A High-Content Screening Assay for the Discovery of Novel Proteasome Inhibitors from Formosan Soft Corals
Abstract
1. Introduction
2. Results
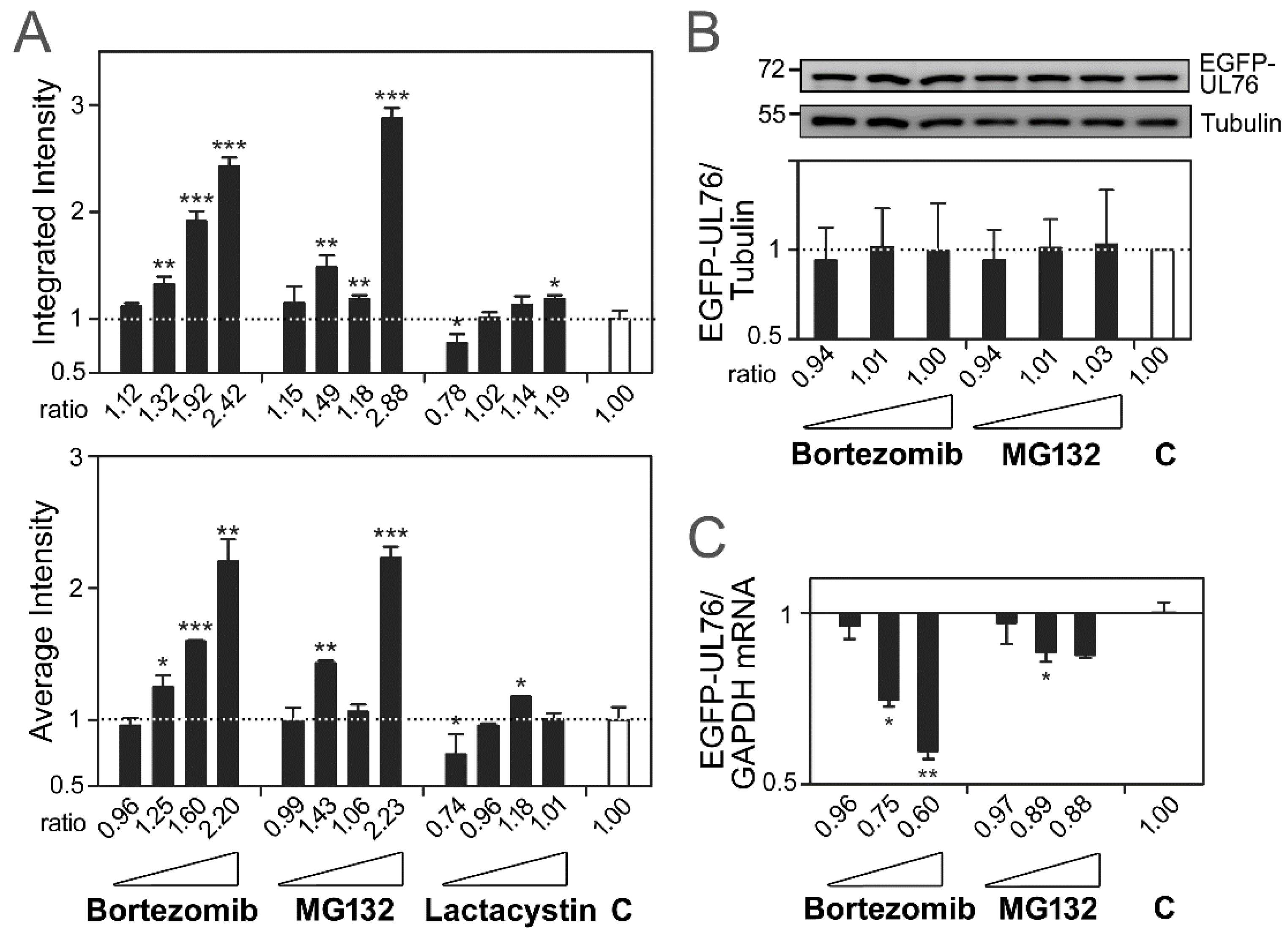
2.1. Validation of EGFP-UL76 High-Content Analytic Measurements by Proteasome Inhibitors
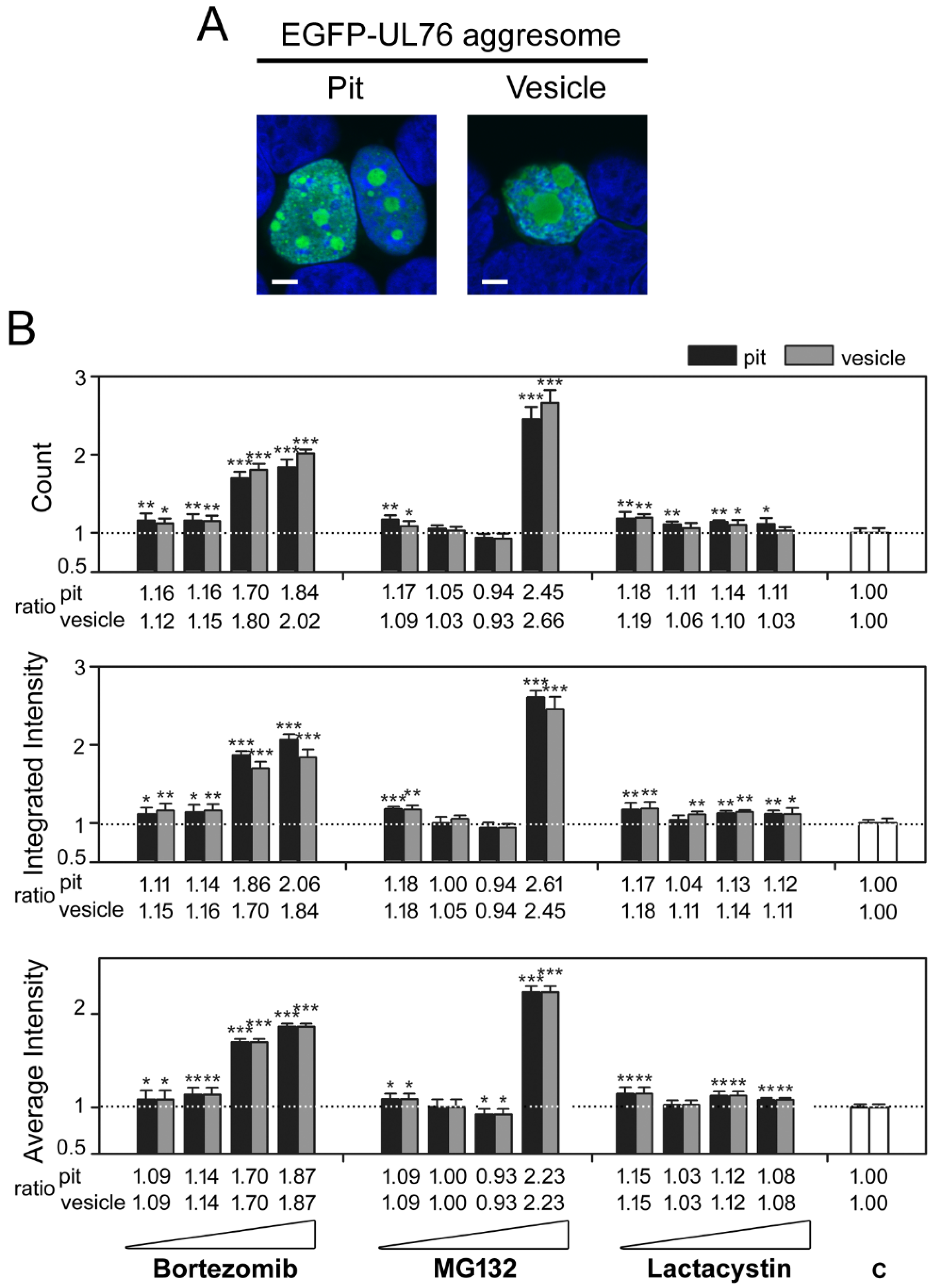
2.2. Identification of Marine Natural Product Showed High-Content Characteristics of Proteasome Inhibition
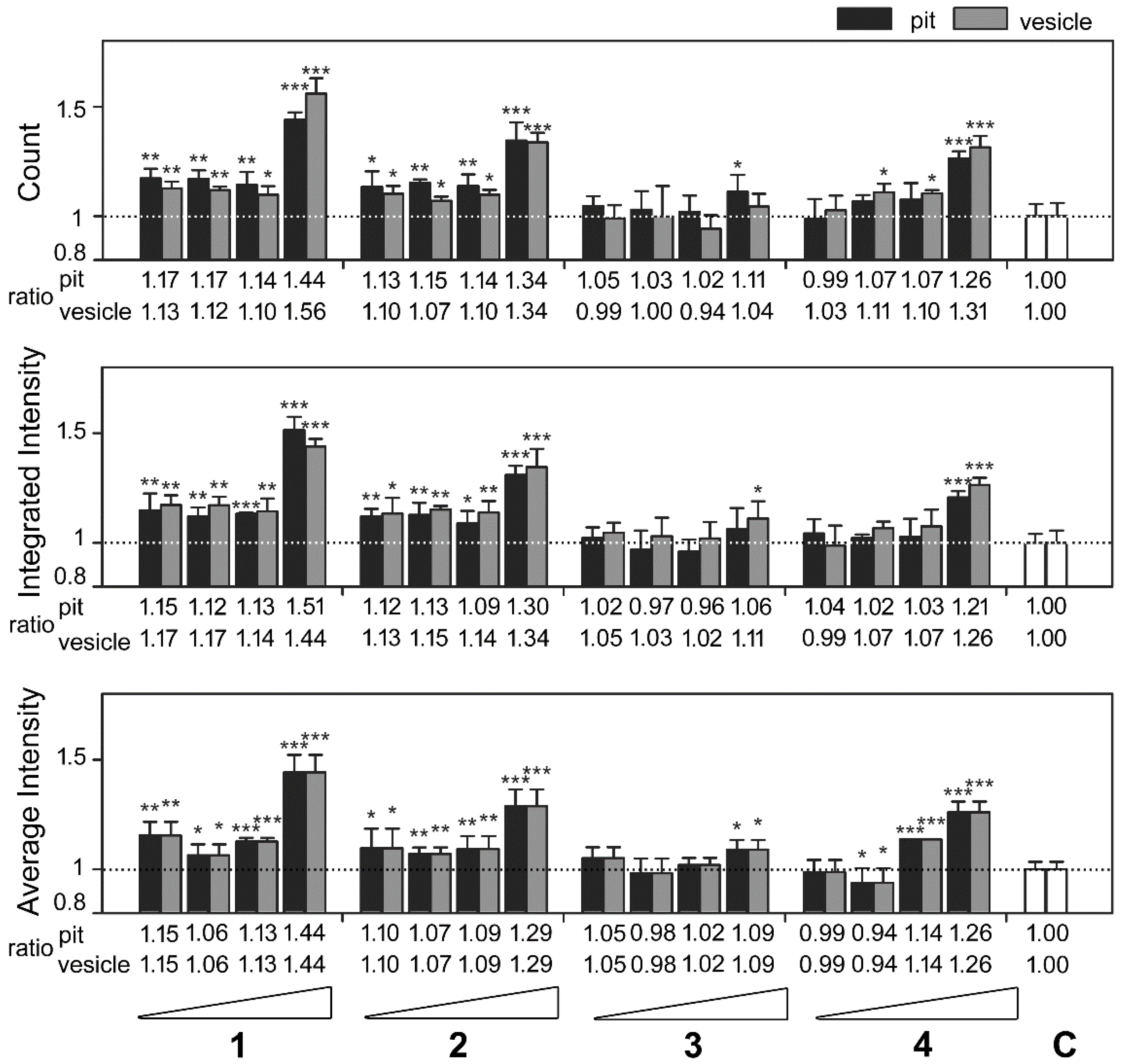
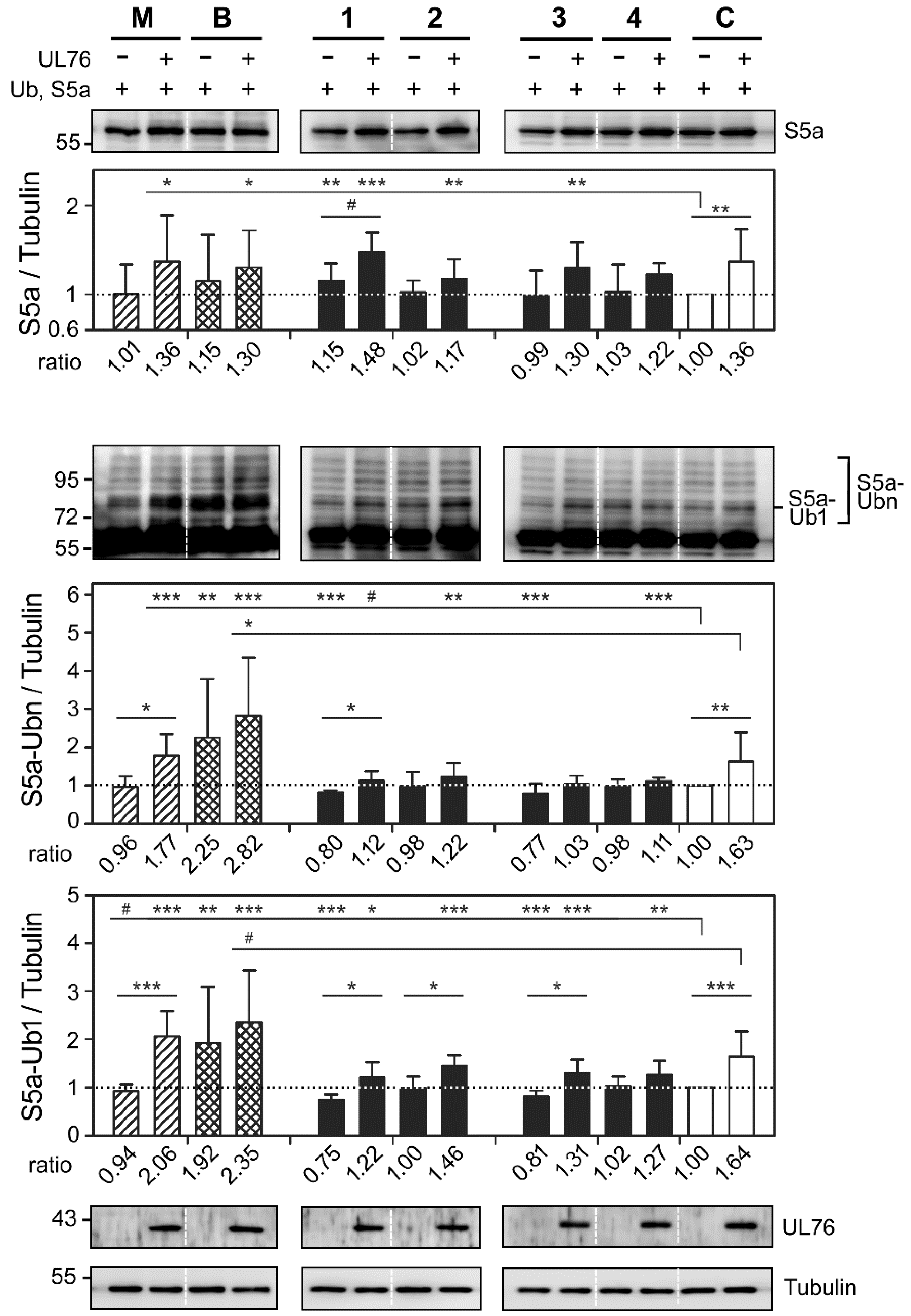
2.3. Cembrane-Based Compounds Exhibited the Accumulation of Ubiquitinated Proteins
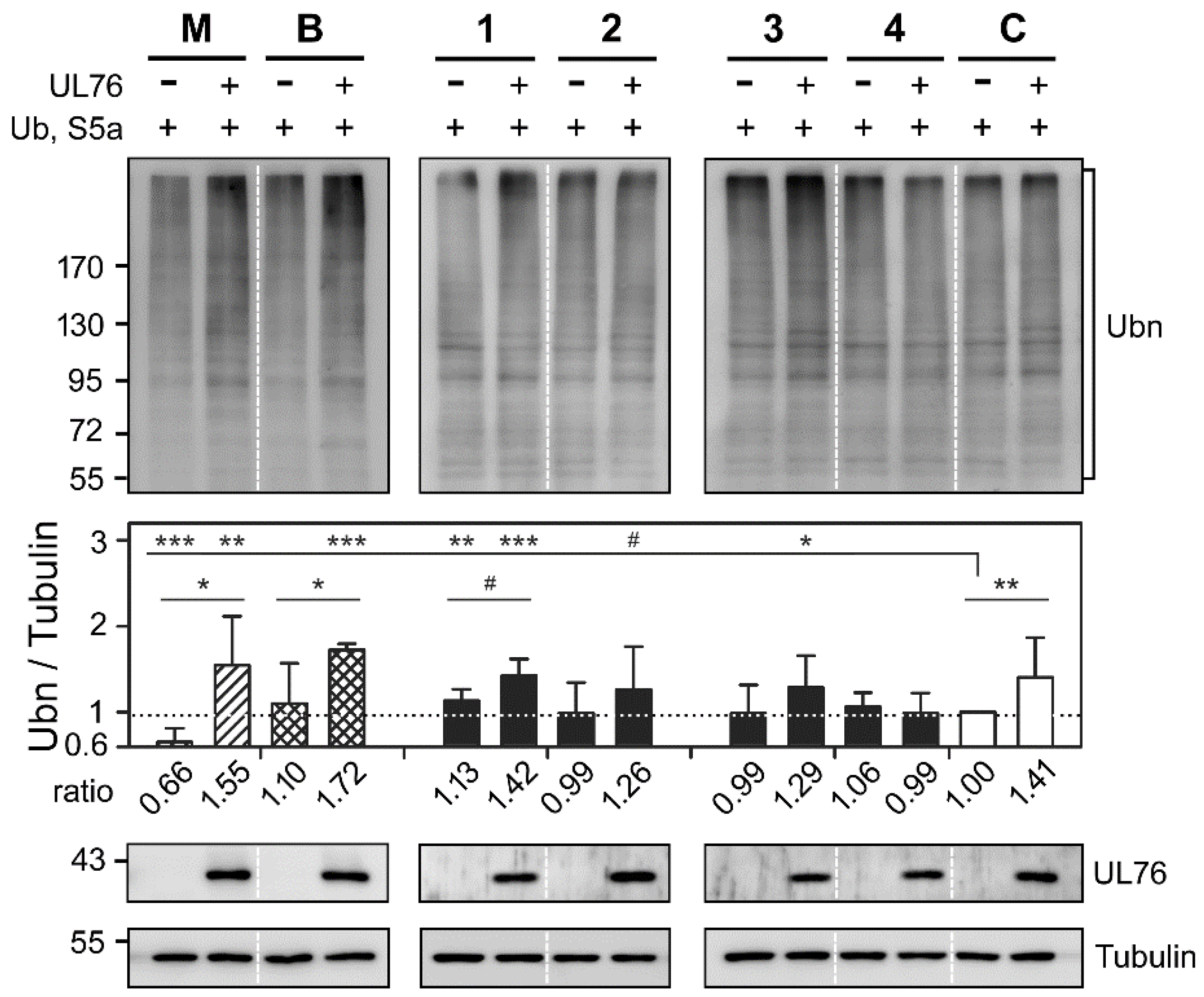
2.4. Novel Proteasome Inhibition Mechanisms
3. Discussion
3.1. Comparison of Assay Efficacy and Sensitivity
3.2. Discovery of a Novel Mechanism of Action
3.3. Roles in Coral Reef Holobiont
4. Materials and Methods
4.1. Compounds
4.2. Cell, Plasmids, and Antibodies
4.3. Cell-Based Screening Protocol
4.4. HCS Analysis
4.5. Western Blotting Imaging and Densitometric Analysis
4.6. RNA Purification and PCR
4.7. Confocal Microscopy
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wegley Kelly, L.; Haas, A.F.; Nelson, C.E. Ecosystem microbiology of coral reefs: Linking genomic, metabolomic, and miogeochemical dynamics from animal symbioses to reefscape processes. mSystems 2018, 3, e00162-17. [Google Scholar] [CrossRef] [PubMed]
- Quinn, R.A.; Vermeij, M.J.; Hartmann, A.C.; Galtier d’Auriac, I.; Benler, S.; Haas, A.; Quistad, S.D.; Lim, Y.W.; Little, M.; Sandin, S.; et al. Metabolomics of reef benthic interactions reveals a bioactive lipid involved in coral defence. Proc. Biol. Sci. 2016, 283, 20160469. [Google Scholar] [CrossRef] [PubMed]
