerted evolution of sea anemone neurotoxin genes is revealed through analysis of the Nematostella vectensis genome. Mol. Biol. Evol. 2008, 25, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Moran, Y.; Weinberger, H.; Lazarus, N.; Gur, M.; Kahn, R.; Gordon, D.; Gurevitz, M. Fusion and retrotransposition events in the evolution of the sea anemone Anemonia viridis neurotoxin genes. J. Mol. Evol. 2009, 69, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Castañeda, O.; Harvey, A.L. Discovery and characterization of cnidarian peptide toxins that affect neuronal potassium ion channels. Toxicon 2009, 54, 1119–1124. [Google Scholar] [CrossRef] [PubMed]
- Nicosia, A.; Maggio, T.; Costa, S.; Salamone, M.; Tagliavia, M.; Mazzola, S.; Gianguzza, F.; Cuttitta, A. Maintenance of a Protein Structure in the Dynamic Evolution of TIMPs over 600 Million Years. Genome Biol. Evol. 2016, 8, 1056–1071. [Google Scholar] [CrossRef] [PubMed]
- Bulati, M.; Longo, A.; Masullo, T.; Vlah, S.; Bennici, C.; Bonura, A.; Salamone, M.; Tagliavia, M.; Nicosia, A.; Mazzola, S.; et al. Partially Purified Extracts of Sea Anemone Anemonia viridis Affect the Growth and Viability of Selected Tumour Cell Lines. Biomed Res. Int. 2016, 2016, 3849897. [Google Scholar] [CrossRef] [PubMed]
- Cuttitta, A.; Ragusa, M.A.; Costa, S.; Bennici, C.; Colombo, P.; Mazzola, S.; Gianguzza, F.; Nicosia, A. Evolutionary conserved mechanisms pervade structure and transcriptional modulation of allograft inflammatory factor-1 from sea anemone Anemonia viridis. Fish Shellfish Immunol. 2017, 67, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Nicosia, A.; Bennici, C.; Biondo, G.; Costa, S.; Di Natale, M.; Masullo, T.; Monastero, C.; Ragusa, M.A.; Tagliavia, M.; Cuttitta, A. Characterization of Translationally Controlled Tumour Protein from the Sea Anemone Anemonia viridis and Transcriptome Wide Identification of Cnidarian Homologues. Genes 2018, 9, 30. [Google Scholar] [CrossRef] [PubMed]
- Frazão, B.; Vasconcelos, V.; Antunes, A. Sea anemone (Cnidaria, Anthozoa, Actiniaria) toxins: An overview. Mar. Drugs 2012, 10, 1812–1851. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, S.; Grishin, E. The mining of toxin-like polypeptides from EST database by single residue distribution analysis. BMC Genom. 2011, 12, 88. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, S.; Grishin, E. Convenient nomenclature of cysteine-rich polypeptide toxins from sea anemones. Peptides 2012, 33, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Macrander, J.; Broe, M.; Daly, M. Tissue-Specific Venom Composition and Differential Gene Expression in Sea Anemones. Genome Biol. Evol. 2016, 8, 2358–2375. [Google Scholar] [CrossRef] [PubMed]
- Sabourault, C.; Ganot, P.; Deleury, E.; Allemand, D.; Furla, P. Comprehensive EST analysis of the symbiotic sea anemone, Anemonia viridis. BMC Genom. 2009, 10, 333. [Google Scholar] [CrossRef] [PubMed]
- Urbarova, I.; Patel, H.; Forêt, S.; Karlsen, B.O.; Jørgensen, T.E.; Hall-Spencer, J.M.; Johansen, S.D. Elucidating the Small Regulatory RNA Repertoire of the Sea Anemone Anemonia viridis Based on Whole Genome and Small RNA Sequencing. Genome Biol. Evol. 2018, 10, 410–426. [Google Scholar] [CrossRef] [PubMed]
- Martens, L.; Hermjakob, H.; Jones, P.; Adamski, M.; Taylor, C.; States, D.; Gevaert, K.; Vandekerckhove, J.; Apweiler, R. PRIDE: The proteomics identifications database. Proteomics 2005, 5, 3537–3545. [Google Scholar] [CrossRef] [PubMed]
