Lindane Bioremediation Capability of Bacteria Associated with the Demosponge Hymeniacidon perlevis
Abstract
:1. Introduction
2. Results
2.1. Screening of Lindane Degradation by Bacterial Isolates
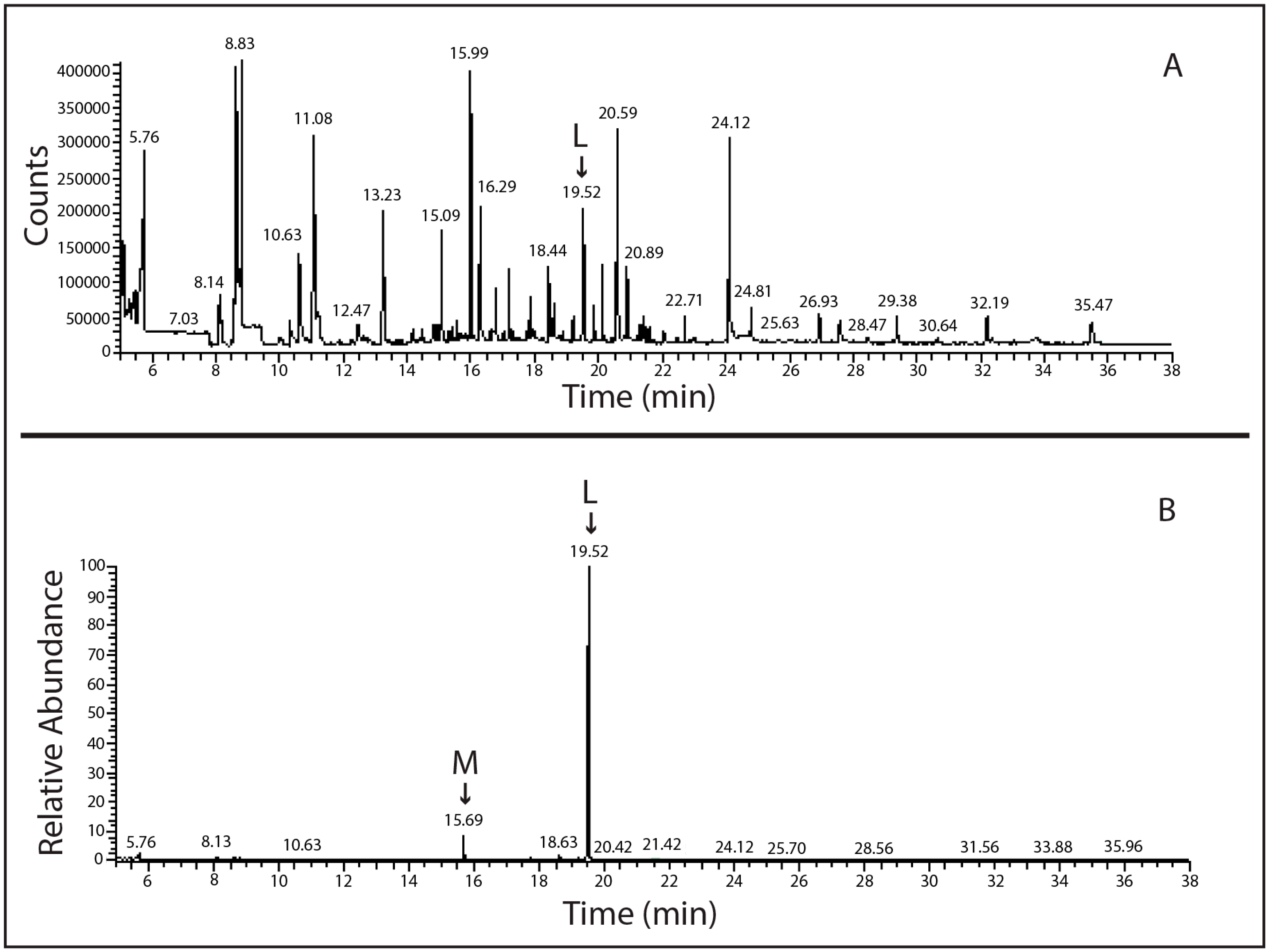
2.2. Molecular Analysis of Lindane-Degrading Bacteria Isolated from Hymeniacidon perlevis
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolates
4.2. Screening of Lindane Degradation by Bacterial Isolates
4.3. SPME-GC-MS Analysis
4.4. 16S rRNA Gene Sequence Analysis of Lindane-Degrading Bacterial Isolates
4.5. 16S rRNA Gene Sequencing and Phylogenetic Analyses
4.6. 16S rRNA GenBank Accession Number
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Bellas, J.; Beiras, R.; Mariño-Balsa, J.C.; Fernández, N. Toxicity of organic compounds to marine invertebrate embryos and larvae: A comparison between the sea urchin embryogenesis bioassay and alternative test species. Ecotoxicology 2005, 14, 337–353. [Google Scholar] [CrossRef] [PubMed]
- United Nations Environment Programme (UNEP). Report of the Conference of the Parties of the Stockholm Convention on Persistent Organic Pollutants on the Work of Its Fourth Meeting; UNEP/POPS/COP.4/38; UNEP: Geneva, Sweden, 2009; pp. 1–112. [Google Scholar]
- Manzetti, S.; van der Spoel, E.R.; van der Spoel, D. Chemical properties, environmental fate, and degradation of seven classes of pollutants. Chem. Res. Toxicol. 2014, 27, 713–737. [Google Scholar] [CrossRef] [PubMed]
- Rieger, P.G.; Meier, H.M.; Gerle, M.; Vogt, U.; Groth, T.; Knackmuss, H.J. Xenobiotics in the environment: Present and future strategies to obviate the problem of biological persistence. J. Biotechnol. 2002, 94, 101–123. [Google Scholar] [CrossRef]
- Nizzetto, L.; Macleod, M.; Borgå, K.; Cabrerizo, A.; Dachs, J.; Guardo, A.D.; Ghirardello, D.; Hansen, K.M.; Jarvis, A.; Lindroth, A.; et al. Past, present, and future controls on levels of persistent organic pollutants in the global environment. Environ. Sci. Technol. 2010, 44, 6526–6531. [Google Scholar] [CrossRef] [PubMed]
- Konuspayeva, G.; Faye, B.; Pauw, E.D.; Focant, J.F. Levels and trends of PCDD/Fs and PCBs in camel milk (Camelus bactrianus and Camelus dromedarius) from Kazakhstan. Chemosphere 2011, 85, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Perugini, M.; Nuñez, E.G.H.; Baldi, L.; Esposito, M.; Serpe, F.P.; Amorena, M. Predicting dioxin-like PCBs soil contamination levels using milk of grazing animal as indicator. Chemosphere 2012, 89, 964–969. [Google Scholar] [CrossRef] [PubMed]
- Vijgen, J. The Legacy of Lindane HCH Isomer Production: A Global Overview of Residue Management, Formulation and Disposal; Main Report; International HCH and Pesticides Association (IHPA): Holte, Denmark, 2006; pp. 1–22. [Google Scholar]
- Li, Y.F.; Macdonald, R.W. Sources and pathways of selected organochlorine pesticides to the Arctic and the effect of pathway divergence on HCH trends in biota: A review. Sci. Total Environ. 2005, 342, 87–106. [Google Scholar] [CrossRef] [PubMed]