- Aratake, S.; Tomura, T.; Saitoh, S.; Yokokura, R.; Kawanishi, Y.; Shinjo, R.; Reimer, J.D.; Tanaka, J.; Maekawa, H. Soft coral Sarcophyton (Cnidaria: Anthozoa: Octocorallia) species diversity and chemotypes. PLoS ONE 2012, 7, e30410. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2018, 35, 8–53. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, G.; Udasin, R.G.; Livneh, I.; Ciechanover, A. Identification of UBact, a ubiquitin-like protein, along with other homologous components of a conjugation system and the proteasome in different gram-negative bacteria. Biochem. Biophys. Res. Commun. 2017, 483, 946–950. [Google Scholar] [CrossRef] [PubMed]
- Budenholzer, L.; Cheng, C.L.; Li, Y.; Hochstrasser, M. Proteasome structure and assembly. J. Mol. Biol. 2017, 429, 3500–3524. [Google Scholar] [CrossRef] [PubMed]
- Collins, G.A.; Goldberg, A.L. The logic of the 26S proteasome. Cell 2017, 169, 792–806. [Google Scholar] [CrossRef] [PubMed]
- Fort, P.; Kajava, A.V.; Delsuc, F.; Coux, O. Evolution of proteasome regulators in eukaryotes. Genome Biol. Evol. 2015, 7, 1363–1379. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, A.C.D.; Maldoner, L.; Hipp, K.; Hartmann, M.D.; Martin, J. Structural characterization of the bacterial proteasome homolog BPH reveals a tetradecameric double-ring complex with unique inner cavity properties. J. Biol. Chem. 2018, 293, 920–930. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, A.C.D.; Alva, V.; Maldoner, L.; Albrecht, R.; Hartmann, M.D.; Martin, J. The architecture of the Anbu complex reflects an evolutionary intermediate at the origin of the proteasome system. Structure 2017, 25, 834–845.e5. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Finley, D. Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta 2014, 1843, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Ravid, T.; Hochstrasser, M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell. Biol. 2008, 9, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Woulfe, J.M. Abnormalities of the nucleus and nuclear inclusions in neurodegenerative disease: A work in progress. Neuropathol. Appl. Neurobiol. 2007, 33, 2–42. [Google Scholar] [CrossRef] [PubMed]
- Cloos, J.; Roeten, M.S.; Franke, N.E.; van Meerloo, J.; Zweegman, S.; Kaspers, G.J.; Jansen, G. (Immuno)proteasomes as therapeutic target in acute leukemia. Cancer Metastasis Rev. 2017, 36, 599–615. [Google Scholar] [CrossRef] [PubMed]
- Popovic, D. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 2014, 20, 1242–1253. [Google Scholar] [CrossRef] [PubMed]
- Burslem, G.M.; Crews, C.M. Small-molecule modulation of protein homeostasis. Chem. Rev. 2017, 117, 11269–11301. [Google Scholar] [CrossRef] [PubMed]
- Raedler, L.A. Ninlaro (Ixazomib): First oral proteasome inhibitor approved for the treatment of patients with relapsed or refractory multiple myeloma. Am. Health Drug Benefits 2016, 9, 102–105. [Google Scholar] [PubMed]
- Kisselev, A.F. A novel bullet hits the proteasome. Cancer Cell 2013, 24, 691–693. [Google Scholar] [CrossRef] [PubMed]
- Gelman, J.S.; Sironi, J.; Berezniuk, I.; Dasgupta, S.; Castro, L.M.; Gozzo, F.C.; Ferro, E.S.; Fricker, L.D. Alterations of the intracellular peptidome in response to the proteasome inhibitor bortezomib. PLoS ONE 2013, 8, e53263. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.R.; Jiang, M.J.; Wang, H.H.; Hu, C.H.; Hsu, M.S.; Hsi, E.; Duh, C.Y.; Wang, S.K. Human cytomegalovirus UL76 elicits novel aggresome formation via interaction with S5a of the ubiquitin proteasome system. J. Virol. 2013, 87, 11562–11578. [Google Scholar] [CrossRef] [PubMed]