- Farrah, T.; Deutsch, E.W.; Kreisberg, R.; Sun, Z.; Campbell, D.S.; Mendoza, L.; Kusebauch, U.; Brusniak, M.-Y.; Hüttenhain, R.; Schiess, R.; et al. PASSEL: The PeptideAtlas SRMexperiment library. Proteomics 2012, 12, 1170–1175. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, M.; Schlegl, J.; Hahne, H.; Gholami, A.M.; Lieberenz, M.; Savitski, M.M.; Ziegler, E.; Butzmann, L.; Gessulat, S.; Marx, H.; et al. Mass-spectrometry-based draft of the human proteome. Nature 2014, 509, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Martens, L.; Vizcaíno, J.A. A Golden Age for Working with Public Proteomics Data. Trends Biochem. Sci. 2017, 42, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Salamone, M.; Nicosia, A.; Bennici, C.; Quatrini, P.; Catania, V.; Mazzola, S.; Ghersi, G.; Cuttitta, A. Comprehensive Analysis of a Vibrio parahaemolyticus Strain Extracellular Serine Protease VpSP37. PLoS ONE 2015, 10, e0126349. [Google Scholar] [CrossRef] [PubMed]
- Himaya, S.W.A.; Lewis, R.J. Venomics-Accelerated Cone Snail Venom Peptide Discovery. Int. J. Mol. Sci. 2018, 19, 788. [Google Scholar] [CrossRef] [PubMed]
- Salgado, V.L.; Kem, W.R. Actions of three structurally distinct sea anemone toxins on crustacean and insect sodium channels. Toxicon 1992, 30, 1365–1381. [Google Scholar] [CrossRef]
- Norton, R.S. Structure and structure-function relationships of sea anemone proteins that interact with the sodium channel. Toxicon 1991, 29, 1051–1084. [Google Scholar] [CrossRef]
- Molgó, J.; Mallart, A. Effects of Anemonia sulcata toxin II on presynaptic currents and evoked transmitter release at neuromuscular junctions of the mouse. Pflugers Arch. 1985, 405, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Schweitz, H.; Vincent, J.P.; Barhanin, J.; Frelin, C.; Linden, G.; Hugues, M.; Lazdunski, M. Purification and pharmacological properties of eight sea anemone toxins from Anemonia sulcata, Anthopleura xanthogrammica, Stoichactis giganteus, and Actinodendron plumosum. Biochemistry 1981, 20, 5245–5252. [Google Scholar] [CrossRef] [PubMed]
- Warashina, A.; Ogura, T.; Fujita, S. Binding properties of sea anemone toxins to sodium channels in the crayfish giant axon. Comp. Biochem. Physiol. C, Comp. Pharmacol. Toxicol. 1988, 90, 351–359. [Google Scholar] [PubMed]
- Wunderer, G.; Fritz, H.; Wachter, E.; Machleidt, W. Amino-acid sequence of a coelenterate toxin: Toxin II from Anemonia sulcata. Eur. J. Biochem. 1976, 68, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Mikov, A.N.; Kozlov, S.A. Structural Features of Cysteine-Stabilized Polypeptides from Sea Anemones Venoms. Bioorg. Khim. 2015, 41, 511–523. [Google Scholar] [PubMed]
- Hartung, K.; Rathmayer, W. Anemonia sulcata toxins modify activation and inactivation of Na+ currents in a crayfish neurone. Pflugers Arch. 1985, 404, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Widmer, H.; Wagner, G.; Schweitz, H.; Lazdunski, M.; Wüthrich, K. The secondary structure of the toxin ATX Ia from Anemonia sulcata in aqueous solution determined on the basis of complete sequence-specific 1H-NMR assignments. Eur. J. Biochem. 1988, 171, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Widmer, H.; Billeter, M.; Wüthrich, K. Three-dimensional structure of the neurotoxin ATX Ia from Anemonia sulcata in aqueous solution determined by nuclear magnetic resonance spectroscopy. Proteins 1989, 6, 357–371. [Google Scholar] [CrossRef] [PubMed]
- Wunderer, G. The disulfide bridges of toxin II from Anemonia sulcata. Hoppe-Seyler’s Z. Physiol. Chem. 1978, 359, 1193–1201. [Google Scholar] [CrossRef] [PubMed]
- Chahine, M.; Plante, E.; Kallen, R.G. Sea anemone toxin (ATX II) modulation of heart and skeletal muscle sodium channel alpha-subunits expressed in tsA201 cells. J. Membr. Biol. 1996, 152, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Scheffler, J.J.; Tsugita, A.; Linden, G.; Schweitz, H.; Lazdunski, M. The amino acid sequence of toxin V from Anemonia sulcata. Biochem. Biophys. Res. Commun. 1982, 107, 272–278. [Google Scholar] [CrossRef]
