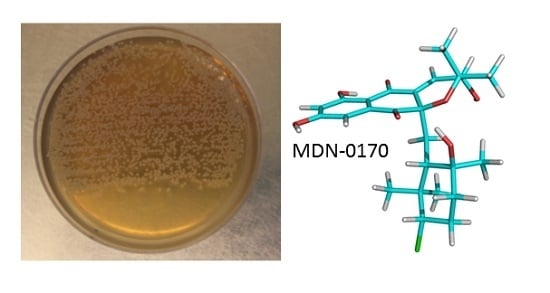
MDN-0170, a New Napyradiomycin from Streptomyces sp. Strain CA-271078
Abstract
:1. Introduction
2. Results and Discussion
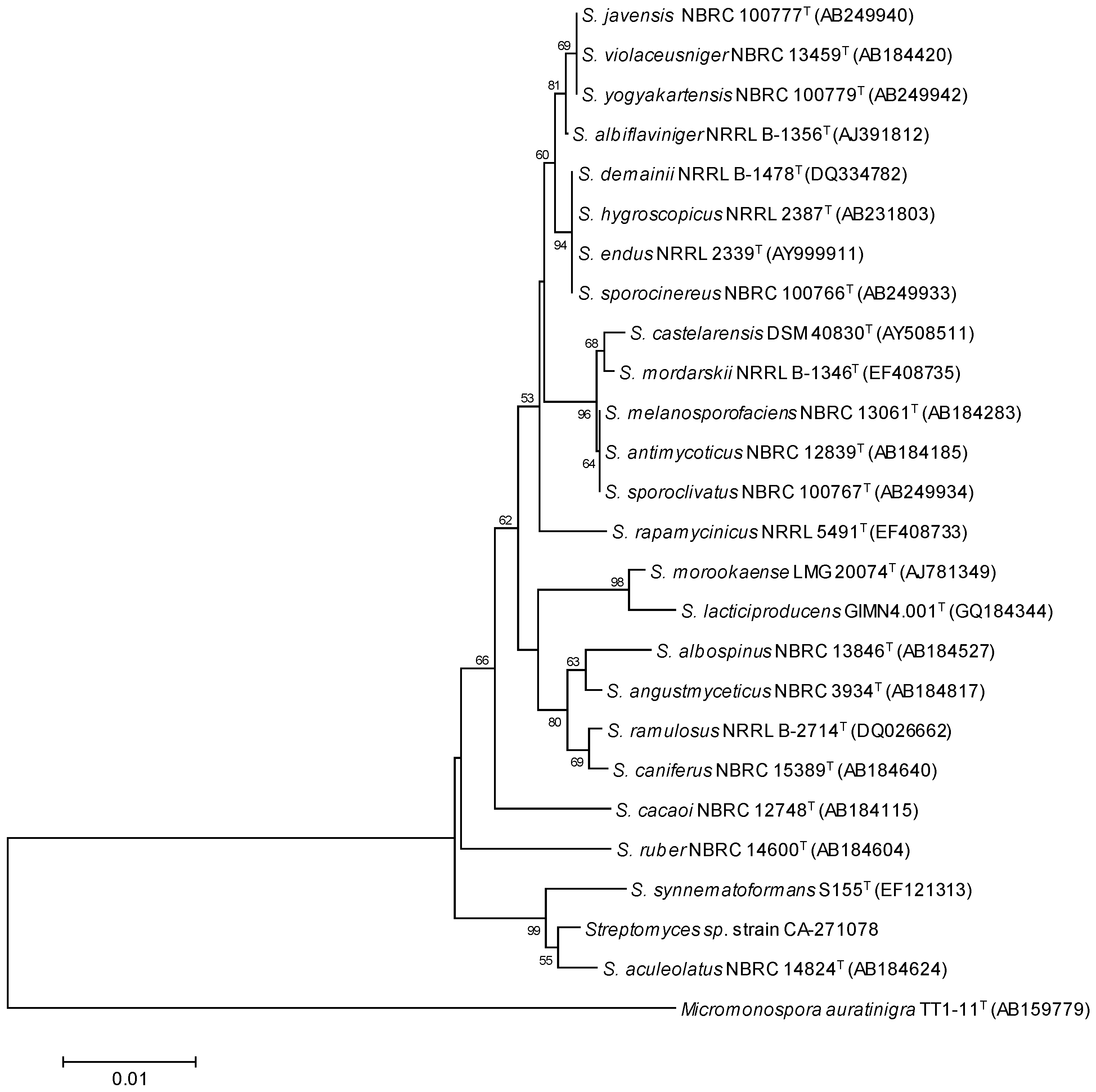
2.1. Isolation and Taxonomy of the Producing Microorganism
2.2. Extraction, Dereplication and Bioassay-Guided Isolation
2.3. Structural Determination of MDN-0170 (1)
2.4. Evaluation of Antimicrobial Activity
Antibacterial and Antifungal Activities
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Taxonomical Identification of the Producing Microorganism
3.3. Fermentation of the Producing Microorganism
3.4. Extraction and Bioassay Guided Isolation
3.5. Antibacterial and Antifungal Assays
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Shiomi, K.; Nakamura, H.; Iinuma, H.; Naganawa, H.; Isshiki, K.; Takeuchi, T.; Umezawa, H.; Iitaka, Y. Structures of new antibiotics napyradiomycins. J. Antibiot. 1986, 39, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Shiomi, K.; Nakamura, H.; Iinuma, H.; Naganawa, H.; Takeuchi, T.; Umezawa, H.; Iitaka, Y. New antibiotic napyradiomycins A2 and B4 and stereochemistry of napyradiomycins. J. Antibiot. 1987, 40, 1213–1219. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, M.; Williams, S.T.; Alderson, G. Transfer of Chainia species to the genus Streptomyces with emended description of species. Syst. Appl. Microbiol. 1986, 8, 55–60. [Google Scholar] [CrossRef]
