Spotlight on Antimicrobial Metabolites from the Marine Bacteria Pseudoalteromonas: Chemodiversity and Ecological Significance
Abstract
:1. Introduction
2. Antimicrobial Metabolites from Pseudoalteromonas Species
2.1. Alkaloids
2.2. Polyketides
2.3. Non Ribosomally Synthesized Peptides
2.4. Bacteriocins and Bacteriocin-Like Inhibitory Substances (BLIS)
2.5. Uncharacterized Chemistry of Antimicrobial Metabolites Produced by Pseudoalteromonas Species
2.6. Genome Mining Strategies as a Tool to Discover Antibiotics in Pseudoalteromonas
3. Ecological Significance in Marine Life
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Gauthier, G.; Gauthier, M.; Christen, R. Phylogenetic analysis of the genera Alteromonas, Shewanella, and Moritella using genes coding for small-subunit rRNA sequences and division of the genus Alteromonas into two genera, Alteromonas (emended) and Pseudoalteromonas gen. nov., and proposal of twelve new species combinations. Int. J. Syst. Bacteriol. 1995, 45, 755–761. [Google Scholar] [PubMed]
- Ivanova, E.P.; Kiprianova, E.A.; Mikhailov, V.V.; Levanova, G.F.; Garagulya, A.D.; Gorshkova, N.M.; Vysotskii, M.V.; Nicolau, D.V.; Yumoto, N.; Taguchi, T.; et al. Phenotypic diversity of Pseudoalteromonas citrea from different marine habitats and emendation of the description. Int. J. Syst. Evol. Microbiol. 1998, 48, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Austin, B. Environmental issues in the control of bacterial diseases of farmed fish. In Environment and Aquaculture in Developing Countries; ICLARM: Penang, Malaysia, 1993; pp. 237–251. [Google Scholar]
- Choudhury, J.D.; Pramanik, A.; Webster, N.S.; Llewellyn, L.E.; Gachhui, R.; Mukherjee, J. The Pathogen of the Great Barrier Reef Sponge Rhopaloeides odorabile is a New Strain of Pseudoalteromonas agarivorans Containing Abundant and Diverse Virulence-Related Genes. Mar. Biotechnol. 2015, 17, 463–478. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.P. Bioactive Compound Synthetic Capacity and Ecological Significance of Marine Bacterial Genus Pseudoalteromonas. Mar. Drugs 2007, 5, 220–241. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, E.; Sharon, G.; Zilber-Rosenberg, I. The hologenome theory of evolution contains Lamarckian aspects within a Darwinian framework. Environ. Microbiol. 2009, 11, 2959–2962. [Google Scholar] [CrossRef] [PubMed]
- Zilber-Rosenberg, I.; Rosenberg, E. Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution. FEMS Microbiol. Rev. 2008, 32, 723–735. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, E.; Zilber-Rosenberg, I. Microbes Drive Evolution of Animals and Plants: The Hologenome Concept. mBio 2016, 7, e01395-15. [Google Scholar] [CrossRef] [PubMed]
- Theis, K.R.; Dheilly, N.M.; Klassen, J.L.; Brucker, R.M.; Baines, J.F.; Bosch, T.C.G.; Cryan, J.F.; Gilbert, S.F.; Goodnight, C.J.; Lloyd, E.A.; et al. Getting the Hologenome Concept Right: An Eco-Evolutionary Framework for Hosts and Their Microbiomes. mSystems 2016, 1, e00028-16. [Google Scholar] [CrossRef]
- Brucker, R.M.; Bordenstein, S.R. The capacious hologenome. Zool. Jena Ger. 2013, 116, 260–261. [Google Scholar] [CrossRef] [PubMed]
- Kelecom, A. Secondary metabolites from marine microorganisms. An. Acad. Bras. Ciênc. 2002, 74, 151–170. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, D.J. Marine natural products. Nat. Prod. Rep. 2001, 18, 1–49. [Google Scholar] [CrossRef] [PubMed]
- Mansson, M.; Gram, L.; Larsen, T.O. Production of bioactive secondary metabolites by marine Vibrionaceae. Mar. Drugs 2011, 9, 1440–1468. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-L.; Li, M.; Yu, Z.; Qian, P.-Y. Correlation between pigmentation and larval settlement deterrence by Pseudoalteromonas sp. sf57. Biofouling 2011, 27, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Vynne, N.G.; Mansson, M.; Nielsen, K.F.; Gram, L. Bioactivity, chemical profiling, and 16S rRNA-based phylogeny of Pseudoalteromonas strains collected on a global research cruise. Mar. Biotechnol. 2011, 13, 1062–1073. [Google Scholar] [CrossRef] [PubMed]
- Fehér, D.; Barlow, R.S.; Lorenzo, P.S.; Hemscheidt, T.K. A 2-Substituted Prodiginine, 2-(p-Hydroxybenzyl)prodigiosin, from Pseudoalteromonas rubra. J. Nat. Prod. 2008, 71, 1970–1972. [Google Scholar] [CrossRef] [PubMed]
- Lattasch, H.; Thomson, R.H. A revised structure for cycloprodigiosin. Tetrahedron Lett. 1983, 24, 2701–2704. [Google Scholar] [CrossRef]
- Gerber, N.N. Prodigiosin-like Pigments from Actinomadura (Nocardia) pelletieri and Actinomadura madurae. Appl. Microbiol. 1969, 18, 1–3. [Google Scholar] [PubMed]
- Gerber, N.N. Prodigiosin-like pigments. CRC Crit. Rev. Microbiol. 1975, 3, 469–485. [Google Scholar] [CrossRef] [PubMed]
- Franks, A.; Haywood, P.; Holmström, C.; Egan, S.; Kjelleberg, S.; Kumar, N. Isolation and structure elucidation of a novel yellow pigment from the marine bacterium Pseudoalteromonas tunicata. Molecules 2005, 10, 1286–1291. [Google Scholar] [CrossRef] [PubMed]
- Holmström, C.; James, S.; Egan, S.; Kjelleberg, S. Inhibition of common fouling organisms by marine bacterial isolates ith special reference to the role of pigmented bacteria. Biofouling 1996, 10, 251–259. [Google Scholar] [CrossRef] [PubMed]
- De Luca, V.; Salim, V.; Thamm, A.; Masada, S.A.; Yu, F. Making iridoids/secoiridoids and monoterpenoid indole alkaloids: Progress on pathway elucidation. Curr. Opin. Plant Biol. 2014, 19, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Gribble, G.W. The diversity of naturally occurring organobromine compounds. Chem. Soc. Rev. 1999, 28, 335–346. [Google Scholar] [CrossRef]
- Neumann, C.S.; Fujimori, D.G.; Walsh, C.T. Halogenation strategies in natural product biosynthesis. Chem. Biol. 2008, 15, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Runguphan, W.; Qu, X.; O’Connor, S.E. Integrating carbon-halogen bond formation into medicinal plant metabolism. Nature 2010, 468, 461–464. [Google Scholar] [CrossRef] [PubMed]
- Fer, W. Natural Products Chemistry the Marine Environme. SCIENCE 1982, 215, 19. [Google Scholar]