- Isasa, M.; Katz, E.J.; Kim, W.; Yugo, V.; Gonzalez, S.; Kirkpatrick, D.S.; Thomson, T.M.; Finley, D.; Gygi, S.P.; Crosas, B. Monoubiquitination of RPN10 regulates substrate recruitment to the proteasome. Mol. Cell 2010, 38, 733–745. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.K.; Duh, C.Y.; Wu, C.W. Human cytomegalovirus UL76 encodes a novel virion-associated protein that is able to inhibit viral replication. J. Virol. 2004, 78, 9750–9762. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.K.; Duh, C.Y.; Chang, T.T. Cloning and identification of regulatory gene UL76 of human cytomegalovirus. J. Gen. Virol. 2000, 81, 2407–2416. [Google Scholar] [CrossRef] [PubMed]
- Kabayashi, M.; Hirase, T. Marine terpenes and terpenoids. XI. Structures of new dihydrofuranocembranoids isolated from a Sarcophyton sp. soft coral of Okinawa. Chem. Pharm. Bull. 1990, 38, 2442–2445. [Google Scholar] [CrossRef]
- Quang, T.H.; Ha, T.T.; Minh, C.V.; Kiem, P.V.; Huong, H.T.; Ngan, N.T.; Nhiem, N.X.; Tung, N.H.; Tai, B.H.; Thuy, D.T.; et al. Cytotoxic and anti-inflammatory cembranoids from the Vietnamese soft coral Lobophytum laevigatum. Bioorg. Med. Chem. 2011, 19, 2625–2632. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.Y.; Wang, S.K.; Chiou, S.F.; Hsu, C.H.; Dai, C.F.; Chiang, M.Y.; Duh, C.Y. Cembranoids from the octocoral Sarcophyton ehrenbergi. J. Nat. Prod. 2010, 73, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Nakagawa, T.; Mitsuhashi, H. Marine terpenes and terpenoids. I. Structures of four cembrane-type diterpenes; sarcophytol-A, sarcophytol-A acetate, sarcophytol-B, and sarcophytonin- A, from the soft coral, Sarcophyton glaueum. Chem. Pharm. Bull. 1979, 27, 2382–2387. [Google Scholar] [CrossRef]