- Fukuda, D.S.; Mynderse, J.S.; Baker, P.J.; Berry, D.M.; Boeck, L.D.; Yao, R.C.; Mertz, F.P.; Nakatsukasa, W.M.; Mabe, J.; Ott, J.; et al. A80915, A new antibiotic complex produced by Streptomyces aculeolatus. Discovery, taxonomy, fermentation, isolation, characterization, and antibacterial evaluation. J. Antibiot. 1990, 43, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Soria-Mercado, I.E.; Prieto-Davo, A.; Jensen, P.R.; Fenical, W. Antibiotic terpenoid chloro-dihydroquinones from a new marine actinomycete. J. Nat. Prod. 2005, 68, 904–910. [Google Scholar] [CrossRef] [PubMed]
- Farnaes, L.; Coufal, N.G.; Kauffman, C.A.; Rheingold, A.L.; DiPasquale, A.G.; Jensen, P.R.; Fenical, W. Napyradiomycin derivatives, produced by a marine-derived actinomycete, illustrate cytotoxicity by induction of apoptosis. J. Nat. Prod. 2014, 77, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, K.; Sue, M.; Furihata, K.; Ito, S.; Seto, H. Terpenoids produced by actinomycetes: Napyradiomycins from Streptomyces antimycoticus NT17. J. Nat. Prod. 2008, 71, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.B.; Jensen, P.R.; Fenical, W. Cytotoxic and antimicrobial napyradiomycins from two marine-derived Streptomyces strains. Eur. J. Org. Chem. 2013, 3751–3757. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, K.; Masuoka, S.; Ohse, T.; Naganawa, H.; Kondo, S.; Ikeda, Y.; Kinoshita, N.; Hamada, M.; Sawa, T.; Takeuchi, T. Isolation from Streptomyces of a novel naphthoquinone compound, naphthablin, that inhibits abl oncogene functions. J. Antibiot. 1995, 48, 604–607. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.Y.; Kwon, H.C.; Williams, P.G.; Jensen, P.R.; Fenical, W. Azamerone, a terpenoid phthalazinone from a marine-derived bacterium related to the genus Streptomyces (Actinomycetales). Org. Lett. 2006, 8, 2471–2474. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, K.; Irie, K.; Toda, T.; Matsuo, Y.; Kasai, H.; Sue, M.; Furihata, K.; Seto, H. Studies on terpenoids produced by actinomycetes. 5-dimethylallylindole-3-carboxylic acid and A80915G-8′′-acid produced by marine-derived Streptomyces sp. MS239. J. Antibiot. 2008, 61, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Li, S.; Li, J.; Chen, Y.; Saurav, K.; Zhang, Q.; Zhang, H.; Zhang, W.; Zhang, W.; Zhang, S.; et al. Antibacterial and cytotoxic new napyradiomycins from the marine-derived Streptomyces sp. SCSIO 10428. Mar. Drugs 2013, 11, 2113–2125. [Google Scholar] [CrossRef] [PubMed]