- Tebben, J.; Motti, C.; Tapiolas, D.; Thomas-Hall, P.; Harder, T. A Coralline Algal-Associated Bacterium, Pseudoalteromonas Strain J010, Yields Five New Korormicins and a Bromopyrrole. Mar. Drugs 2014, 12, 2802–2815. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, K.; Takadera, T.; Adachi, K.; Nishijima, M.; Sanc, H. Korormicin, a novel antibiotic specifically active against marine gram-negative bacteria, produced by a marine bacterium. J. Antibiot. 1997, 50, 949–953. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, K.; Nakayama, Y.; Hayashi, M.; Unemoto, T.; Mochida, K. Korormicin, an antibiotic specific for gram-negative marine bacteria, strongly inhibits the respiratory chain-linked Na+-translocating NADH: quinone reductase from the marine Vibrio alginolyticus. J. Antibiot. 1999, 52, 182–185. [Google Scholar] [CrossRef] [PubMed]
- Egan, S.; James, S.; Holmström, C.; Kjelleberg, S. Correlation between pigmentation and antifouling compounds produced by Pseudoalteromonas tunicata. Environ. Microbiol. 2002, 4, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Egan, S.; James, S.; Kjelleberg, S. Identification and Characterization of a Putative Transcriptional Regulator Controlling the Expression of Fouling Inhibitors in Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 2002, 68, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Holmström, C.; James, S.; Neilan, B.A.; White, D.C.; Kjelleberg, S. Pseudoalteromonas tunicata sp. nov., a bacterium that produces antifouling agents. Int. J. Syst. Bacteriol. 1998, 48, 1205–1212. [Google Scholar] [CrossRef] [PubMed]
- Holmström, C.; Kjelleberg, S. Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol. Ecol. 1999, 30, 285–293. [Google Scholar] [CrossRef]
- Pinkerton, D.M.; Banwell, M.G.; Garson, M.J.; Kumar, N.; de Moraes, M.O.; Cavalcanti, B.C.; Barros, F.W.A.; Pessoa, C. Antimicrobial and Cytotoxic Activities of Synthetically Derived Tambjamines C and E–J, BE-18591, and a Related Alkaloid from the Marine Bacterium Pseudoalteromonas tunicata. Chem. Biodivers. 2010, 7, 1311–1324. [Google Scholar] [CrossRef] [PubMed]
- Burke, C.; Thomas, T.; Egan, S.; Kjelleberg, S. The use of functional genomics for the identification of a gene cluster encoding for the biosynthesis of an antifungal tambjamine in the marine bacterium Pseudoalteromonas tunicata. Environ. Microbiol. 2007, 9, 814–818. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, F.; Zhu, X.; Yan, Y.; Yu, X.; Jiang, P.; Xing, X.-H. Biosynthesis and characterization of violacein, deoxyviolacein and oxyviolacein in heterologous host, and their antimicrobial activities. Biochem. Eng. J. 2012, 67, 148–155. [Google Scholar] [CrossRef]
- Yang, L.H.; Xiong, H.; Lee, O.O.; Qi, S.-H.; Qian, P.-Y. Effect of agitation on violacein production in Pseudoalteromonas luteoviolacea isolated from a marine sponge. Lett. Appl. Microbiol. 2007, 44, 625–630. [Google Scholar] [CrossRef] [PubMed]
- Hurdle, J.G.; O’Neill, A.J.; Chopra, I. Anti-staphylococcal activity of indolmycin, a potential topical agent for control of staphylococcal infections. J. Antimicrob. Chemother. 2004, 54, 549–552. [Google Scholar] [CrossRef] [PubMed]
- Wratten, S.J.; Wolfe, M.S.; Andersen, R.J.; Faulkner, D.J. Antibiotic Metabolites from a Marine Pseudomonad. Antimicrob. Agents Chemother. 1977, 11, 411–414. [Google Scholar] [CrossRef] [PubMed]
- Del Castillo, C.S.; Wahid, M.I.; Takeshi, Y.; Taizo, S. Isolation and inhibitory effect of anti-Vibrio substances from Pseudoalteromonas sp. A1-J11 isolated from the coastal sea water of Kagoshima Bay. Fish. Sci. 2008, 74, 174–179. [Google Scholar] [CrossRef]
- Sakata, T.; Sakaguchi, K.; Kakimoto, D. Antiobiotic production by marine pigmented bacteria. I. Antibacterial effect of Alteromonas luteoviolaceus. Mem. Fac. Fish. Kagoshima Univ. 1982, 31, 243–250. [Google Scholar]
- Sakata, T.; Sakaguchi, K.; Kakimota, D. Antibiotic production by marine pigmented bacteria. II. Purification and characterization of antibiotic substance of Alteromonas luteoviolacea. Mem. Fac. Fish. 1986, 35, 29–37. [Google Scholar]
- Zheng, L.; Chen, H.; Han, X.; Lin, W.; Yan, X. Antimicrobial screening and active compound isolation from marine bacterium NJ6-3-1 associated with the sponge Hymeniacidon perleve. World J. Microbiol. Biotechnol. 2005, 21, 201–206. [Google Scholar] [CrossRef]
- Yu, M.; Wang, J.; Tang, K.; Shi, X.; Wang, S.; Zhu, W.-M.; Zhang, X.-H. Purification and characterization of antibacterial compounds of Pseudoalteromonas flavipulchra JG1. Microbiol. Read. Engl. 2012, 158, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, M.; Wada, H. The bacterial oxidation of indole. Biochim. Biophys. Acta 1968, 158, 70–78. [Google Scholar] [CrossRef]
- Glover, V.; Halket, J.M.; Watkins, P.J.; Clow, A.; Goodwin, B.L.; Sandler, M. Isatin: Identity with the purified endogenous monoamine oxidase inhibitor tribulin. J. Neurochem. 1988, 51, 656–659. [Google Scholar] [CrossRef] [PubMed]
- Kalinovskaya, N.I.; Ivanova, E.P.; Alexeeva, Y.V.; Gorshkova, N.M.; Kuznetsova, T.A.; Dmitrenok, A.S.; Nicolau, D.V. Low-molecular-weight, biologically active compounds from marine Pseudoalteromonas species. Curr. Microbiol. 2004, 48, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Moree, W.J.; McConnell, O.J.; Nguyen, D.D.; Sanchez, L.M.; Yang, Y.-L.; Zhao, X.; Liu, W.-T.; Boudreau, P.D.; Srinivasan, J.; Atencio, L.; et al. Microbiota of Healthy Corals Are Active against Fungi in a Light-Dependent Manner. ACS Chem. Biol. 2014, 9, 2300–2308. [Google Scholar] [CrossRef] [PubMed]
- Andersen, R.J.; Wolfe, M.S.; Faulkner, D.J. Autotoxic antibiotic production by a marine Chromobacterium. Mar. Biol. 1974, 27, 281–285. [Google Scholar] [CrossRef]
- Agarwal, V.; El Gamal, A.A.; Yamanaka, K.; Poth, D.; Kersten, R.D.; Schorn, M.; Allen, E.E.; Moore, B.S. Biosynthesis of polybrominated aromatic organic compounds by marine bacteria. Nat. Chem. Biol. 2014, 10, 640–647. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Hansen, P.E.; Lin, X. Bromophenols in Marine Algae and Their Bioactivities. Mar. Drugs 2011, 9, 1273–1292. [Google Scholar] [CrossRef] [PubMed]
- Zsolnai, T. Versuche zur entdeckung neuer fungistatika-I: Phenol-derivate. Biochem. Pharmacol. 1960, 5, 1–19. [Google Scholar] [CrossRef]
- Burkholder, P.R.; Pfister, R.M.; Leitz, F.H. Production of a pyrrole antibiotic by a marine bacterium. Appl. Microbiol. 1966, 14, 649–653. [Google Scholar] [PubMed]
- Lovell, F.M. The Structure of a Bromine-Rich Marine Antibiotic. J. Am. Chem. Soc. 1966, 88, 4510–4511. [Google Scholar] [CrossRef]