- Vijgen, J.; Abhilash, P.C.; Li, Y.F.; Lal, R.; Forter, M.; Torres, J.; Singh, N.; Yunus, M.; Tian, C.; Schäffer, A.; et al. Hexachlorocyclohexane (HCH) as new Stockholm Convention POPs—A global perspective on the management of lindane and its waste isomers. Environ. Sci. Pollut. Res. 2011, 18, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Peither, A.; Jüttner, I.; Kettrup, A.; Lay, J.P. A pond mesocosm study to determine direct and indirect effects of lindane on a natural zooplankton community. Environ. Pollut. 1996, 93, 49–56. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Lindane, Environmental Health Criteria No. 124; World Health Organization: Geneva, Switzerland, 1991. [Google Scholar]
- Picard, A.; Palavan, G.; Robert, S.; Pesando, D.; Ciapa, B. Effect of organochlorine pesticides on maturation of starfish and mouse oocytes. Toxicol. Sci. 2003, 73, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Görge, G.; Nagel, R. Toxicity of lindane, atrazine, and deltamethrin to early life stages of zebrafish (Brachydanio rerio). Ecotoxicol. Environ. Saf. 1990, 20, 246–255. [Google Scholar] [CrossRef]
- Pesando, D.; Robert, S.; Huitorel, P.; Gutknecht, E.; Pereira, L.; Girard, J.P.; Ciapa, B. Effects of methoxychlor, dieldrin and lindane on sea urchin fertilization and early development. Aquat. Toxicol. 2004, 66, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.G. Chlorinated hydrocarbon insecticides. In Handbook of Pesticide Toxicology; Academic Press Inc.: New York, NY, USA, 1991; pp. 731–915. [Google Scholar]
- Walker, K.; Vallero, D.A.; Lewis, R.G. Factors Influencing the distribution of lindane and other hexachlorocyclohexanes in the environment. Environ. Sci. Technol. 1999, 33, 4373–4378. [Google Scholar] [CrossRef]
- Humphreys, E.H.; Janssen, S.; Heil, A.; Hiatt, P.; Solomon, G.; Miller, M.D. Outcomes of the California Ban on pharmaceutical lindane: Clinical and ecologic impacts. Environ. Health Perspect. 2007, 116, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, D.F. Linking environmental cancer with occupational epidemiology research: The role of the International Agency for Research on Cancer (IARC). J. Environ. Pathol. Toxicol. 2000, 19, 171–175. [Google Scholar]
- Rambo, I.M. Aerobic Degradation of α-, β-, γ-Hexachlorocyclohexane by Narragansett Bay Bacterioplankton; Senior Honors Projects, Honors Program at the University of Rhode Island; Berkeley Electronic Press: Berkeley, CA, USA, 2013; Volume 334, pp. 1–10. [Google Scholar]
- Díaz, E. Bacterial degradation of aromatic pollutants: A paradigm of metabolic versatility. Int. Microbiol. 2004, 7, 173–180. [Google Scholar] [PubMed]
- Dua, M.; Singh, A.; Sethunathan, N.; Johri, A.K. Biotechnology and bioremediation: Successes and limitations. Appl. Microbiol. Biotechnol. 2002, 59, 143–152. [Google Scholar] [PubMed]
- Hagger, J.A.; Jones, M.B.; Leonard, D.R.P.; Owen, R.; Galloway, T.S. Biomarkers and integrated environmental risk assessment: Are there more questions than answers? Integr. Environ. Assess. Manag. 2006, 2, 312–329. [Google Scholar] [CrossRef] [PubMed]
- Crawford, R.L.; Crawford, D.L. Bioremediation Principles and Applications; Cambridge University Press: New York, NY, USA, 1996. [Google Scholar]
- Alexander, M. Biodegradation and Bioremediation; Academic Press Inc.: San Diego, CA, USA, 1999. [Google Scholar]