- Haste, N.M.; Farnaes, L.; Perera, V.R.; Fenical, W.; Nizet, V.; Hensler, M.E. Bactericidal kinetics of marine-derived napyradiomycins against contemporary methicillin-resistant Staphylococcus aureus. Mar. Drugs 2011, 9, 680–689. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Tashiro, E.; Motohashi, K.; Seto, H.; Imoto, M. Napyradiomycin A1, an inhibitor of mitochondrial complexes I and II. J. Antibiot. 2012, 65, 211–214. [Google Scholar] [CrossRef] [PubMed]
- Farnaes, L.; La Clair, J.J.; Fenical, W. Napyradiomycins CNQ525.510B and A80915C target the hsp90 paralogue grp94. Org. Biomol. Chem. 2014, 12, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Lacret, R.; Oves-Costales, D.; Gómez, C.; Díaz, C.; de la Cruz, M.; Pérez-Victoria, I.; Vicente, F.; Genilloud, O.; Reyes, F. New ikarugamycin derivatives with antifungal and antibacterial properties from Streptomyces zhaozhouensis. Mar. Drugs 2015, 13, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Kim, O.S.; Cho, Y.J.; Lee, K.; Yoon, S.H.; Kim, M.; Na, H.; Park, S.C.; Jeon, Y.S.; Lee, J.H.; Yi, H.; et al. Introducing EzTaxon-e: A prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 2012, 62, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Jukes, T.H.; Cantor, C. Evolution of proteins molecules. In Mammalian Protein Metabolism; Munro, H.N., Ed.; Academic Press: New York, NY, USA, 1969; pp. 121–132. [Google Scholar]