- Duh, C.Y.; Hou, R.S. Cytotoxic cembranoids from the soft corals Sinularia gibberosa and Sarcophyton trocheliophorum. J. Nat. Prod. 1996, 59, 595–598. [Google Scholar] [CrossRef]
- Katsuyama, I.; Fahmy, H.; Zjawiony, J.K.; Khalifa, S.I.; Kilada, R.W.; Konoshima, T.; Takasaki, M.; Tokuda, H. Semisynthesis of new sarcophine derivatives with chemopreventive activity. J. Nat. Prod. 2002, 65, 1809–1814. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C.; Chevet, E.; Harding, H.P. Targeting the unfolded protein response in disease. Nat. Rev. Drug Discov. 2013, 12, 703–719. [Google Scholar] [CrossRef] [PubMed]
- Cuong, N.X.; Tuan, T.A.; Kiem, P.V.; Minh, C.V.; Choi, E.M.; Kim, Y.H. New cembranoid diterpenes from the Vietnamese soft coral Sarcophyton mililatensis stimulate osteoblastic differentiation in MC3T3-E1 cells. Chem. Pharm. Bull. 2008, 56, 988–992. [Google Scholar] [CrossRef] [PubMed]
- Ne’eman, I.; Fishelson, L.; Kashman, Y. Sarcophine--a new toxin from the soft coral Sarcophyton glaucum (Alcyonaria). Toxicon 1974, 12, 593–598. [Google Scholar] [CrossRef]
- Erman, A.; Neeman, I. Inhibition of phosphofructokinase by the toxic cembranolide sarcophine isolated from the soft-bodied coral Sarcophyton glaucum. Toxicon 1977, 15, 207–215. [Google Scholar] [CrossRef]
- El Sayed, K.A.; Hamann, M.T.; Waddling, C.A.; Jensen, C.; Lee, S.K.; Dunstan, C.A.; Pezzuto, J.M. Structurally novel bioconversion products of the marine natural product sarcophine effectively inhibit JB6 cell transformation. J. Org. Chem. 1998, 63, 7449–7455. [Google Scholar] [CrossRef] [PubMed]