- Gifford, S.; Dunstan, R.H.; O’Connor, W.; Koller, C.E.; MacFarlane, G.R. Aquatic zooremediation: Deploying animals to remediate contaminated aquatic environments. Trends Biotechnol. 2007, 25, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Stabili, L.; Licciano, M.; Giangrande, A.; Longo, C.; Mercurio, M.; Nonnis-Marzano, C.; Corriero, G. Filtering activity of Spongia officinalis var. adriatica (Schmidt) (Porifera, Demospongiae) on bacterioplankton: Implications for bioremediation of polluted seawater. Water Res. 2006, 40, 3083–3090. [Google Scholar] [CrossRef] [PubMed]
- Stabili, L.; Licciano, M.; Longo, C.; Corriero, G.; Mercurio, M. Evaluation of microbiological accumulation capability of the commercial sponge Spongia officinalis var. adriatica (Schmidt) (Porifera, Demospongiae). Water Res. 2008, 42, 2499–2506. [Google Scholar] [CrossRef] [PubMed]
- Longo, C.; Corriero, G.; Licciano, M.; Stabili, L. Bacterial accumulation by the Demospongiae Hymeniacidon perlevis: A tool for the bioremediation of polluted seawater. Mar. Pollut. Bull. 2010, 60, 1182–1187. [Google Scholar] [CrossRef] [PubMed]
- Cebrian, E.; Martı́, R.; Uriz, J.; Turon, X. Sublethal effects of contamination on the Mediterranean sponge Crambe crambe: Metal accumulation and biological responses. Mar. Pollut. Bull. 2003, 46, 1273–1284. [Google Scholar] [CrossRef]
- Perez, T.; Longet, D.; Schembri, T.; Rebouillon, P.; Vacelet, J. Effects of 12 years’ operation of a sewage treatment plant on trace metal occurrence within a Mediterranean commercial sponge (Spongia officinalis, Demospongiae). Mar. Pollut. Bull. 2005, 50, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Hill, M.; Hill, A.; Lopez, N.; Harriott, O. Sponge-specific bacterial symbionts in the Caribbean sponge, Chondrilla nucula (Demospongiae, Chondrosida). Mar. Biol. 2006, 148, 1221–1230. [Google Scholar] [CrossRef]
- Hill, R.T. Microbes from marine sponges: A treasure trove of biodiversity for natural products discovery. In Microbial Diversity and Bioprospecting; Bull, A.T., Ed.; ASM Press: Washington, DC, USA, 2004; pp. 177–190. [Google Scholar]
- Hentschel, U.; Usher, K.M.; Taylor, M.W. Marine sponges as microbial fermenters: Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 2006, 55, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Burja, A.M.; Hill, R.T. Microbial symbionts of the Australian Great Barrier Reef sponge, Candidaspongia flabellate. Hydrobiologia 2001, 461, 41–47. [Google Scholar] [CrossRef]
- Enticknap, J.J.; Kelly, M.; Peraud, O.; Hill, R.T. Characterization of a culturable Alphaproteobacterial symbiont common to many marine sponges and evidence for vertical transmission via sponge larvae. Appl. Environ. Microb. 2006, 72, 3724–3732. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.K.; Garson, M.J.; Fuerst, J.A. Marine actinomycetes related to the “Salinospora” group from the Great Barrier Reef sponge Pseudoceratina clavata. Environ. Microbiol. 2005, 7, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Lee, O.O. Gillisia myxillae sp. nov., a novel member of the family Flavobacteriaceae, isolated from the marine sponge Myxilla incrustans. Int. J. Syst. Evol. Microbiol. 2006, 56, 1795–1799. [Google Scholar] [CrossRef] [PubMed]
- Montalvo, N.F.; Mohamed, N.M.; Enticknap, J.J.; Hill, R.T. Novel actinobacteria from marine sponges. Antonie van Leeuwenhoek 2005, 87, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Webster, N.S.; Wilson, K.J.; Blackall, L.L.; Hill, R.T. Phylogenetic diversity of bacteria associated with the marine sponge Rhopaloeides odorabile. Appl. Environ. Microbiol. 2001, 67, 434–444. [Google Scholar] [CrossRef] [PubMed]