- Pérez-Victoria, I.; Martín, J.; Reyes, F. Combined LC/UV/MS and NMR strategies for the dereplication of marine natural products. Planta Med. 2016, 82, 857–871. [Google Scholar] [CrossRef] [PubMed]
- Dictionary of Natural Products (DPN 2016). Chapman & Hall/CRC. Available online: http://dnp.Chemnetbase.Com/intro/index.jsp (accessed on 8 August 2016).
- Baker, G.H.; Dorgan, R.J.; Everett, J.R.; Hood, J.D.; Poulton, M.E. A novel series of milbemycin antibiotics from Streptomyces strain E225. II. Isolation, characterization, structure elucidation and solution conformations. J. Antibiot. 1990, 43, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Winter, J.M.; Moffitt, M.C.; Zazopoulos, E.; McAlpine, J.B.; Dorrestein, P.C.; Moore, B.S. Molecular basis for chloronium-mediated meroterpene cyclization: Cloning, sequencing, and heterologous expression of the napyradiomycin biosynthetic gene cluster. J. Biol. Chem. 2007, 282, 16362–16368. [Google Scholar] [CrossRef] [PubMed]
- Soria-Mercado, I.E.; Jensen, P.R.; Fenical, W.; Kassel, S.; Golen, J. 3,4a-Dichloro-10a-(3-chloro-6-hydroxy-2,2,6-trimethylcyclohexylmethyl)-6,8-dihydroxy-2,2,7-trimethyl-3,4,4a,10a-tetrahydro-2h-benzo[g]chromene-5,10-dione. Acta Crystallogr. Sect. E 2004, 60, O1627–O1629. [Google Scholar] [CrossRef]