- Fehér, D.; Barlow, R.; McAtee, J.; Hemscheidt, T.K. Highly Brominated Antimicrobial Metabolites from a Marine Pseudoalteromonas sp. J. Nat. Prod. 2010, 73, 1963–1966. [Google Scholar] [CrossRef] [PubMed]
- Ridley, C.P.; Lee, H.Y.; Khosla, C. Evolution of polyketide synthases in bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 4595–4600. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Cheng, Y.-Q.; Christenson, S.D.; Jiang, H.; Ju, J.; Kwon, H.-J.; Lim, S.-K.; Liu, W.; Nonaka, K.; Seo, J.-W.; et al. Polyketide Biosynthesis beyond the Type I, II, and III Polyketide Synthase Paradigms: A Progress Report. In Polyketides; American Chemical Society: Washington, DC, USA, 2007; Volume 955, pp. 154–166. [Google Scholar]
- Katsuyama, Y.; Ohnishi, Y. Type III polyketide synthases in microorganisms. Methods Enzymol. 2012, 515, 359–377. [Google Scholar] [PubMed]
- Zhang, H.; Wang, Y.; Wu, J.; Skalina, K.; Pfeifer, B.A. Complete Biosynthesis of Erythromycin A and Designed Analogs Using E. coli as a Heterologous Host. Chem. Biol. 2010, 17, 1232–1240. [Google Scholar] [CrossRef] [PubMed]
- Pickens, L.B.; Tang, Y. Decoding and Engineering Tetracycline Biosynthesis. Metab. Eng. 2009, 11, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Murphy, A.C.; Fukuda, D.; Song, Z.; Hothersall, J.; Cox, R.J.; Willis, C.L.; Thomas, C.M.; Simpson, T.J. Engineered Thiomarinol Antibiotics Active against MRSA Are Generated by Mutagenesis and Mutasynthesis of Pseudoalteromonas SANK73390. Angew. Chem. Int. Ed. 2011, 50, 3271–3274. [Google Scholar] [CrossRef] [PubMed]
- Isnansetyo, A.; Kamei, Y. MC21-A, a Bactericidal Antibiotic Produced by a New Marine Bacterium, Pseudoalteromonas phenolica sp. nov. O-BC30T, against Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2003, 47, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Boyd, K.G.; Mearns-spragg, A.; Adams, D.R.; Wright, P.C.; Burgess, J.G. Two Diketopiperazines and One Halogenated Phenol from Cultures of the Marine Bacterium, Pseudoalteromonas luteoviolacea. Nat. Prod. Lett. 2006, 14, 435–440. [Google Scholar] [CrossRef]
- Shiozawa, H.; Kagasaki, T.; Kinoshita, T.; Haruyama, H.; Domon, H.; Utsui, Y.; Kodama, K.; Takahashi, S. Thiomarinol, a new hybrid antimicrobial antibiotic produced by a marine bacterium. Fermentation, isolation, structure, and antimicrobial activity. J. Antibiot. 1993, 46, 1834–1842. [Google Scholar] [CrossRef] [PubMed]
- Shiozawa, H.; Shimada, A.; Takahashi, S. Thiomarinols D, E, F and G, new hybrid antimicrobial antibiotics produced by a marine bacterium; isolation, structure, and antimicrobial activity. J. Antibiot. 1997, 50, 449–452. [Google Scholar] [CrossRef] [PubMed]
- Shiozawa, H.; Kagasaki, T.; Torikata, A.; Tanaka, N.; Fujimoto, K.; Hata, T.; Furukawa, Y.; Takahashi, S. Thiomarinols B and C, new antimicrobial antibiotics produced by a marine bacterium. J. Antibiot. 1995, 48, 907–909. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Huang, S.; Yu, Y.; Deng, H. Dithiolopyrrolone Natural Products: Isolation, Synthesis and Biosynthesis. Mar. Drugs 2013, 11, 3970–3997. [Google Scholar] [CrossRef] [PubMed]
- Strieker, M.; Tanović, A.; Marahiel, M.A. Nonribosomal peptide synthetases: Structures and dynamics. Curr. Opin. Struct. Biol. 2010, 20, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Challis, G.L.; Naismith, J.H. Structural aspects of non-ribosomal peptide biosynthesis. Curr. Opin. Struct. Biol. 2004, 14, 748–756. [Google Scholar] [CrossRef] [PubMed]
- Koglin, A.; Walsh, C.T. Structural insights into nonribosomal peptide enzymatic assembly lines. Nat. Prod. Rep. 2009, 26, 987–1000. [Google Scholar] [CrossRef] [PubMed]
- Desriac, F.; Jégou, C.; Balnois, E.; Brillet, B.; Chevalier, P.L.; Fleury, Y. Antimicrobial Peptides from Marine Proteobacteria. Mar. Drugs 2013, 11, 3632–3660. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.D.; Wu, C.-H.; Moree, W.J.; Lamsa, A.; Medema, M.H.; Zhao, X.; Gavilan, R.G.; Aparicio, M.; Atencio, L.; Jackson, C.; et al. MS/MS networking guided analysis of molecule and gene cluster families. Proc. Natl. Acad. Sci. USA 2013, 110, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Speitling, M.; Smetanina, O.F.; Kuznetsova, T.A.; Laatsch, H. Bromoalterochromides A and A’, unprecedented chromopeptides from a marine Pseudoalteromonas maricaloris strain KMM 636T. J. Antibiot. 2007, 60, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Sobolevskaya, M.P.; Smetanina, O.F.; Speitling, M.; Shevchenko, L.S.; Dmitrenok, P.S.; Laatsch, H.; Kuznetsova, T.A.; Ivanova, E.P.; Elyakov, G.B. Controlling production of brominated cyclic depsipeptides by Pseudoalteromonas maricaloris KMM 636T. Lett. Appl. Microbiol. 2005, 40, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Rungprom, W.; Siwu, E.R.O.; Lambert, L.K.; Dechsakulwatana, C.; Barden, M.C.; Kokpol, U.; Blanchfield, J.T.; Kita, M.; Garson, M.J. Cyclic tetrapeptides from marine bacteria associated with the seaweed Diginea sp. and the sponge Halisarca ectofibrosa. Tetrahedron 2008, 64, 3147–3152. [Google Scholar] [CrossRef]
- Nakano, M.; Danowski, T.S.; Weitzel, W. The technical assistance of D. R. Crystalline Mammalian l-Amino Acid Oxidase from Rat Kidney Mitochondria. J. Biol. Chem. 1966, 241, 2075–2083. [Google Scholar] [PubMed]
- Yu, Z.; Wang, J.; Lin, J.; Zhao, M.; Qiu, J. Exploring regulation genes involved in the expression of l-amino acid oxidase in Pseudoalteromonas sp. Rf-1. PLoS ONE 2015, 10, e0122741. [Google Scholar] [CrossRef] [PubMed]
- Gómez, D.; Espinosa, E.; Bertazzo, M.; Lucas-Elío, P.; Solano, F.; Sanchez-Amat, A. The macromolecule with antimicrobial activity synthesized by Pseudoalteromonas luteoviolacea strains is an l-amino acid oxidase. Appl. Microbiol. Biotechnol. 2008, 79, 925–930. [Google Scholar] [CrossRef] [PubMed]
- James, S.G.; Holmström, C.; Kjelleberg, S. Purification and characterization of a novel antibacterial protein from the marine bacterium D2. Appl. Environ. Microbiol. 1996, 62, 2783–2788. [Google Scholar] [PubMed]
- Chen, W.M.; Lin, C.Y.; Chen, C.A.; Wang, J.T.; Sheu, S.Y. Involvement of an l-amino acid oxidase in the activity of the marine bacterium Pseudoalteromonas flavipulchra against methicillin-resistant Staphylococcus aureus. Enzyme Microb. Technol. 2010, 47, 52–58. [Google Scholar] [CrossRef]