- Hentschel, U.; Hopke, J.; Horn, M.; Friedrich, A.B.; Wagner, M.; Hacker, J.; Moore, B.S. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 2002, 68, 4431–4440. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.R.; Kavlekar, D.P.; LokaBharathi, P.A. Drugs from sponge-microbe association—A review. Mar. Drugs 2010, 8, 1417–1468. [Google Scholar] [CrossRef] [PubMed]
- Aresta, A.; Nonnis Marzano, C.; Lopane, C.; Corriero, G.; Longo, C.; Zambonin, C.; Stabili, L. Analytical investigations on the lindane bioremediation capability of the demosponge Hymeniacidon perlevis. Mar. Pollut. Bull. 2015, 90, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Quintero, J.C.; Moreira, M.T.; Feijoo, G.; Lema, J.M. Anaerobic degradation of hexachlorocyclohexane isomers in liquid and soil slurry systems. Chemosphere 2005, 61, 528–536. [Google Scholar] [CrossRef] [PubMed]
- Quintero, J.C.; Moreira, M.T.; Feijoo, G.; Lema, J.M. Screening of white rot fungal species for their capacity to degrade lindane and other isomers of hexachlorocyclohexane (HCH). Cienc. Investig. Agrar. 2008, 35, 159–167. [Google Scholar] [CrossRef]
- Guillén-Jiménez, F.D.M.; Cristiani-Urbina, E.; Cancino-Díaz, J.C.; Flores-Moreno, J.L.; Barragán-Huerta, B.E. Lindane biodegradation by the Fusarium verticillioides AT-100 strain, isolated from Agave tequilana leaves: Kinetic study and identification of metabolites. Int. Biodeter. Biodegrad. 2012, 74, 36–47. [Google Scholar] [CrossRef]
- Kim, O.S.; Cho, Y.J.; Lee, K.; Yoon, S.H.; Kim, M.; Na, H.; Park, S.C. Introducing EzTaxon-e: A prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 2012, 62, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, E.P.; Ng, H.J.; Webb, H.K.; Kurilenko, V.V.; Zhukova, N.V.; Mikhailov, V.V.; Ponamoreva, O.N.; Crawford, R.J. Alteromonas australica sp. nov., isolated from the Tasman Sea. Antonie van Leeuwenhoek 2013, 103, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhang, J.; Lei, X.; Lai, Q.; Yang, L.; Zhang, H.; Li, Y.; Zheng, W.; Tian, Y.; Yu, Z.; et al. Mameliella phaeodactyli sp. nov., a member of the family Rhodobacteraceae isolated from the marine algae Phaeodactylum tricornutum. Int. J. Syst. Evol. Microbiol. 2015, 65, 1617–1621. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, Y.; Kurahashi, M.; Tanaka, K.; Yanagi, K.; Yokota, A.; Harayama, S. Pseudovibrio ascidiaceicola sp. nov., isolated from ascidians (sea squirts). Int. J. Syst. Evol. Microbiol. 2006, 56, 343–347. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.H.; Sheu, S.Y.; Chen, C.A.; Wang, J.T.; Chen, W.M. Oceanicaulis stylophorae sp. nov., isolated from the reef-building coral Stylophora pistillata. Int. J. Syst. Evol. Microbiol. 2012, 62, 2241–2246. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, E.P.; Gorshkova, N.M.; Sawabe, T.; Zhukova, N.V.; Hayashi, K.; Kurilenko, V.V.; Alexeeva, Y.; Buljan, V.; Nicolau, D.V.; Mikhailov, V.V.; et al. Sulfitobacter delicatus sp. nov. and Sulfitobacter dubius sp. nov., respectively from a starfish (Stellaster equestris) and sea grass (Zostera marina). Int. J. Syst. Evol. Microbiol. 