- Martín, J.; Crespo, G.; González-Menéndez, V.; Pérez-Moreno, G.; Sánchez-Carrasco, P.; Pérez-Victoria, I.; Ruiz-Pérez, L.M.; González-Pacanowska, D.; Vicente, F.; Genilloud, O.; et al. MDN-0104, an antiplasmodial betaine lipid from Heterospora chenopodii. J. Nat. Prod. 2014, 77, 2118–2123. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.; da S Sousa, T.; Crespo, G.; Palomo, S.; González, I.; Tormo, J.R.; de la Cruz, M.; Anderson, M.; Hill, R.T.; Vicente, F.; et al. Kocurin, the true structure of PM181104, an anti-methicillin-resistant Staphylococcus aureus (MRSA) thiazolyl peptide from the marine-derived bacterium Kocuria palustris. Mar. Drugs 2013, 11, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, M.C.; de la Cruz, M.; Cantizani, J.; Moreno, C.; Tormo, J.R.; Mellado, E.; De Lucas, J.R.; Asensio, F.; Valiante, V.; Brakhage, A.A.; et al. A new approach to drug discovery: High-throughput screening of microbial natural extracts against Aspergillus fumigatus using resazurin. J. Biomol. Screen. 2012, 17, 542–549. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ravipati, A.S.; Koyyalamudi, S.R.; Jeong, S.C.; Reddy, N.; Bartlett, J.; Smith, P.T.; de la Cruz, M.; Monteiro, M.C.; Melguizo, A.; et al. Anti-fungal and anti-bacterial activities of ethanol extracts of selected traditional Chinese medicinal herbs. Asian Pac. J. Trop. Med. 2013, 6, 673–681. [Google Scholar] [CrossRef]