- Martinez, G.; Kopeyan, C. Toxin III from Anemonia sulcata: Primary structure. FEBS Lett. 1977, 84, 247–252. [Google Scholar] [CrossRef]
- Moran, Y.; Kahn, R.; Cohen, L.; Gur, M.; Karbat, I.; Gordon, D.; Gurevitz, M. Molecular analysis of the sea anemone toxin Av3 reveals selectivity to insects and demonstrates the heterogeneity of receptor site-3 on voltage-gated Na+ channels. Biochem. J. 2007, 406, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Osmakov, D.I.; Kozlov, S.A.; Andreev, Y.A.; Koshelev, S.G.; Sanamyan, N.P.; Sanamyan, K.E.; Dyachenko, I.A.; Bondarenko, D.A.; Murashev, A.N.; Mineev, K.S.; et al. Sea Anemone Peptide with Uncommon β-Hairpin Structure Inhibits Acid-sensing Ion Channel 3 (ASIC3) and Reveals Analgesic Activity. J. Biol. Chem. 2013, 288, 23116–23127. [Google Scholar] [CrossRef] [PubMed]
- Honma, T.; Hasegawa, Y.; Ishida, M.; Nagai, H.; Nagashima, Y.; Shiomi, K. Isolation and molecular cloning of novel peptide toxins from the sea anemone Antheopsis maculata. Toxicon 2005, 45, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Honma, T.; Shiomi, K. Peptide toxins in sea anemones: Structural and functional aspects. Mar. Biotechnol. 2006, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Honma, T.; Kawahata, S.; Ishida, M.; Nagai, H.; Nagashima, Y.; Shiomi, K. Novel peptide toxins from the sea anemone Stichodactyla haddoni. Peptides 2008, 29, 536–544. [Google Scholar] [CrossRef] [PubMed]
- Tudor, J.E.; Pallaghy, P.K.; Pennington, M.W.; Norton, R.S. Solution structure of ShK toxin, a novel potassium channel inhibitor from a sea anemone. Nat. Struct. Biol. 1996, 3, 317–320. [Google Scholar] [CrossRef] [PubMed]
- Pennington, M.W.; Kem, W.R.; Mahnir, V.M.; Byrnes, M.E.; Zaydenberg, I.; Khaytin, I.; Krafte, D.S.; Hill, R. Identification of essential residues in the potassium channel inhibitor ShK toxin: Analysis of monosubstituted analogs. In Peptides: Chemistry, Structure and Biology; Kaumaya, P.T.P., Hodges, R.S., Eds.; Escom: Leiden, The Netherlands, 1995; pp. 14–16. [Google Scholar]
- Schweitz, H.; Bruhn, T.; Guillemare, E.; Moinier, D.; Lancelin, J.-M.; Béress, L.; Lazdunski, M. Kalicludines and Kaliseptine Two different classes of sea anemone toxins for voltage sensitive K+ channels. J. Biol. Chem. 1995, 270, 25121–25126. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Orts, D.J.B.; Peigneur, S.; Madio, B.; Cassoli, J.S.; Montandon, G.G.; Pimenta, A.M.C.; Bicudo, J.E.P.W.; Freitas, J.C.; Zaharenko, A.J.; Tytgat, J. Biochemical and electrophysiological characterization of two sea anemone type 1 potassium toxins from a geographically distant population of Bunodosoma caissarum. Mar. Drugs 2013, 11, 655–679. [Google Scholar] [CrossRef] [PubMed]
- Scheidig, A.J.; Hynes, T.R.; Pelletier, L.A.; Wells, J.A.; Kossiakoff, A.A. Crystal structures of bovine chymotrypsin and trypsin complexed to the inhibitor domain of Alzheimer’s amyloid beta-protein precursor (APPI) and basic pancreatic trypsin inhibitor (BPTI): Engineering of inhibitors with altered specificities. Protein Sci. 1997, 6, 1806–1824. [Google Scholar] [CrossRef] [PubMed]
- Zweckstetter, M.; Czisch, M.; Mayer, U.; Chu, M.L.; Zinth, W.; Timpl, R.; Holak, T.A. Structure and multiple conformations of the kunitz-type domain from human type VI collagen alpha3(VI) chain in solution. Structure 1996, 4, 195–209. [Google Scholar] [CrossRef]
- Chen, C.; Hsu, C.H.; Su, N.Y.; Lin, Y.C.; Chiou, S.H.; Wu, S.H. Solution structure of a Kunitz-type chymotrypsin inhibitor isolated from the elapid snake Bungarus fasciatus. J. Biol. Chem. 2001, 276, 45079–45087. [Google Scholar] [CrossRef] [PubMed]