2004, 54, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Kim, I.G.; Kang, K.H.; Oh, T.K.; Park, Y.H. Bacillus marisflavi sp. nov. and Bacillus aquimaris sp. nov., isolated from sea water of a tidal flat of the Yellow Sea in Korea. Int. J. Syst. Evol. Microbiol. 2003, 53, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Uchino, Y.; Hirata, A.; Yokota, A.; Sugiyama, J. Reclassification of marine Agrobacterium species: Proposals of Stappia stellulata gen. nov., comb. nov., Stappia aggregata sp. nov., nom. rev., Ruegeria atlantica gen. nov., comb. nov., Ruegeria gelatinovora comb. nov., Ruegeria algicola comb. nov., and Ahrensia kieliense gen. nov., sp. nov., nom. rev. J. Gen. Appl. Microbiol. 1998, 44, 201–210. [Google Scholar] [PubMed]
- Perez, T.; Vacelet, J.; Rebouillon, P. In situ comparative study of several Mediterranean sponges as potential biomonitors of heavy metals. In Sponge Science in the New Millenium; Pansini, M., Pronzato, R., Bavestrello, G., Manconi, R., Eds.; Bollettino dei Musei degli Instituti Biologici dell’Universita di Genova; Officine Grafiche Canessa Rapallo: Genova, Italy, 2004; Volume 68, pp. 517–525. [Google Scholar]
- Hansen, V.; Weeks, J.M.; Depledge, M.H. Accumulation of copper, zinc, cadmium and chromium by the marine sponge Halichondria panicea Pallas and the implications for biomonitoring. Mar. Pollut. Bull. 1995, 31, 133–138. [Google Scholar] [CrossRef]
- Gaino, E.; Cardone, F.; Corriero, G. Reproduction of the intertidal sponge Hymeniacidon perlevis (Montagu) along a bathymetric gradient. Open Mar. Biol. J. 2010, 4, 47–56. [Google Scholar] [CrossRef]
- Taylor, M.W.; Radax, R.; Steger, D.; Wagner, M. Sponge-associated microorganisms: Evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 2007, 71, 295–347. [Google Scholar] [CrossRef] [PubMed]
- Camacho-Pérez, B.; Ríos-Leal, E.; Rinderknecht-Seijas, N.; Poggi-Varaldo, H.M. Enzymes involved in the biodegradation of hexachlorocyclohexane: A mini review. J. Environ. Manag. 2012, 95, 306–318. [Google Scholar] [CrossRef] [PubMed]
- Timmis, K.N.; Pieper, D.H. Bacteria designed for bioremediation. Trends Biotechnol. 1999, 17, 200–204. [Google Scholar] [CrossRef]
- Castle, D.M.; Montgomery, M.T.; Kirchman, D.L. Effects of naphthalene on microbial community composition in the Delaware estuary. FEMS Microbiol. Ecol. 2006, 56, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tam, N. Natural attenuation of contaminated marine sediments from an old floating dock Part II: Changes of sediment microbial community structure and its relationship with environmental variables. Sci. Total Environ. 2012, 423, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Cordova-Kreylos, A.L.; Cao, Y.; Green, P.G.; Hwang, H.M.; Kuivila, K.M.; LaMontagne, M.G.; van de Werfhorst, L.C.; Holden, P.A.; Scow, K.M. Diversity, composition, and geographical distribution of microbial communities in California salt marsh sediments. Appl. Environ. Microbiol. 2006, 72, 3357–3366. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Chaudhary, A.; Kaur, A.; Choudhary, R.; Kaushik, R. Phospholipid fatty acid—A bioindicator of environment monitoring and assessment in soil ecosystem. Curr. Sci. 2005, 89, 1103–1112. [Google Scholar]
- Suzuki, M.T.; Rappé, M.S.; Haimberger, Z.W.; Winfield, H.; Adair, N.; Ströbel, J.; Giovannoni, S.J. Bacterial diversity among small subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl. Environ. Microbiol. 1997, 63, 983–989. [Google Scholar] [PubMed]