- Yu, M.; Tang, K.; Shi, X.; Zhang, X.-H. Genome Sequence of Pseudoalteromonas flavipulchra JG1, a Marine Antagonistic Bacterium with Abundant Antimicrobial Metabolites. J. Bacteriol. 2012, 194, 3735. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, S.A.; Johnson, R.M.; Kakimoto, D. Characterization of an antibiotic produced by Alteromonas luteoviolacea Gauthier 1982, 85 isolated from Kinko Bay, Japan. J. Appl. Bacteriol. 1994, 77, 426–432. [Google Scholar] [CrossRef] [PubMed]
- Defer, D.; Bourgougnon, N.; Fleury, Y. Screening for antibacterial and antiviral activities in three bivalve and two gastropod marine molluscs. Aquaculture 2009, 293, 1–7. [Google Scholar] [CrossRef]
- Aranda, C.P.; Valenzuela, C.; Barrientos, J.; Paredes, J.; Leal, P.; Maldonado, M.; Godoy, F.A.; Osorio, C.G. Bacteriostatic anti-Vibrio parahaemolyticus activity of Pseudoalteromonas sp. strains DIT09, DIT44 and DIT46 isolated from Southern Chilean intertidal Perumytilus purpuratus. World J. Microbiol. Biotechnol. 2012, 28, 2365–2374. [Google Scholar] [CrossRef] [PubMed]
- Barja, J.L.; Lemos, M.L.; Toranzo, A.E. Purification and characterization of an antibacterial substance produced by a marine Alteromonas species. Antimicrob. Agents Chemother. 1989, 33, 1674–1679. [Google Scholar] [CrossRef] [PubMed]
- Chelossi, E.; Milanese, M.; Milano, A.; Pronzato, R.; Riccardi, G. Characterisation and antimicrobial activity of epibiotic bacteria from Petrosia ficiformis (Porifera, Demospongiae). J. Exp. Mar. Biol. Ecol. 2004, 309, 21–33. [Google Scholar] [CrossRef]
- Hentschel, U.; Schmid, M.; Wagner, M.; Fieseler, L.; Gernert, C.; Hacker, J. Isolation and phylogenetic analysis of bacteria with antimicrobial activities from the Mediterranean sponges Aplysina aerophoba and Aplysina cavernicola. FEMS Microbiol. Ecol. 2001, 35, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Holmström, C.; Rittschof, D.; Kjelleberg, S. Inhibition of Settlement by Larvae of Balanus amphitrite and Ciona intestinalis by a Surface-Colonizing Marine Bacterium. Appl. Environ. Microbiol. 1992, 58, 2111–2115. [Google Scholar] [PubMed]
- Egan, S.; James, S.; Holmström, C.; Kjelleberg, S. Inhibition of algal spore germination by the marine bacterium Pseudoalteromonas tunicata. FEMS Microbiol. Ecol. 2001, 35, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Buck, J.D.; Meyers, S.P. In vitro inhibition of Rhodotorula minuta by a variant of the marine bacterium, Pseudomonas piscicida. Helgoländer Wiss. Meeresunters. 1966, 13, 171–180. [Google Scholar] [CrossRef]
- Longeon, A.; Peduzzi, J.; Barthelemy, M.; Corre, S.; Nicolas, J.-L.; Guyot, M. Purification and Partial Identification of Novel Antimicrobial Protein from Marine Bacterium Pseudoalteromonas Species Strain X153. Mar. Biotechnol. 2004, 6, 633–641. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, M.J. Alteromonas citrea, a New Gram-Negative, Yellow-Pigmented Species from Seawater. Int. J. Syst. Bacteriol. 1977, 27, 349–354. [Google Scholar] [CrossRef]
- Gauthier, M.J.; Breittmayer, V.A. A New Antibiotic-Producing Bacterium from Seawater: Alteromonas aurantia sp. nov. Int. J. Syst. Bacteriol. 1979, 29, 366–372. [Google Scholar] [CrossRef]
- Scheffler, R.J.; Colmer, S.; Tynan, H.; Demain, A.L.; Gullo, V.P. Antimicrobials, drug discovery, and genome mining. Appl. Microbiol. Biotechnol. 2013, 97, 969–978. [Google Scholar] [CrossRef] [PubMed]
- Katz, L.; Baltz, R.H. Natural product discovery: past, present, and future. J. Ind. Microbiol. Biotechnol. 2016, 43, 155–176. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. antiSMASH 3.0—A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Ziemert, N.; Podell, S.; Penn, K.; Badger, J.H.; Allen, E.; Jensen, P.R. The Natural Product Domain Seeker NaPDoS: A Phylogeny Based Bioinformatic Tool to Classify Secondary Metabolite Gene Diversity. PLoS ONE 2012, 7, e34064. [Google Scholar] [CrossRef] [PubMed]
- Anand, S.; Prasad, M.V.R.; Yadav, G.; Kumar, N.; Shehara, J.; Ansari, M.Z.; Mohanty, D. SBSPKS: Structure based sequence analysis of polyketide synthases. Nucleic Acids Res. 2010. [Google Scholar] [CrossRef] [PubMed]
- Li, M.H.; Ung, P.M.; Zajkowski, J.; Garneau-Tsodikova, S.; Sherman, D.H. Automated genome mining for natural products. BMC Bioinform. 2009, 10, 185. [Google Scholar] [CrossRef] [PubMed]
- Stachelhaus, T.; Mootz, H.D.; Marahiel, M.A. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 1999, 6, 493–505. [Google Scholar] [CrossRef]
- Röttig, M.; Medema, M.H.; Blin, K.; Weber, T.; Rausch, C.; Kohlbacher, O. NRPSpredictor2—A web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 2011. [Google Scholar] [CrossRef] [PubMed]
- Khayatt, B.I.; Overmars, L.; Siezen, R.J.; Francke, C. Classification of the Adenylation and Acyl-Transferase Activity of NRPS and PKS Systems Using Ensembles of Substrate Specific Hidden Markov Models. PLoS ONE 2013, 8, e62136. [Google Scholar] [CrossRef] [PubMed]
- Van Heel, A.J.; de Jong, A.; Montalbán-López, M.; Kok, J.; Kuipers, O.P. BAGEL3: Automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res. 2013, 41, W448–W453. [Google Scholar] [CrossRef] [PubMed]
- Amoutzias, G.D.; Chaliotis, A.; Mossialos, D. Discovery Strategies of Bioactive Compounds Synthesized by Nonribosomal Peptide Synthetases and Type-I Polyketide Synthases Derived from Marine Microbiomes. Mar. Drugs 2016, 14, 80. [Google Scholar] [CrossRef] [PubMed]
- Chávez, R.; Fierro, F.; García-Rico, R.O.; Vaca, I. Filamentous fungi from extreme environments as a promising source of novel bioactive secondary metabolites. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Tietz, J.; Mitchell, D. Using Genomics for Natural Product Structure Elucidation. Curr. Top. Med. Chem. 2015, 16, 1645–1694. [Google Scholar] [CrossRef]
- Zerikly, M.; Challis, G.L. Strategies for the Discovery of New Natural Products by Genome Mining. ChemBioChem 2009, 10, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Challis, G.L. Genome Mining for Novel Natural Product Discovery. J. Med. Chem. 2008, 51, 2618–2628. [Google Scholar] [CrossRef] [PubMed]
- Zarins-Tutt, J.S. Gene Mining of Biosynthesis Genes and Biosynthetic Manipulation of Marine Bacteria for the Production of New Antibiotic Candidates. Ph.D. Thesis, University of St Andrews, Scotland, UK, 2015. [Google Scholar]