- Antuch, W.; Berndt, K.D.; Chávez, M.A.; Delfín, J.; Wüthrich, K. The NMR solution structure of a Kunitz-type proteinase inhibitor from the sea anemone Stichodactyla helianthus. Eur. J. Biochem. 1993, 212, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Diochot, S.; Loret, E.; Bruhn, T.; Béress, L.; Lazdunski, M. APETx1, a new toxin from the sea anemone Anthopleura elegantissima, blocks voltage-gated human ether-a-go-go-related gene potassium channels. Mol. Pharmacol. 2003, 64, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Diochot, S.; Baron, A.; Rash, L.D.; Deval, E.; Escoubas, P.; Scarzello, S.; Salinas, M.; Lazdunski, M. A new sea anemone peptide, APETx2, inhibits ASIC3, a major acid-sensitive channel in sensory neurons. EMBO J. 2004, 23, 1516–1525. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, M.G.; Rash, L.D.; Kellenberger, S. Inhibition of voltage-gated Na(+) currents in sensory neurones by the sea anemone toxin APETx2. Br. J. Pharmacol. 2012, 165, 2167–2177. [Google Scholar] [CrossRef] [PubMed]
- Béress, L.; Doppelfeld, I.S.; Etschenberg, E.; Graf, E.; Henschen, A.; Zwick, J. Polypeptides, Methods of Production and Their Uses as Antihypertensives. German Patent DE3324689A1, 17 January 1985. [Google Scholar]
- Diochot, S.; Schweitz, H.; Béress, L.; Lazdunski, M. Sea anemone peptides with a specific blocking activity against the fast inactivating potassium channel Kv3.4. J. Biol. Chem. 1998, 273, 6744–6749. [Google Scholar] [CrossRef] [PubMed]
- Yeung, S.Y.M.; Thompson, D.; Wang, Z.; Fedida, D.; Robertson, B. Modulation of Kv3 subfamily potassium currents by the sea anemone toxin BDS: Significance for CNS and biophysical studies. J. Neurosci. 2005, 25, 8735–8745. [Google Scholar] [CrossRef] [PubMed]
- Abbott, G.W.; Butler, M.H.; Bendahhou, S.; Dalakas, M.C.; Ptacek, L.J.; Goldstein, S.A. MiRP2 forms potassium channels in skeletal muscle with Kv3.4 and is associated with periodic paralysis. Cell 2001, 104, 217–231. [Google Scholar] [CrossRef]
- Liu, P.; Jo, S.; Bean, B.P. Modulation of neuronal sodium channels by the sea anemone peptide BDS-I. J. Neurophysiol. 2012, 107, 3155–3167. [Google Scholar] [CrossRef] [PubMed]
- Driscoll, P.C.; Gronenborn, A.M.; Beress, L.; Clore, G.M. Determination of the three-dimensional solution structure of the antihypertensive and antiviral protein BDS-I from the sea anemone Anemonia sulcata: A study using nuclear magnetic resonance and hybrid distance geometry-dynamical simulated annealing. Biochemistry 1989, 28, 2188–2198. [Google Scholar] [CrossRef] [PubMed]
- Chagot, B.; Diochot, S.; Pimentel, C.; Lazdunski, M.; Darbon, H. Solution structure of APETx1 from the sea anemone Anthopleura elegantissima: A new fold for an HERG toxin. Proteins 2005, 59, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Chagot, B.; Escoubas, P.; Diochot, S.; Bernard, C.; Lazdunski, M.; Darbon, H. Solution structure of APETx2, a specific peptide inhibitor of ASIC3 proton-gated channels. Protein Sci. 2005, 14, 2003–2010. [Google Scholar] [CrossRef] [PubMed]
- Shiomi, K.; Honma, T.; Ide, M.; Nagashima, Y.; Ishida, M.; Chino, M. An epidermal growth factor-like toxin and two sodium channel toxins from the sea anemone Stichodactyla gigantea. Toxicon 2003, 41, 229–236. [Google Scholar] [CrossRef]