- García, M.T.; Ventosa, A.; Mellado, E. Catabolic versatility of aromatic compound-degrading halophilic bacteria. FEMS Microbiol. Ecol. 2005, 54, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.M.; Kim, J.M.; Lee, H.J.; Madsen, E.L.; Jeon, C.O. italic>Alteromonas as a key agent of polycyclic aromatic hydrocarbon biodegradation in crude oil-contaminated coastal sediment. Environ. Sci. Technol. 2012, 14, 7731–7740. [Google Scholar] [CrossRef] [PubMed]
- Catania, V.; Santisi, S.; Signa, G.; Vizzini, S.; Mazzola, A.; Cappello, S.; Yakimov, M.M.; Quatrini, P. Intrinsic bioremediation potential of a chronically polluted marine coastal area. Mar. Pollut. Bull. 2015, 99, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Selvin, J.; Shanmugha Priya, S.; Seghal Kiran, G.; Thangavelu, T.; Sapna Bai, N. Sponge-associated marine bacteria as indicators of heavy metal pollution. Microbiol. Res. 2009, 164, 352–363. [Google Scholar] [CrossRef] [PubMed]
- Buchan, A.; González, J.M.; Moran, M.A. Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 2005, 71, 5665–5677. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Tan, T.; Shao, Z. Roseovarius pacificus sp. nov., isolated from deep-sea sediment. Int. J. Syst. Evol. Microbiol. 2009, 59, 1116–1121. [Google Scholar] [CrossRef] [PubMed]
- Brakstad, O.; Lødeng, A. Microbial diversity during biodegradation of crude oil in seawater from the North Sea. Microb. Ecol. 2005, 49, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Harwati, T.U.; Kasai, Y.; Kodama, Y.; Susilaningsih, D.; Watanabe, K. Tropicibacter naphthalenivorans gen. nov., sp. nov., a polycyclic aromatic hydrocarbon-degrading bacterium isolated from Semarang Port in Indonesia. Int. J. Syst. Evol. Microbiol. 2009, 59, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.J.; Morgan, P.; Weightman, A.J.; Fry, J.C. Cultivation-dependent and independent approaches for determining bacterial diversity in heavy metal contaminated soil. Appl. Environ. Microbiol. 2003, 69, 3223–3230. [Google Scholar] [CrossRef] [PubMed]
- Shivaji, S.; Reddy, G.S.N.; Aduri, R.P.; Kutty, R.; Ravenschlag, K. Bacterial diversity of a soil sample from Schirmacher Oasis, Antarctica. Cell. Mol. Biol. 2004, 50, 525–536. [Google Scholar] [PubMed]
- Lamendella, R.; Strutt, S.; Borglin, S.; Chakraborty, R.; Tas, N.; Mason, O.U.; Hultman, J.; Prestat, E.; Hazen, T.C.; Jansson, J.K. Assessment of the deep-water horizon oil spill impact on gulf coast microbial communities. Front. Microbiol. 2014, 5, 130. [Google Scholar] [CrossRef] [PubMed]
- Janssen, D.B. Evolving haloalkane dehalogenases. Curr. Opin. Chem. Biol. 2004, 8, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Koudelakova, T.; Bidmanova, S.; Dvorak, P.; Pavelka, A.; Chaloupkova, R.; Prokop, Z.; Damborsky, J. Haloalkane dehalogenases: Biotechnology applications. Biotechnol. J. 2013, 8, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Labrenz, M.; Tindall, B.J.; Lawson, P.A.; Collins, M.D.; Schumann, P.; Hirsch, P. Staleya guttiformis gen. nov., sp. nov. and Sulfitobacter brevis sp. nov., α-3-Proteobacteria from hypersaline, heliothermal and meromictic antarctic Ekho Lake. Int. J. Syst. Evol. Microbiol. 2000, 50, 303–313. [Google Scholar] [CrossRef] [PubMed]