- Machado, H.; Sonnenschein, E.C.; Melchiorsen, J.; Gram, L. Genome mining reveals unlocked bioactive potential of marine Gram-negative bacteria. BMC Genom. 2015, 16, 158. [Google Scholar] [CrossRef] [PubMed]
- Papaleo, M.C.; Fondi, M.; Maida, I.; Perrin, E.; Lo Giudice, A.; Michaud, L.; Mangano, S.; Bartolucci, G.; Romoli, R.; Fani, R. Sponge-associated microbial Antarctic communities exhibiting antimicrobial activity against Burkholderia cepacia complex bacteria. Biotechnol. Adv. 2012, 30, 272–293. [Google Scholar] [CrossRef] [PubMed]
- Maansson, M.; Vynne, N.G.; Klitgaard, A.; Nybo, J.L.; Melchiorsen, J.; Nguyen, D.D.; Sanchez, L.M.; Ziemert, N.; Dorrestein, P.C.; Andersen, M.R.; et al. An Integrated Metabolomic and Genomic Mining Workflow to Uncover the Biosynthetic Potential of Bacteria. mSystems 2016, 1, e00028–15. [Google Scholar] [CrossRef]
- Wang, P.; Yu, Z.; Li, B.; Cai, X.; Zeng, Z.; Chen, X.; Wang, X. Development of an efficient conjugation-based genetic manipulation system for Pseudoalteromonas. Microb. Cell Fact. 2015, 14, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Duilio, A.; Tutino, M.L.; Marino, G. Recombinant protein production in Antarctic Gram-negative bacteria. Recomb. Gene Expr. Rev. Protoc. 2004. [Google Scholar] [CrossRef]
- Giuliani, M.; Parrilli, E.; Pezzella, C.; Rippa, V.; Duilio, A.; Marino, G.; Tutino, M.L. A Novel Strategy for the Construction of Genomic Mutants of the Antarctic Bacterium Pseudoalteromonas haloplanktis TAC125. In Recombinant Gene Expression; Lorence, A., Ed.; Humana Press: Totowa, NJ, USA, 2012; Volume 824, pp. 219–233. [Google Scholar]
- Yu, Z.-C.; Zhao, D.-L.; Ran, L.-Y.; Mi, Z.-H.; Wu, Z.-Y.; Pang, X.; Zhang, X.-Y.; Su, H.-N.; Shi, M.; Song, X.-Y.; et al. Development of a genetic system for the deep-sea psychrophilic bacterium Pseudoalteromonas sp. SM9913. Microb. Cell Fact. 2014, 13. [Google Scholar] [CrossRef] [PubMed]
- Miranda, C.D.; Rojas, R.; Geisse, J.; Romero, J.; González-Rocha, G. Scallop larvae hatcheries as source of bacteria carrying genes encoding for non-enzymatic phenicol resistance. Mar. Pollut. Bull. 2015, 95, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Bakunina, I.Y.; Nedashkovskaya, O.I.; Alekseeva, S.A.; Ivanova, E.P.; Romanenko, L.A.; Gorshkova, N.M.; Isakov, V.V.; Zvyagintseva, T.N.; Mikhailov, V.V. Degradation of Fucoidan by the Marine Proteobacterium Pseudoalteromonas citrea. Microbiology 2002, 71, 41–47. [Google Scholar] [CrossRef]
- Ivanova, E.P.; Kurilenko, V.V.; Kurilenko, A.V.; Gorshkova, N.M.; Shubin, F.N.; Nicolau, D.V.; Chelomin, V.P. Tolerance to Cadmium of Free-Living and Associated with Marine Animals and Eelgrass Marine Gamma-Proteobacteria. Curr. Microbiol. 2002, 44, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, E.P.; Gorshkova, N.M.; Kurilenko, V.V. Tolerance of Marine Proteobacteria of the Genera Pseudoalteromonas and Alteromonas to Heavy Metals. Microbiology 2001, 70, 239–241. [Google Scholar] [CrossRef]
- Ivanova, E.P.; Mikhailov, V.V. A New Family, Alteromonadaceae fam. nov., Including Marine Proteobacteria of the Genera Alteromonas, Pseudoalteromonas, Idiomarina, and Colwellia. Microbiology 2001, 70, 10–17. [Google Scholar] [CrossRef]
- Dieckmann, R.; Graeber, I.; Kaesler, I.; Szewzyk, U.; von Döhren, H. Rapid screening and dereplication of bacterial isolates from marine sponges of the Sula Ridge by Intact-Cell-MALDI-TOF mass spectrometry (ICM-MS). Appl. Microbiol. Biotechnol. 2004, 67, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Romanenko, L.A.; Uchino, M.; Kalinovskaya, N.I.; Mikhailov, V.V. Isolation, phylogenetic analysis and screening of marine mollusc-associated bacteria for antimicrobial, hemolytic and surface activities. Microbiol. Res. 2008, 163, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Wang, S.; Yu, M.; Yan, S.; Zhang, X.-H. Identification of a marine antagonistic strain JG1 and establishment of a polymerase chain reaction detection technique based on the gyrB gene. Aquac. Res. 2010, 41, 1867–1874. [Google Scholar] [CrossRef]
- Hayashida-Soiza, G.; Uchida, A.; Mori, N.; Kuwahara, Y.; Ishida, Y. Purification and characterization of antibacterial substances produced by a marine bacterium Pseudoalteromonas haloplanktis strain. J. Appl. Microbiol. 2008, 105, 1672–1677. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, E.P.; Sawabe, T.; Alexeeva, Y.V.; Lysenko, A.M.; Gorshkova, N.M.; Hayashi, K.; Zukova, N.V.; Christen, R.; Mikhailov, V.V. Pseudoalteromonas issachenkonii sp. nov., a bacterium that degrades the thallus of the brown alga Fucus evanescens. Int. J. Syst. Evol. Microbiol. 2002, 52, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Pujalte, M.J.; Ortigosa, M.; Macián, M.C.; Garay, E. Aerobic and facultative anaerobic heterotrophic bacteria associated to Mediterranean oysters and seawater. Int. Microbiol. 2010, 2, 259–266. [Google Scholar]
- Huggett, M.J.; Williamson, J.E.; de Nys, R.; Kjelleberg, S.; Steinberg, P.D. Larval settlement of the common Australian sea urchin Heliocidaris erythrogramma in response to bacteria from the surface of coralline algae. Oecologia 2006, 149, 604–619. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, E.P.; Shevchenko, L.S.; Sawabe, T.; Lysenko, A.M.; Svetashev, V.I.; Gorshkova, N.M.; Satomi, M.; Christen, R.; Mikhailov, V.V. Pseudoalteromonas maricaloris sp. nov., isolated from an Australian sponge, and reclassification of [Pseudoalteromonas aurantia] NCIMB 2033 as Pseudoalteromonas flavipulchra sp. nov. Int. J. Syst. Evol. Microbiol. 2002, 52, 263–271. [Google Scholar] [CrossRef] [PubMed]
- El-Tarabily, K.A.; Youssef, T. Improved growth performance of the mangrove Avicennia marina seedlings using a 1-aminocyclopropane-1-carboxylic acid deaminase-producing isolate of Pseudoalteromonas maricaloris. Plant Growth Regul. 2011, 65, 473–483. [Google Scholar] [CrossRef]
- Microbial Community of Black Band Disease on Infection, Healthy, and Dead Part of Scleractinian Montipora sp. Colony at Seribu Islands, Indonesia. Available online: http://www.academia.edu/11707009/MICROBIAL_COMMUNITY_OF_BLACK_BAND_DISEASE_ON_INFECTION_HEALTHY_AND_DEAD_PART_OF_SCLERACTINIAN_Montipora_sp._COLONY_AT_SERIBU_ISLANDS_INDONESIA (accessed on 15 January 2016).