- Sawant, S.S.; Youssef, D.T.; Reiland, J.; Ferniz, M.; Marchetti, D.; El Sayed, K.A. Biocatalytic and antimetastatic studies of the marine cembranoids sarcophine and 2-epi-16-deoxysarcophine. J. Nat. Prod. 2006, 69, 1010–1013. [Google Scholar] [CrossRef] [PubMed]
- Hassan, H.M.; Sallam, A.A.; Mohammed, R.; Hifnawy, M.S.; Youssef, D.T.; El Sayed, K.A. Semisynthetic analogues of the marine cembranoid sarcophine as prostate and breast cancer migration inhibitors. Bioorg. Med. Chem. 2011, 19, 4928–4934. [Google Scholar] [CrossRef] [PubMed]
- Arif, J.M.; Kunhi, M.; Siddiqui, Y.M.; El Sayed, K.A.; Orabi, K.Y.; Al-Hazzani, A.; Al-Ahdal, M.N.; Al-Khodairy, F.M. Role of intermediary biomarkers in determing the anticancer efficacy of marine compounds. Med. Chem. Res. 2004, 13, 553–562. [Google Scholar] [CrossRef]
- Thao, N.P.; Luyen, B.T.; Brun, R.; Kaiser, M.; Van Kiem, P.; Van Minh, C.; Schmidt, T.J.; Kang, J.S.; Kim, Y.H. Anti-protozoal activities of cembrane-type diterpenes from Vietnamese soft corals. Molecules 2015, 20, 12459–12468. [Google Scholar] [CrossRef] [PubMed]
- Zuin, A.; Bichmann, A.; Isasa, M.; Puig-Sarries, P.; Diaz, L.M.; Crosas, B. Rpn10 monoubiquitination orchestrates the association of the ubiquilin-type DSK2 receptor with the proteasome. Biochem. J. 2015, 472, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Keren-Kaplan, T.; Zeev Peters, L.; Levin-Kravets, O.; Attali, I.; Kleifeld, O.; Shohat, N.; Artzi, S.; Zucker, O.; Pilzer, I.; Reis, N.; et al. Structure of ubiquitylated-Rpn10 provides insight into its autoregulation mechanism. Nat. Commun. 2016, 7, 12960. [Google Scholar] [CrossRef] [PubMed]