- Zhang, J.H.; Chung, T.D.; Oldenburg, K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999, 4, 67–73. [Google Scholar] [CrossRef] [PubMed]
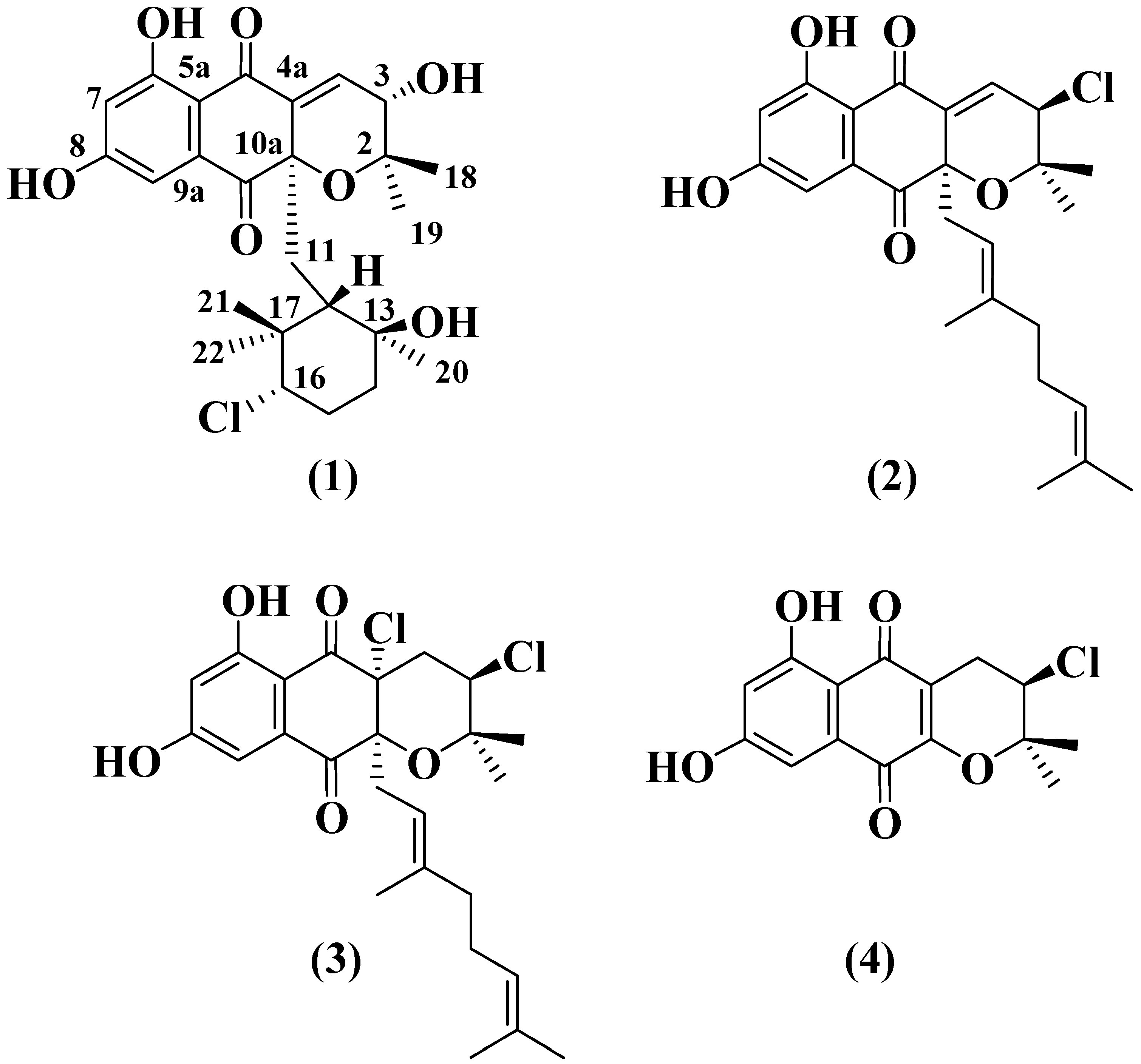
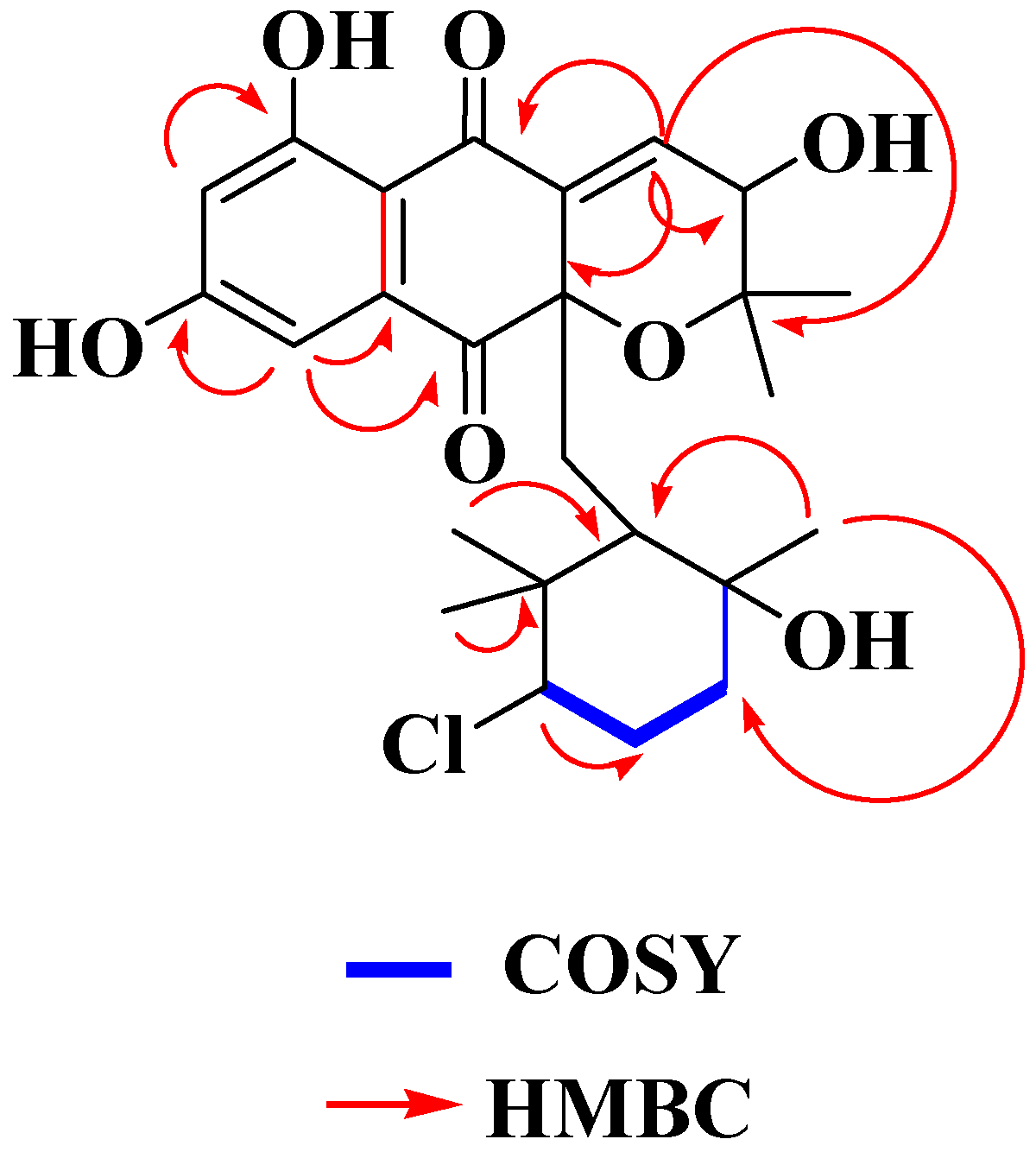

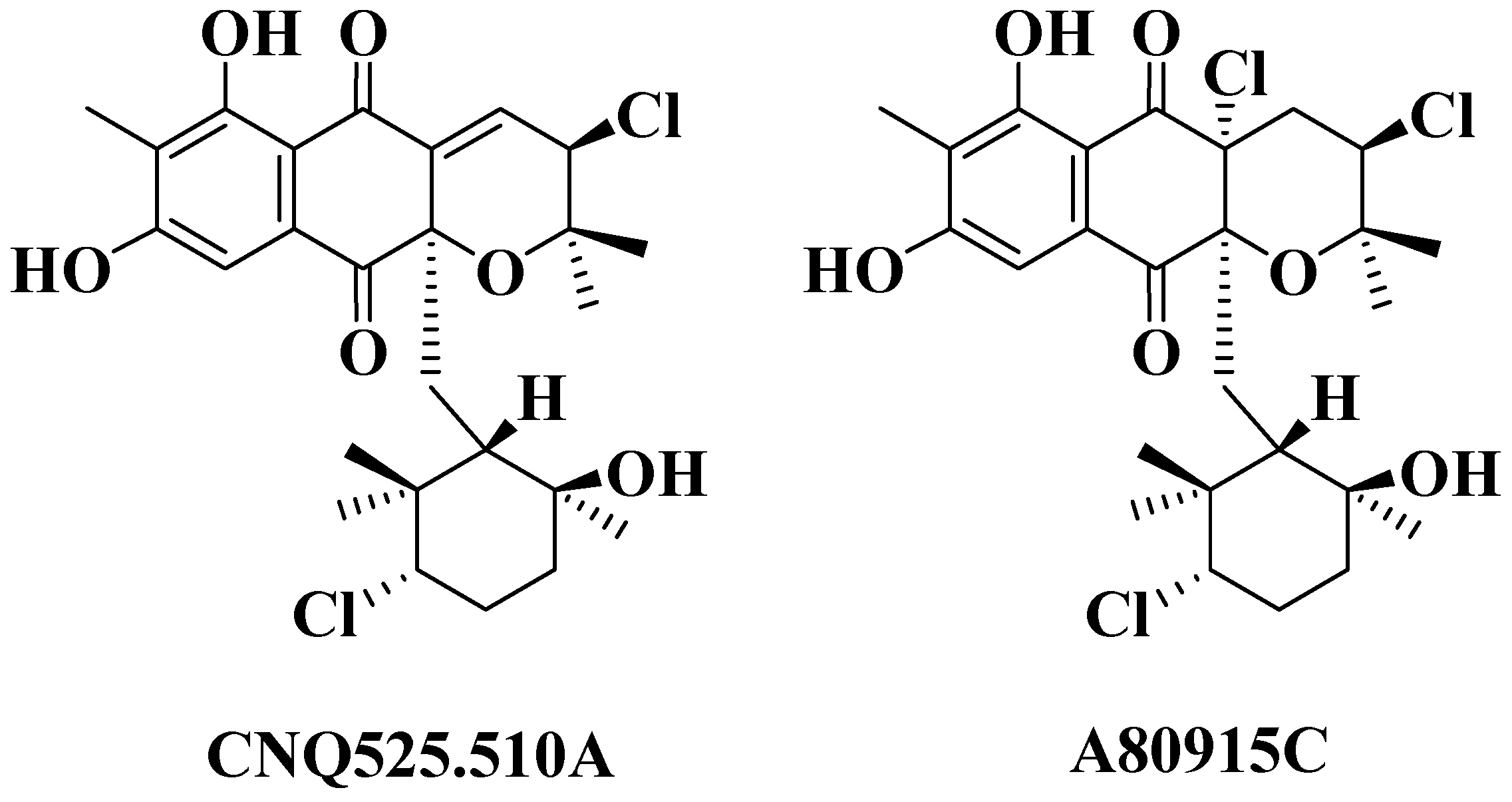
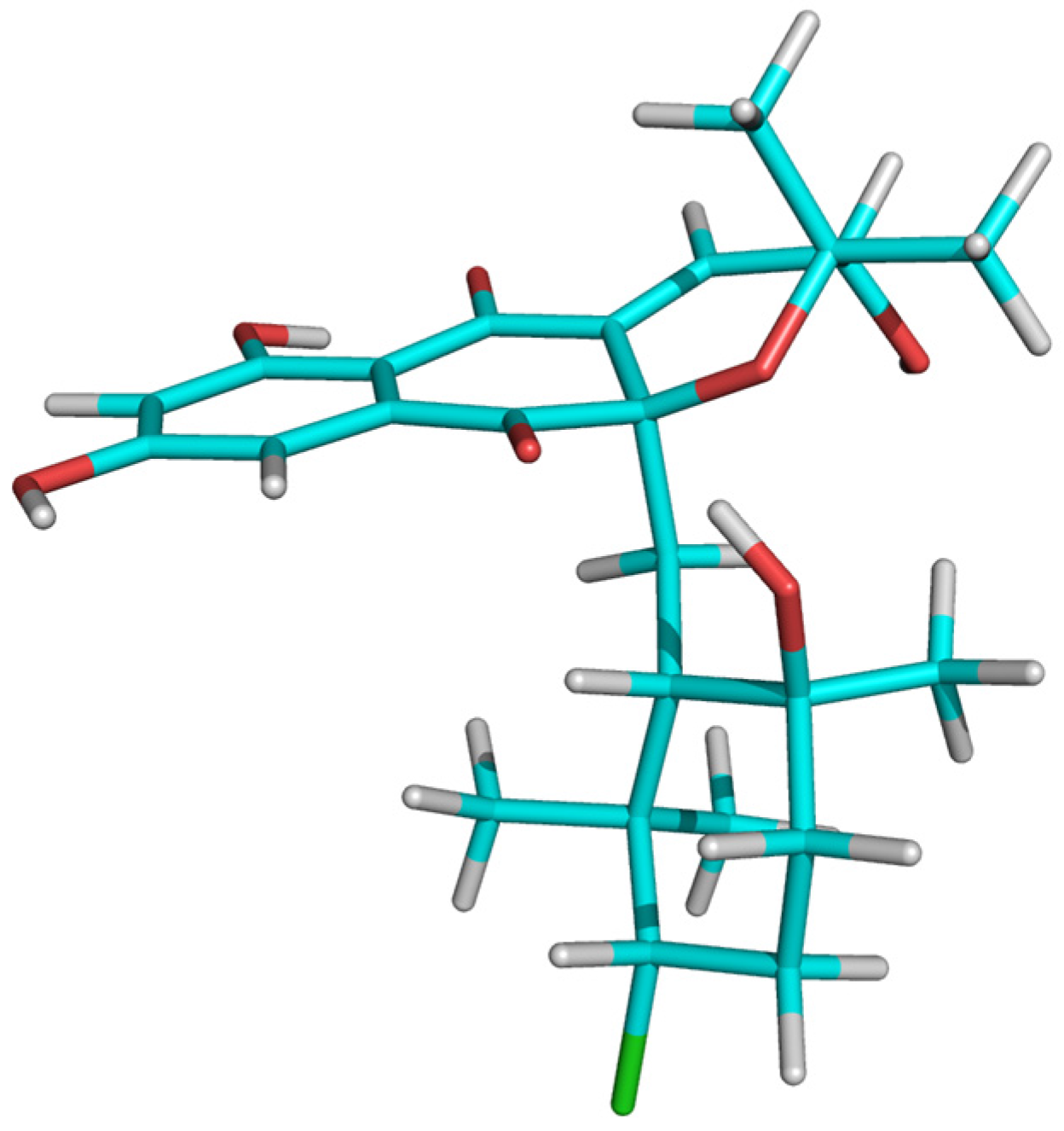
| Position | δ 1H (mult, J, Hz) | δ 13C | Position | δ 1H (mult, J, Hz) | δ 13C |
|---|---|---|---|---|---|
| 2 | ---- | 78.8, s | 11 | 1.96, dd, 15.7, 1.4 2.22, dd, 15.7, 7.8 | 40.9, t |
| 3 | 3.88, d, 6.9 | 67.0, d | 12 | 1.55, d, 7.8 | 52.6, d |
| 4 | 7.10, d, 6.9 | 134.2, d | 13 | ---- | 72.1, s |
| 4a | ---- | 140.2, s | 14 | 1.55, m 1.77, m | 42.2, t |
| 5 | ---- | 190.6, s | 15 | 1.82, m 1.91, dd, 13.7, 4.0 | 31.6, t |
| 5a | ---- | 111.7, s | 16 | 3.70, dd, 12.2, 4.0 | 72.2, d |
| 6 | ---- | 166.3, s | 17 | ---- | 41.8, s |
| 7 | 6.60, d, 2.0 | 109.7, d | 18 | 1.04, s | 25.6, q |
| 8 | ---- | 167.3, s | 19 | 1.44, s | 24.7, q |
| 9 | 6.96, d, 2.0 | 109.5, d | 20 | 1.17, s | 24.4, q |
| 9a | ---- | 137.8, s | 21 | 0.56, s | 29.2, q |
| 10 | ---- | 196.3, s | 22 | 0.71, s | 16.3, q |
| 10a | ---- | 83.4, s |
| MIC (μg/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Microbial Strain | Strain Number | (1) | (2) | (3) | (4) | V | R | A |
| MRSA | MB5393 | >64 | 4–8 | 0.5–1 | >64 | 2–4 | ||
| E. coli | MB2884 | >64 | >64 | >64 | >64 | 6.5–12.5 | ||