- Zheng, L.; Yan, X.; Han, X.; Chen, H.; Lin, W.; Lee, F.S.C.; Wang, X. Identification of norharman as the cytotoxic compound produced by the sponge (Hymeniacidon perleve)-associated marine bacterium Pseudoalteromonas piscicida and its apoptotic effect on cancer cells. Biotechnol. Appl. Biochem. 2006, 44, 135–142. [Google Scholar] [PubMed]
- Nelson, E.J.; Ghiorse, W.C. Isolation and identification of Pseudoalteromonas piscicida strain Cura-d associated with diseased damselfish (Pomacentridae) eggs. J. Fish Dis. 1999, 22, 253–260. [Google Scholar] [CrossRef]
- Zhang, X.; Nakahara, T.; Miyazaki, M.; Nogi, Y.; Taniyama, S.; Arakawa, O.; Inoue, T.; Kudo, T. Diversity and function of aerobic culturable bacteria in the intestine of the sea cucumber Holothuria leucospilota. J. Gen. Appl. Microbiol. 2012, 58, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Kumari, P.; Reddy, C.R.K. Antimicrobial compounds from seaweeds-associated bacteria and fungi. Appl. Microbiol. Biotechnol. 2015, 99, 1571–1586. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, E.P.; Gorshkova, N.M.; Sawabe, T. Pseudoalteromonas ruthenica sp. nov., isolated from marine invertebrates. Int. J. Syst. Evol. Microbiol. 2002, 52, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Tan, B.; Mai, K. Isolation and identification of a bacterium from marine shrimp digestive tract: A new degrader of starch and protein. J. Ocean Univ. China 2011, 10, 287–292. [Google Scholar] [CrossRef]
- Egan, S.; Thomas, T.; Holmström, C.; Kjelleberg, S. Phylogenetic relationship and antifouling activity of bacterial epiphytes from the marine alga Ulva lactuca. Environ. Microbiol. 2000, 2, 343–347. [Google Scholar] [CrossRef] [PubMed]
- Wilson, G.S.; Raftos, D.A.; Corrigan, S.L.; Nair, S.V. Diversity and antimicrobial activities of surface-attached marine bacteria from Sydney Harbour, Australia. Microbiol. Res. 2010, 165, 300–311. [Google Scholar] [CrossRef] [PubMed]
- Pujalte, M.J.; Sitjà-Bobadilla, A.; Macián, M.C.; Álvarez-Pellitero, P.; Garay, E. Occurrence and virulence of Pseudoalteromonas spp. in cultured gilthead sea bream (Sparus aurata L.) and European sea bass (Dicentrarchus labrax L.). Molecular and phenotypic characterisation of P. undina strain U58. Aquaculture 2007, 271, 47–53. [Google Scholar] [CrossRef]
- Radjasa, O.K.; Kencana, D.S.; Sabdono, A.; Hutagalung, R.A.; Lestari, E.S. Antibacterial Activity of Marine Bacteria Associated with sponge Aaptos sp. against Multi Drugs Resistant (MDR) strains. J. Mat. Sains 2009, 12, 147–152. [Google Scholar]
- Flemer, B.; Kennedy, J.; Margassery, L.M.; Morrissey, J.P.; O’Gara, F.; Dobson, A.D.W. Diversity and antimicrobial activities of microbes from two Irish marine sponges, Suberites carnosus and Leucosolenia sp. J. Appl. Microbiol. 2012, 112, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Shnit-Orland, M.; Sivan, A.; Kushmaro, A. Antibacterial Activity of Pseudoalteromonas in the Coral Holobiont. Microb. Ecol. 2012, 64, 851–859. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-H.; Kuo, J.; Sung, P.-J.; Chang, Y.-C.; Lu, M.-C.; Wong, T.-Y.; Liu, J.-K.; Weng, C.-F.; Twan, W.-H.; Kuo, F.-W. Isolation of marine bacteria with antimicrobial activities from cultured and field-collected soft corals. World J. Microbiol. Biotechnol. 2012, 28, 3269–3279. [Google Scholar] [CrossRef] [PubMed]
- Defer, D.; Desriac, F.; Henry, J.; Bourgougnon, N.; Baudy-Floc’h, M.; Brillet, B.; Le Chevalier, P.; Fleury, Y. Antimicrobial peptides in oyster hemolymph: The bacterial connection. Fish Shellfish Immunol. 2013, 34, 1439–1447. [Google Scholar] [CrossRef] [PubMed]
- Desriac, F.; Chevalier, P.; Brillet, B.; Leguerinel, I.; Thuillier, B.; Paillard, C.; Fleury, Y. Exploring the hologenome concept in marine bivalvia: Haemolymph microbiota as a pertinent source of probiotics for aquaculture. FEMS Microbiol. Lett. 2014, 350, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Goulden, E.F.; Hall, M.R.; Pereg, L.L.; Høj, L. Identification of an Antagonistic Probiotic Combination Protecting Ornate Spiny Lobster (Panulirus ornatus) Larvae against Vibrio owensii Infection. PLoS ONE 2012, 7, e39667. [Google Scholar] [CrossRef] [PubMed]
- Lokmer, A.; Wegner, K.M. Hemolymph microbiome of Pacific oysters in response to temperature, temperature stress and infection. ISME J. 2014, 9, 670–682. [Google Scholar] [CrossRef] [PubMed]
- Carillo, S.; Pieretti, G.; Parrilli, E.; Tutino, M.L.; Gemma, S.; Molteni, M.; Lanzetta, R.; Parrilli, M.; Corsaro, M.M. Structural Investigation and Biological Activity of the Lipooligosaccharide from the Psychrophilic Bacterium Pseudoalteromonas haloplanktis TAB 23. Chem. Eur. J. 2011, 17, 7053–7060. [Google Scholar] [CrossRef] [PubMed]