- O’Halloran, J.A.; Barbosa, T.M.; Morrissey, J.P.; Kennedy, J.; Dobson, A.D.W.; O’Gara, F. Pseudovibrio axinellae sp. nov., isolated from an Irish marine sponge. Int. J. Syst. Evol. Microbiol. 2013, 63, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Santos, O.C.; Pontes, P.V.; Santos, J.F.; Muricy, G.; Giambiagi-deMarval, M.; Laport, M.S. Isolation, characterization and phylogeny of sponge-associated bacteria with antimicrobial activities from Brazil. Res. Microbiol. 2010, 161, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Muscholl-Silberhorn, A.; Thiel, V.; Imhoff, J.F. Abundance and bioactivity of cultured sponge-associated bacteria from the Mediterranean Sea. Microbiol. Ecol. 2008, 55, 94–106. [Google Scholar] [CrossRef] [PubMed]
- Bondarev, V.; Richter, M.; Romano, S.; Piel, J.; Schwedt, A.; Schulz-Vogt, H.N. The genus Pseudovibrio contains metabolically versatile bacteria adapted for symbiosis. Environ. Microbiol. 2013, 15, 2095–2113. [Google Scholar] [CrossRef] [PubMed]
- Strömpl, C.; Hold, G.L.; Lünsdorf, H.; Graham, J.; Gallacher, S.; Abraham, W.R.; Moore, E.R.; Timmis, K.N. Oceanicaulis alexandrii gen. nov., sp. nov., a novel stalked bacterium isolated from a culture of the dinoflagellate Alexandrium tamarense (Lebour). Balech. Int. J. Syst. Evol. Microbiol. 2003, 53, 1901–1906. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.B.; Liu, C.T.; Anzai, Y.; Kim, H.; Aono, T.; Oyaizu, H. The hierarchical system of the ‘Alphaproteobacteria’: Description of Hyphomonadaceae fam. nov., Xanthobacteraceae fam. nov. and Erythrobacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 2005, 55, 1907–1919. [Google Scholar] [CrossRef] [PubMed]
- Puspasari, F.; Nurachman, Z.; Noer, A.S.; Radjasa, O.K.; van der Maarel, M.J.E.C.; Dessy, N. Characteristics of raw starch degrading a-amylase from Bacillus aquimaris MKSC 6.2 associated with soft coral Sinularia sp. Starch/Stärke 2011, 63, 461–467. [Google Scholar] [CrossRef]
- Trivedi, P.; Duan, Y.; Wang, N. Huanglongbing, a systemic disease, restructures the bacterial community associated with citrus roots. Appl. Environ. Microbiol. 2010, 76, 3427–3436. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.G.; Vijayakumar, L.; Joshi, G.; Magesh, P.D.; Dharani, G.; Kirubagaran, R. Biodegradation of complex hydrocarbons in spent engine oil by novel bacterial consortium isolated from deep-sea sediment. Bioresour. Technol. 2014, 170, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, C.R. Immunological evidence for the Precambrian origin of bacterial symbioses in marine sponges. Proc. R. Soc. Lond. 1984, 220, 509–517. [Google Scholar] [CrossRef]
- Afsar, M.; Radha, S.; Girish, K.; Manonmani, H.K.; Kunhi, A.A.M. Optimization of carbon sources for the preparation of inoculum of hexachlorocyclohexane-degrading microbial consortium. J. Food Sci. Technol. 2005, 42, 209–213. [Google Scholar]
- Longo, C.; Cardone, F.; Corriero, G.; Licciano, M.; Pierri, C.; Stabili, L. The co-occurrence of the demosponge Hymeniacidon perlevis and the edible mussel Mytilus galloprovincialis as a new tool for bacterial load mitigation in aquaculture. Environ. Sci. Pollut. Res. 2016, 23, 3736–3746. [Google Scholar] [CrossRef] [PubMed]