- Berkers, C.R.; Verdoes, M.; Lichtman, E.; Fiebiger, E.; Kessler, B.M.; Anderson, K.C.; Ploegh, H.L.; Ovaa, H.; Galardy, P.J. Activity probe for in vivo profiling of the specificity of proteasome inhibitor bortezomib. Nat. Methods 2005, 2, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Arastu-Kapur, S.; Anderl, J.L.; Kraus, M.; Parlati, F.; Shenk, K.D.; Lee, S.J.; Muchamuel, T.; Bennett, M.K.; Driessen, C.; Ball, A.J.; et al. Nonproteasomal targets of the proteasome inhibitors bortezomib and carfilzomib: A link to clinical adverse events. Clin. Cancer Res. 2011, 17, 2734–2743. [Google Scholar] [CrossRef] [PubMed]
- Tamas, M.J.; Fauvet, B.; Christen, P.; Goloubinoff, P. Misfolding and aggregation of nascent proteins: A novel mode of toxic cadmium action in vivo. Curr. Genet. 2018, 64, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Fernandez, M.R.; Gragera, M.; Ochoa-Ibarrola, L.; Quintana-Gallardo, L.; Valpuesta, J.M. Hsp70—A master regulator in protein degradation. FEBS Lett. 2017, 591, 2648–2660. [Google Scholar] [CrossRef] [PubMed]
- Van de Water, J.; Allemand, D.; Ferrier-Pages, C. Host-microbe interactions in octocoral holobionts—Recent advances and perspectives. Microbiome 2018, 6, 64. [Google Scholar] [CrossRef] [PubMed]
- Bogdanov, A.; Hertzer, C.; Kehraus, S.; Nietzer, S.; Rohde, S.; Schupp, P.J.; Wagele, H.; Konig, G.M. Secondary metabolome and its defensive role in the aeolidoidean Phyllodesmium longicirrum, (Gastropoda, Heterobranchia, Nudibranchia). Beilstein J. Org. Chem. 2017, 13, 502–519. [Google Scholar] [CrossRef] [PubMed]
- Siew, V.K.; Duh, C.Y.; Wang, S.K. Human cytomegalovirus UL76 induces chromosome aberrations. J. Biomed. Sci. 2009, 16, 107. [Google Scholar] [CrossRef] [PubMed]