- Maaetoft-Udsen, K.; Vynne, N.; Heegaard, P.M.; Gram, L.; Frøkiær, H. Pseudoalteromonas strains are potent immunomodulators owing to low-stimulatory LPS. Innate Immun. 2013, 19, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Corsaro, M.M.; Piaz, F.D.; Lanzetta, R.; Parrilli, M. Lipid A structure of Pseudoalteromonas haloplanktis TAC 125: Use of electrospray ionization tandem mass spectrometry for the determination of fatty acid distribution. J. Mass Spectrom. 2002, 37, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Krasikova, I.N.; Kapustina, N.V.; Isakov, V.V.; Gorshkova, N.M.; Solov’eva, T.F. Elucidation of Structure of Lipid A from the Marine Gram-Negative Bacterium Pseudoalteromonas haloplanktis ATCC 14393T. Russ. J. Bioorg. Chem. 2004, 30, 367–373. [Google Scholar] [CrossRef]
- Gardères, J.; Bedoux, G.; Koutsouveli, V.; Crequer, S.; Desriac, F.; Pennec, G.L. Lipopolysaccharides from Commensal and Opportunistic Bacteria: Characterization and Response of the Immune System of the Host Sponge Suberites domuncula. Mar. Drugs 2015, 13, 4985–5006. [Google Scholar] [CrossRef] [PubMed]
- Silipo, A.; Molinaro, A.; Nazarenko, E.L.; Gorshkova, R.P.; Ivanova, E.P.; Lanzetta, R.; Parrilli, M. The O-chain structure from the LPS of marine halophilic bacterium Pseudoalteromonas carrageenovora-type strain IAM 12662T. Carbohydr. Res. 2005, 340, 2693–2697. [Google Scholar] [CrossRef] [PubMed]
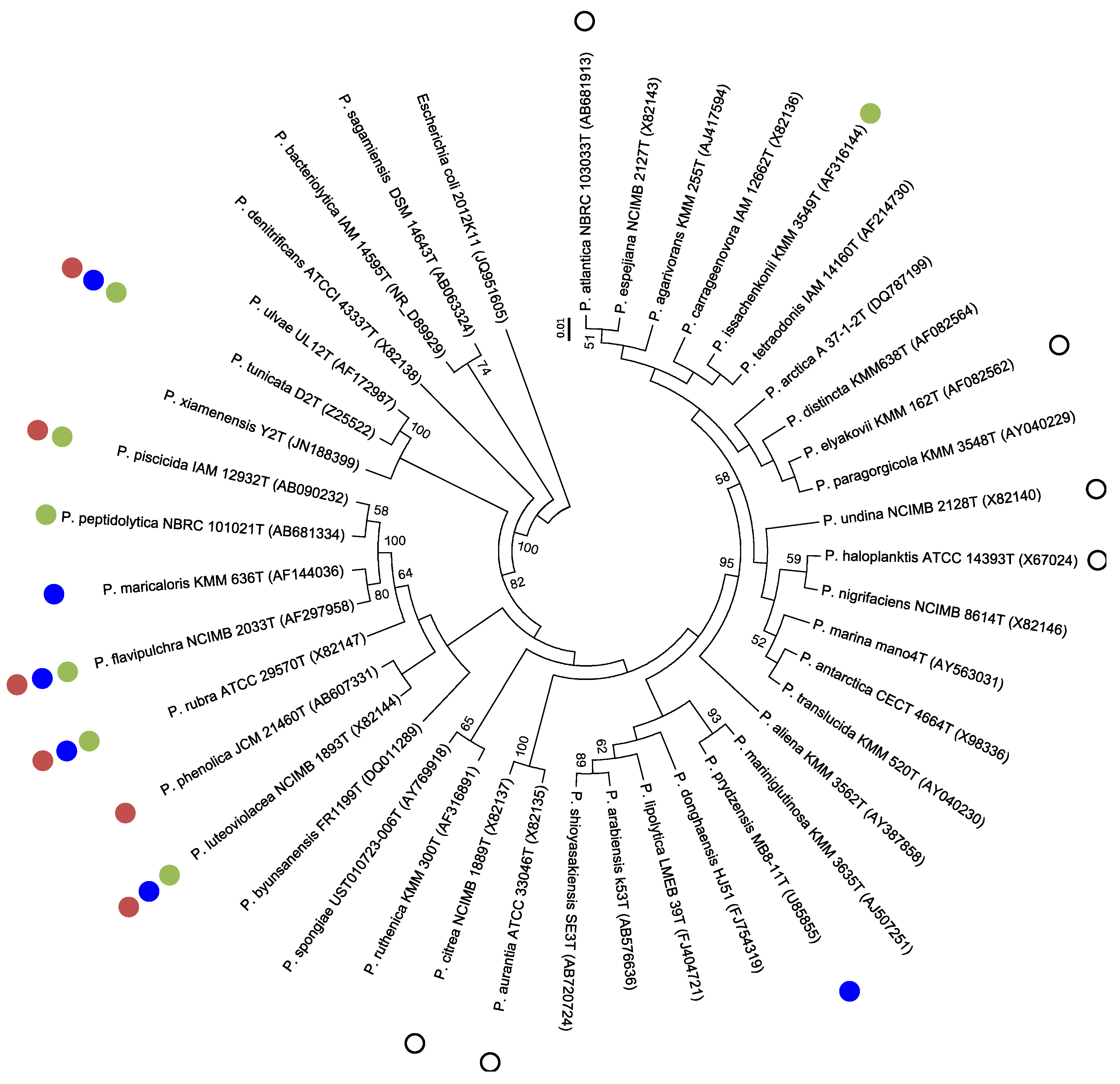
| Producers | Products | Sensitive Microorganisms | Ref. | ||
|---|---|---|---|---|---|
| Affiliated Species | Strain | Name | Structure | ||
| P. peptidolytica | J010 | Korormicin, R: (CH2)7CH3 | 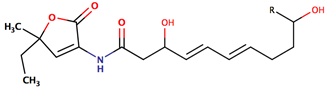 | Gram-negative | [27,28,29] |
| Korormicin G | 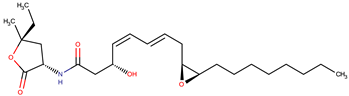 | ||||
| Korormicin J R = CH2CH3 | 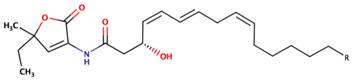 | ||||
| Korormicin K R = (CH2)2CH3 | |||||
| P. tunicata | CCUG 267547 | Tambjamine |  | Staphylococcus aureus, Escherichia coli, Candida albicans, Malassezia furfur | [20,21,30,31,32,33,34,35] |
| Tambjamine YP1 | 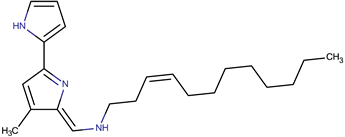 | ||||
| P. rubra | DSM 6842 | Prodigiosin | 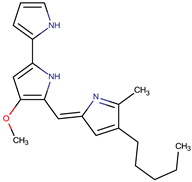 | Staphylococcus aureus, Escherichia coli, Candida albicans | [15,16,17] |
| Cycloprodigiosin | 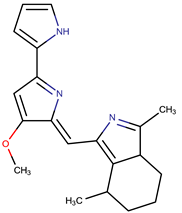 | ||||
| 2-(p-H-benzyl) prodigiosin | 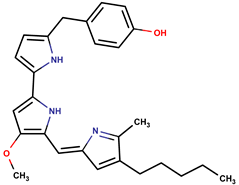 | ||||
| P. luteoviolacea | NCIMB 2035 | Violacein R1 = OH, R2 = H | 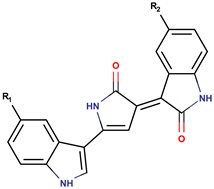 | Staphylococcus aureus, Bacilus subtilis, Bacillus megaterium, Photobacterium sp., fungi | [15,36,37] |
| Oxyviolacein R1 = R2 = OH | |||||
| Deoxyviolacein R1 = R2 = H | |||||
| Indolmycin | 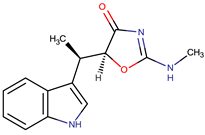 | Staphylococcus aureus | [15,38] | ||
| P. rubra, P. piscicida | DSM 6842 A1-J11 | pentyl-quinolinone R: (CH2)3CH3 |  | Staphylococcus aureus, Vibrio anguillarum, Vibrio harveyi, Candida albicans | [15,39,40,41,42] |
| heptyl-quinolinone R: (CH2)5CH3 | |||||
| nonyl-quinolinone R: (CH2)7CH3 | |||||
| P. piscicida | NJ6-3-1 | Norharman | 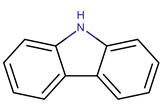 | Bacillus subtilis, Staphylococcus aureus, Agrobacterium tumefaciens, Escherichia coli, Saccharomyces cerevisiae | [43] |
| P. flavipulchra | JG1 | n-hydroxy benzoisoxazolone, 2′-deoxyadenosine | 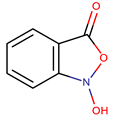 | Gram-positive and Gram-negative | [44] |
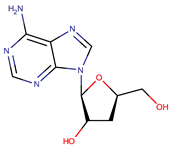 | |||||
| P. issachenkonni | non-referenced | Isatin |  | Candida albicans | [45,46,47] |
| P. piscicida | OT59 | Alteramide A | 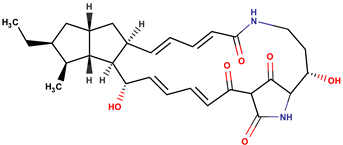 | Fungi | [48] |
| Producers | Products | Sensitive Microorganisms | Ref. | ||
|---|---|---|---|---|---|
| Affiliated Species | Strain | Name | Structure | ||
| P. luteoviolacea P. peptidolytica | I-L-33 J010 | Tetrabromopyrrole |  | Staphylococcus aureus, Enterobacter aerogenes, Escherichia coli, Photobacterium fisheri, Photobacterium mandapamensis, Photobacterium phosphoreum, Pseudomonas aeruginosa, Candida albicans | [27,49] |
| P. luteoviolacea | I-L-33 | Hexa-bromo-2,2′-bipyrrole | 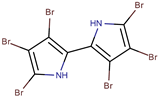 | Photobacterium sp. | [23,49] |
| 4′-((3,4,5-tribromo-1H-pyrrol-2-yl)methyl) phenol |  | Staphylococcus aureus | [27] | ||
| P. luteoviolacea | 2ta16 | 2,4,6-Tribromophenol | 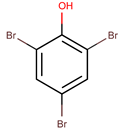 | Bacteria and fungi | [50,51] |
| 2,6-dibromophenol | 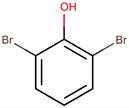 | Fungi | [50,52] | ||
| P. luteoviolacea P. phenolica | I-L-33 D5047 | Pentabromopseudilin | 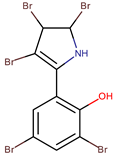 | Staphylococcus aureus Photobacterium phosophoreum | [23,49,53,54,55] |
| P. phenolica | D5047 | 2,3,5,7-tetrabromobenzofuro [3,2-b]pyrrole | 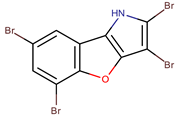 | Staphylococcus aureus, Candida albicans | [55] |
| P. peptidolytica | J010 | Korormicin H, F, I H: R1: (CH2)2CH3 R2: OH R3: Cl F: R1: (CH2)2CH3 R2: Br R3: OH I: R1: (CH2)2CH3 R2: Cl R3: OH | 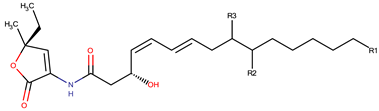 | Gram-negative | [27] |