- Corriero, G.; Longo, C.; Mercurio, M.; Nonnis-Marzano, C.; Lembo, G.; Spedicato, M.T. Rearing performance of Spongia officinalis on suspended ropes off the Southern Italian coast (Central Mediterranean Sea). Aquaculture 2004, 238, 195–205. [Google Scholar] [CrossRef]
- Talà, A.; Lenucci, M.S.; Gaballo, A.; Durante, M.; Tredici, S.M.; Debowles, D.A.; Pizzolante, G.; Marcuccio, C.; Carata, E.; Piro, G.; et al. Sphingomonas cynarae sp. nov., a Proteobacterium that produces an unusual type of sphingan. Int. J. Syst. Evol. Microbiol. 2013, 63, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russel, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Vigliotta, G.; Nutricati, E.; Carata, E.; Tredici, S.M.; De Stefano, M.; Pontieri, P.; Massardo, D.R.; Prati, M.V.; De Bellis, L.; Alifano, P. Clonothrix fusca Roze 1896 a filamentous, sheathed, methanotrophic Gamma-Proteobacterium. Appl. Environ. Microbiol. 2007, 73, 3556–3565. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J.; Pace, B.; Olsen, G.J.; Stahl, D.A.; Sogin, M.L.; Pace, N.R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 1985, 82, 6955–6959. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView Version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbour-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.K. Bootstrap hypothesis tests for evolutionary trees and other dendrograms. Proc. Natl. Acad. Sci. USA 1994, 91, 12293–122937. [Google Scholar] [CrossRef] [PubMed]
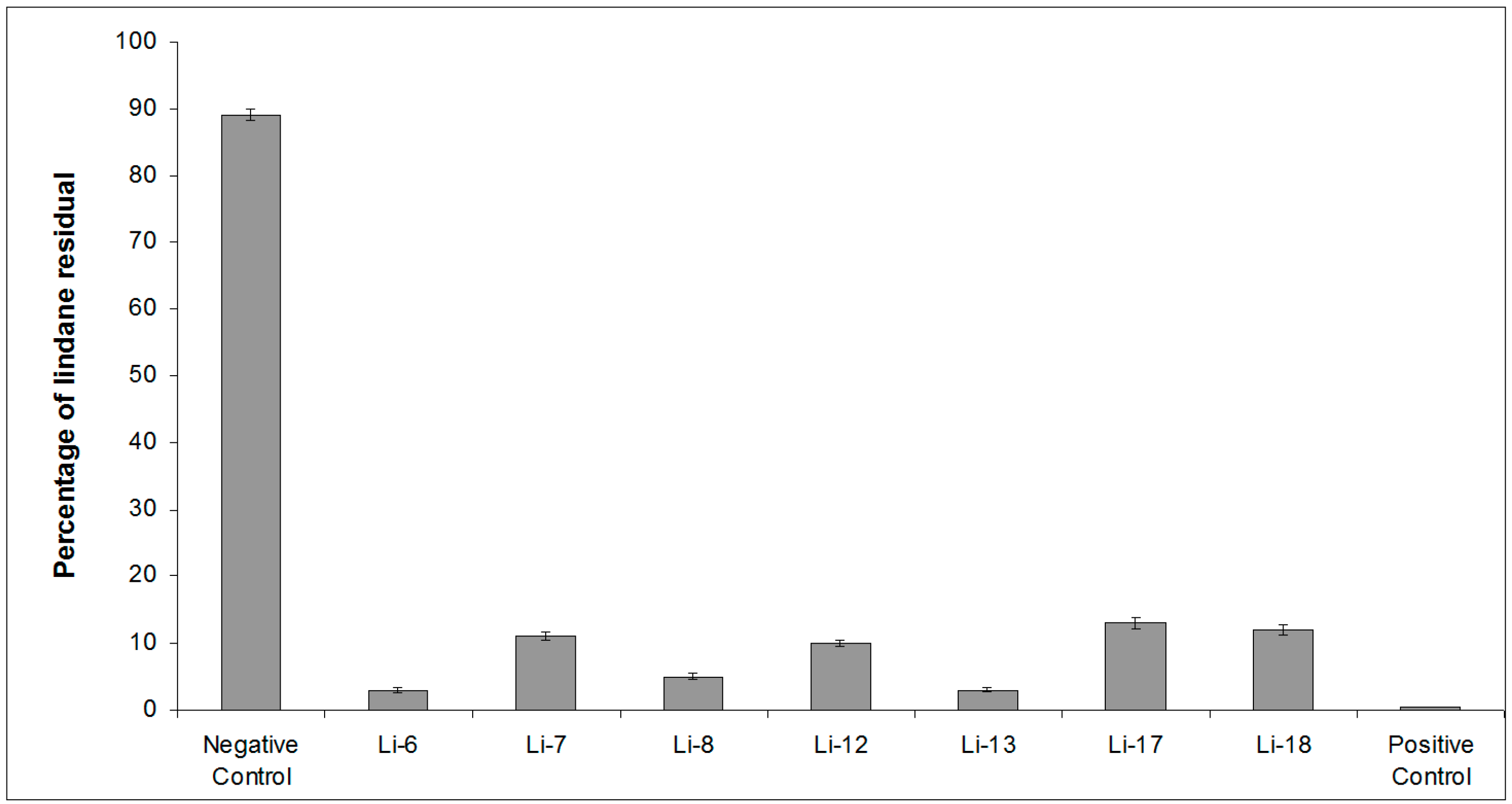
| Bacterial Isolate | Closest Species | Similarity (%) | Lindane Residual (%) |
|---|---|---|---|
| (12 Days) | |||
| Control | 89 ± 4 | ||
| Li-6 | Alteromonas australica CIP 109921T | 97.23 | 3 ± 0.4 |
| Li-7 | Mameliella phaeodactyli KCTC 42178T | 99.63 | 11 ± 0.6 |
| Li-8 | Pseudovibrio ascidiaceicola NBRC 100514T | 100 | 5 ± 0.5 |
| Li-12 | Oceanicaulis stylophorae LMG 2723T | 99.34 | 10 ± 0.5 |
| Li-13 | Sulfitobacter dubius KMM 3584T | 98.34 | 3 ± 0.3 |
| Li-17 | Bacillus aquimaris JCM 11544T | 98.67 | 13 ± 0.7 |
| Li-18 | Ruegeria atlantica NBRC 15792T | 99.12 | 12 ± 0.7 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loredana, S.; Graziano, P.; Antonio, M.; Carlotta, N.M.; Caterina, L.; Maria, A.A.; Carlo, Z.; Giuseppe, C.; Pietro, A. Lindane Bioremediation Capability of Bacteria Associated with the Demosponge Hymeniacidon perlevis. Mar. Drugs 2017, 15, 108. https://doi.org/10.3390/md15040108
Loredana S, Graziano P, Antonio M, Carlotta NM, Caterina L, Maria AA, Carlo Z, Giuseppe C, Pietro A. Lindane Bioremediation Capability of Bacteria Associated with the Demosponge Hymeniacidon perlevis. Marine Drugs. 2017; 15(4):108. https://doi.org/10.3390/md15040108
Chicago/Turabian StyleLoredana, Stabili, Pizzolante Graziano, Morgante Antonio, Nonnis Marzano Carlotta, Longo Caterina, Aresta Antonella Maria, Zambonin Carlo, Corriero Giuseppe, and Alifano Pietro. 2017. "Lindane Bioremediation Capability of Bacteria Associated with the Demosponge Hymeniacidon perlevis" Marine Drugs 15, no. 4: 108. https://doi.org/10.3390/md15040108
APA StyleLoredana, S., Graziano, P., Antonio, M., Carlotta, N. M., Caterina, L., Maria, A. A., Carlo, Z., Giuseppe, C., & Pietro, A. (2017). Lindane Bioremediation Capability of Bacteria Associated with the Demosponge Hymeniacidon perlevis. Marine Drugs, 15(4), 108. https://doi.org/10.3390/md15040108