| A. fumigatus | ATCC46645 | >64 | >64 | >64 | >64 | 4 | ||
| C. albicans | MY1055 | >64 | >64 | >64 | >64 | 2–4 | ||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lacret, R.; Pérez-Victoria, I.; Oves-Costales, D.; De la Cruz, M.; Domingo, E.; Martín, J.; Díaz, C.; Vicente, F.; Genilloud, O.; Reyes, F. MDN-0170, a New Napyradiomycin from Streptomyces sp. Strain CA-271078. Mar. Drugs 2016, 14, 188. https://doi.org/10.3390/md14100188
Lacret R, Pérez-Victoria I, Oves-Costales D, De la Cruz M, Domingo E, Martín J, Díaz C, Vicente F, Genilloud O, Reyes F. MDN-0170, a New Napyradiomycin from Streptomyces sp. Strain CA-271078. Marine Drugs. 2016; 14(10):188. https://doi.org/10.3390/md14100188
Chicago/Turabian StyleLacret, Rodney, Ignacio Pérez-Victoria, Daniel Oves-Costales, Mercedes De la Cruz, Elizabeth Domingo, Jesús Martín, Caridad Díaz, Francisca Vicente, Olga Genilloud, and Fernando Reyes. 2016. "MDN-0170, a New Napyradiomycin from Streptomyces sp. Strain CA-271078" Marine Drugs 14, no. 10: 188. https://doi.org/10.3390/md14100188
APA StyleLacret, R., Pérez-Victoria, I., Oves-Costales, D., De la Cruz, M., Domingo, E., Martín, J., Díaz, C., Vicente, F., Genilloud, O., & Reyes, F. (2016). MDN-0170, a New Napyradiomycin from Streptomyces sp. Strain CA-271078. Marine Drugs, 14(10), 188. https://doi.org/10.3390/md14100188