| P. flavipulchra | JG1 | 6-bromoindolyl-3-acetic acid |  | Gram-positive and Gram-negative | [44] |
| Producers | Products | Sensitive Microorganisms | Ref. | ||
|---|---|---|---|---|---|
| Affiliated Species | Strain | Name | Structure | ||
| P. phenolica | O-BC30T | MC21-A | 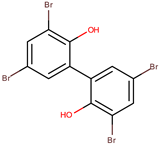 | Staphylococcus aureus, Escherichia coli | [55,62] |
| D5047 | 4,4′,6-tribromo-2,2′-biphenol | 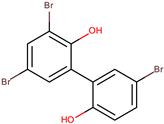 | |||
| P. luteoviolacea | Non-referenced | 2,4-dibromo-6-chlorophenol | 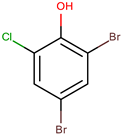 | Staphylococcus aureus | [63] |
| I-L-33 | 4-hydroxy benzaldehyde (2) |  | Gram-positive Gram-negative | [49] | |
| n-propyl-3-hydroxybenzoate (3) | 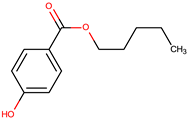 | ||||
| P. luteoviolacea | SANK 73390 | Thiomarinols A,C,D,E,F | 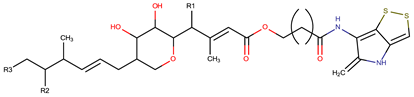 | Gram-positive Gram-negative | [61,64,65,66,67] |
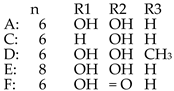 | |||||
| Thiomarinol B |  | ||||
| Thiomarinol G |  | ||||
| Xenorhabdin | 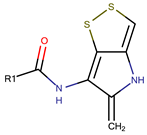 | ||||
| R1: decanoyl | |||||
| R1: dodecanoyl | |||||
| R1: E-dec-3-enoyl | |||||
| R1: Z-dec-4-enoyl | |||||
| R1: E-tetradecenoyl | |||||
| R1: Z-hexadecenoyl | |||||
| P. flavipulchra | JG1 | p-hydroxybenzoic acid |  | Bacillus subtilis, Aeromonas hydrophila, Photobacterium damselae, Vibrio anguillarum, Vibrio harveyi | [44] |
| trans-cinnamic acid | 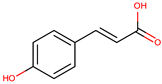 | ||||
| Producer | Compound | Sensitive Microorganisms | Ref. | ||
|---|---|---|---|---|---|
| Affiliated Species | Strain | Name | Structure | ||
| P. rubra P. flavipulchra P. maricaloris | DSM 6842 NCIMB 2033 KMM 636 | Bromoalterochromide A, R = i B, R = i Dibromoalterochromide A, R = ii B, R = ii |  | Bacillus subtilis, Staphylococcus aureus, Enterococcus faecium, Vibrio anguillarum, Candida albicans | [15,74] |
 | 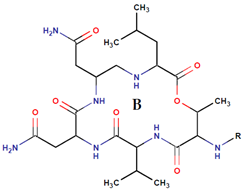 | ||||
| P. maricaloris | KMM 636 | cyclo-(isoleucyl-prolyl-leucyl-alanyl) | Bacteria and fungi | [75] | |
| Producer | Compound | Sensitive Microorganisms | Ref. | |||
|---|---|---|---|---|---|---|
| Affiliated Species | Strain | Name | Structural Information | |||
| P. luteoviolacea | CPMOR-1 | l-amino acid oxidase | 110 kDa | Bacillus subtilis, Staphylococcus | [78] | |
| epidermidis, Escherichia coli | ||||||
| P. tunicata | D2 | AlpP | l-Lysine oxidase | 190 kDa | Gram positive Gram negative | [79] |
| P. flavipulchra | C2 | l-amino acid oxidase | 60 kDa | Staphylococcus aureus | [80] | |
| JG1 | PfaP | 694 amino acids | 77 kDa | Vibrio anguillarum | [81] | |
| P. luteoviolacea | 9k-V10 | 100 kDa | Gram positive Gram negative | [82] | ||
| 9k-V9 | 50 kDa | Vibrio parahaemolyticus | [41,42] | |||
| P. prydzensis | hCg-6 | BLIS | Gram negative | |||
| hCg-42 | BLIS | Gram negative | [71,83] | |||
| DIT44, DIT46, DIT9 | Amphiphilic compounds | <3 kDa | Vibrio parahaemolyticus | [84] | ||
| Antimicrobial Pseudoalteromonas sp. | Marine Host | Ref. |
|---|---|---|
| P. atlantica | [87,117] | |
| P. citrea | [118,119,120] | |
| P. elyakovii | [117,121,122,123] | |
| P. flavipulchra | [80,124] | |
| P. haloplanktis | [125] | |
| P. issachenkonii | [126] | |
| P. luteoviolacea | [37,78,127,128] | |
| P. maricaloris | [129,130] | |
| P. peptidolytica | [131] | |
| P. phenolica | Unassociated marine macroorganisms | |
| P. piscicida | [90,132,133] | |
| P. prydzensis | [83,117,134] | |
| P. rubra | [16,135] | |
| P. ruthenica | [136,137] | |
| P. tunicata | [32,138,139] | |
| P. undina | Dicentrarchus labrax, Sparus aurata | [140] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Offret, C.; Desriac, F.; Le Chevalier, P.; Mounier, J.; Jégou, C.; Fleury, Y. Spotlight on Antimicrobial Metabolites from the Marine Bacteria Pseudoalteromonas: Chemodiversity and Ecological Significance. Mar. Drugs 2016, 14, 129. https://doi.org/10.3390/md14070129
Offret C, Desriac F, Le Chevalier P, Mounier J, Jégou C, Fleury Y. Spotlight on Antimicrobial Metabolites from the Marine Bacteria Pseudoalteromonas: Chemodiversity and Ecological Significance. Marine Drugs. 2016; 14(7):129. https://doi.org/10.3390/md14070129
Chicago/Turabian StyleOffret, Clément, Florie Desriac, Patrick Le Chevalier, Jérôme Mounier, Camille Jégou, and Yannick Fleury. 2016. "Spotlight on Antimicrobial Metabolites from the Marine Bacteria Pseudoalteromonas: Chemodiversity and Ecological Significance" Marine Drugs 14, no. 7: 129. https://doi.org/10.3390/md14070129
APA StyleOffret, C., Desriac, F., Le Chevalier, P., Mounier, J., Jégou, C., & Fleury, Y. (2016). Spotlight on Antimicrobial Metabolites from the Marine Bacteria Pseudoalteromonas: Chemodiversity and Ecological Significance. Marine Drugs, 14(7), 129. https://doi.org/10.3390/md14070129