- Honma, T.; Minagawa, S.; Nagai, H.; Ishida, M.; Nagashima, Y.; Shiomi, K. Novel peptide toxins from acrorhagi, aggressive organs of the sea anemone Actinia equina. Toxicon 2005, 46, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Morabito, R.; La Spada, G.; Crupi, R.; Esposito, E.; Marino, A. Crude Venom from Nematocysts of the Jellyfish Pelagia noctiluca as a Tool to Study Cell Physiology. Cent. Nerv. Syst. Agents Med. Chem. 2015, 15, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Prentis, P.J.; Pavasovic, A.; Norton, R.S. Sea Anemones: Quiet Achievers in the Field of Peptide Toxins. Toxins 2018, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Chi, V.; Pennington, M.W.; Norton, R.S.; Tarcha, E.J.; Londono, L.M.; Sims-Fahey, B.; Upadhyay, S.K.; Lakey, J.T.; Iadonato, S.; Wulff, H.; et al. Development of a sea anemone toxin as an immunomodulator for therapy of autoimmune diseases. Toxicon 2012, 59, 529–546. [Google Scholar] [CrossRef] [PubMed]
- Baranauskas, G.; Tkatch, T.; Nagata, K.; Yeh, J.Z.; Surmeier, D.J. Kv3.4 subunits enhance the repolarizing efficiency of Kv3.1 channels in fast-spiking neurons. Nat. Neurosci. 2003, 6, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Angulo, E.; Noe, V.; Casado, V.; Mallol, J.; Gomez-Isla, T.; Lluis, C.; Ferrer, I.; Ciudad, C.J.; Franco, R. Up-regulation of the Kv3.4 potassium channel subunit in early stages of Alzheimer’s disease. J. Neurochem. 2004, 91, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Chabbert, C.; Chambard, J.M.; Sans, A.; Desmadryl, G. Three types of depolarization-activated potassium currents in acutely isolated mouse vestibular neurons. J. Neurophysiol. 2001, 85, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, T.; Teruyama, R.; Armstrong, W.E. High-threshold, Kv3-like potassium currents in magnocellular neurosecretory neurons and their role in spike repolarization. J. Neurophysiol. 2004, 92, 3043–3055. [Google Scholar] [CrossRef] [PubMed]
| Accession | Experiment Title | Platform | Submitter | Amount |
|---|---|---|---|---|
| ERX1926108 ERX1926107 ERX1926106 ERX1926105 | Study of mitogenome and corresponding transcriptome of sea anemones | Ion Torrent PGM and Sanger technology | The Arctic University of Norway (UiT) | unspecified |
SRX3049371 SRX3049370 SRX3049369 SRX3049368 SRX3049367 SRX3049366 SRX3049365 SRX3049364 SRX3049363 | Small RNA sequencing of Anemonia viridis | AB 5500Xl Genetic Analyzer | Urbarova et al., 2018 | 2.9 × 103 Mb |
| SRX699624 | Anemonia viridis Transcriptome or Gene expression | Illumina HiSeq 2000 | University of Haifa | 5.4 × 103 Mb |
| Under different accession | Symbiotic sea anemone A. viridis cDNA library | ABI-3730 Genetic Analyze (Sanger Technology) | Sabourault et al., 2010 | 39,939 ESTs |
| SRX971460 | Tissue specific transcriptomes of the emerging model organism Anemonia sulcata | Illumina HiSeq 1500 | The Ohio State University | 8.8 × 103 Mb |
| SRX971488 | Tissue specific transcriptomes | Illumina HiSeq 2000 | The Ohio State University | 33.9 × 103 Mb |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nicosia, A.; Mikov, A.; Cammarata, M.; Colombo, P.; Andreev, Y.; Kozlov, S.; Cuttitta, A. The Anemonia viridis Venom: Coupling Biochemical Purification and RNA-Seq for Translational Research. Mar. Drugs 2018, 16, 407. https://doi.org/10.3390/md16110407
Nicosia A, Mikov A, Cammarata M, Colombo P, Andreev Y, Kozlov S, Cuttitta A. The Anemonia viridis Venom: Coupling Biochemical Purification and RNA-Seq for Translational Research. Marine Drugs. 2018; 16(11):407. https://doi.org/10.3390/md16110407
Chicago/Turabian StyleNicosia, Aldo, Alexander Mikov, Matteo Cammarata, Paolo Colombo, Yaroslav Andreev, Sergey Kozlov, and Angela Cuttitta. 2018. "The Anemonia viridis Venom: Coupling Biochemical Purification and RNA-Seq for Translational Research" Marine Drugs 16, no. 11: 407. https://doi.org/10.3390/md16110407
APA StyleNicosia, A., Mikov, A., Cammarata, M., Colombo, P., Andreev, Y., Kozlov, S., & Cuttitta, A. (2018). The Anemonia viridis Venom: Coupling Biochemical Purification and RNA-Seq for Translational Research. Marine Drugs, 16(11), 407. https://doi.org/10.3390/md16110407