- Bres, V.; Kiernan, R.E.; Linares, L.K.; Chable-Bessia, C.; Plechakova, O.; Treand, C.; Emiliani, S.; Peloponese, J.M.; Jeang, K.T.; Coux, O.; et al. A non-proteolytic role for ubiquitin in Tat-mediated transactivation of the HIV-1 promoter. Nat. Cell Biol. 2003, 5, 754–761. [Google Scholar] [CrossRef] [PubMed]
- Gus, Y.; Karni, R.; Levitzki, A. Subunit S5a of the 26S proteasome is regulated by antiapoptotic signals. FEBS J. 2007, 274, 2815–2831. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.C.; Berkelman, T.; Yadav, G.; Hammond, M. A defined methodology for reliable quantification of Western blot data. Mol. Biotechnol. 2013, 55, 217–226. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ling, X.-H.; Wang, S.-K.; Huang, Y.-H.; Huang, M.-J.; Duh, C.-Y. A High-Content Screening Assay for the Discovery of Novel Proteasome Inhibitors from Formosan Soft Corals. Mar. Drugs 2018, 16, 395. https://doi.org/10.3390/md16100395
Ling X-H, Wang S-K, Huang Y-H, Huang M-J, Duh C-Y. A High-Content Screening Assay for the Discovery of Novel Proteasome Inhibitors from Formosan Soft Corals. Marine Drugs. 2018; 16(10):395. https://doi.org/10.3390/md16100395
Chicago/Turabian StyleLing, Xue-Hua, Shang-Kwei Wang, Yun-Hsuan Huang, Min-Jay Huang, and Chang-Yih Duh. 2018. "A High-Content Screening Assay for the Discovery of Novel Proteasome Inhibitors from Formosan Soft Corals" Marine Drugs 16, no. 10: 395. https://doi.org/10.3390/md16100395
APA StyleLing, X.-H., Wang, S.-K., Huang, Y.-H., Huang, M.-J., & Duh, C.-Y. (2018). A High-Content Screening Assay for the Discovery of Novel Proteasome Inhibitors from Formosan Soft Corals. Marine Drugs, 16(10), 395. https://doi.org/10.3390/md16100395



