Abstract
The A. salmonicida A450 LPS O-antigen, encoded by the wbsalmo gene cluster, is exported through an ABC-2 transporter-dependent pathway. It represents the first example of an O-antigen LPS polysaccharide with three different monosaccharides in their repeating unit assembled by this pathway. Until now, only repeating units with one or two different monosaccharides have been described. Functional genomic analysis of this wbsalmo region is mostly in agreement with the LPS O-antigen structure of acetylated l-rhamnose (Rha), d-glucose (Glc), and 2-amino-2-deoxy-d-mannose (ManN). Between genes of the wbsalmo we found the genes responsible for the biosynthesis and assembly of the S-layer (named A-layer in these strains). Through comparative genomic analysis and in-frame deletions of some of the genes, we concluded that all the A. salmonicida typical and atypical strains, other than A. salmonicida subsp. pectinolytica strains, shared the same wbsalmo and presence of A-layer. A. salmonicida subsp. pectinolytica strains lack wbsalmo and A-layer, two major virulence factors, and this could be the reason they are the only ones not found as fish pathogens.
1. Introduction
Currently, there are five accepted subspecies of Aeromonas salmonicida: A. salmonicida subsp. salmonicida, masoucida, achromogenes, pectinolytica and smithia [1]. Aeromonas salmonicida subsp. salmonicida is the etiological agent producing the systemic disease named furunculosis, being then an important fish pathogen [2]. This pathogen has been subjected to considerable investigation due to its importance in the farmed fish industry. Its major virulence factor is an S-layer (named A-layer), which principally consists of a unique two-dimensional crystalline tetragonal protein (A-protein with a molecular weight of 49 kDa) array [3], which is tethered to the cell by the lipopolysaccharide (LPS) [4].
Immunolabeling studies have been able to show that the A-layer appears to cover most of the surface of the virulent A. salmonicida [5], nevertheless some LPS molecules are still surface exposed [6]. Both LPS O-antigen and the A-layer are required to fully protect this bacterium from serum killing [7]. Although the A-layer is not completely necessary for the bacterium’s resistance to serum killing, it is an important barrier against opsonophagocytosis [8].
In Gram-negative bacteria, the LPS are large amphiphilic molecules consisting of a hydrophilic polysaccharide part, and a covalently bound hydrophobic and highly conserved lipid component, termed lipid A (the bioactive endotoxin subunit). The polysaccharide part can be conceptually divided into two sub-domains: one more internal and conserved, the core region, and one more external and highly variable, the O-specific chain, named also O-antigen for its immunogenic properties. These three regions have been differentiated and formally classified by their chemical structure, degree of conservation, biosynthetic pathways and genetic determination (see general review, [9]).
Some studies have chemically characterized structures of the O-antigen polysaccharide and the core oligosaccharide regions of the LPS from A. salmonicida strain SJ-15 [10,11]. More recent studies describe the structural elucidation of the O-antigen LPS of the A. salmonicida subsp. salmonicida from strains A449 and 80204-1 [12], and their core oligosaccharide region [13]. We studied the functional genetics of the O-antigen of the LPS from A. salmonicida subsp. salmonicida strain A450, whose chemical structure is similar to the previously described for other strains [12]. Furthermore, we found genes encoding for the production and export/assembly of the A-layer characteristic from A. salmonicida subsp. salmonicida strains, between the biosynthetic genes for the LPS O-antigen production (cluster named wbsalmo). We also studied the wbsalmo and genes encoding for the production and export/assembly of the A-layer in different strains of subspecies masoucida, achromogenes, pectinolytica and smithia.
2. Results
LPS was extracted from enzymatically digested A. salmonicida A450 cells by the Westphal procedure [14] and the O-polysaccharide isolated after mild acid degradation. Sugar analysis by gas-liquid chromatography (GLC) of resultant monosaccharides as alditol acetates and (S)-octyl glycosides revealed that it was composed of l-rhamnose (Rha), d-glucose (Glc), and 2-amino-2-deoxy-d-mannose (ManN) in the approximate molar ratio of 1:0.95:0.9. The methylation analysis of the O-polysaccharide revealed the presence of 2,3-di-O-methylrhamnose, 2-O-methylrhamnose, 2-deoxy-4,6-di-O-methyl-2-(N-methylacetamido) mannose and 2,3,4,6-tetra-O-methylglucose (Table 1), suggesting that the O-antigen polysaccharide contained 3,4-substituted Rha, 3-substituted ManNAc and terminal Glc.

Table 1.
A. salmonicida A450 O-antigen LPS methylation analysis.
| Sugar Linkage | RtGM a (min) | Relative Molar Ratios b |
|---|---|---|
| 4-Substituted Rha | 5.19 | 0.07 |
| Terminal Glc | 6.18 | 1.00 |
| 3,4-Substituted Rha | 7.03 | 0.85 |
| 3-Substituted ManNAc | 34.81 | 0.60 |
a Retention time of the derived alditol acetate derivative adjusted to that of 1,5-di-O-acetyl-2,3,4,6-tetra-O-methylglucitol-1-d. b Total ion count based on the detector response.
The high-resolution electrospray ionization mass spectrum results obtained were consistent with those of compositional and methylation analyses. We found the presence of the fragment ion at m/z 392.5 suggesting that HexNAc was attached to RhaOAc, and a fragment ion at m/z 554.5 was consistent with the consecutive addition of Hex (+162). Additional ions (m/z 757.5, 946.5 and 1108.5) corresponded to the consecutive addition of sugar residues HexNAc, RhaOAc and Hex, respectively. From these initial studies, we concluded that the A. salmonicida A450 LPS O-antigen seems to be identical to the one described for strains A449 and 80204 [12], which can be depicted as follows:
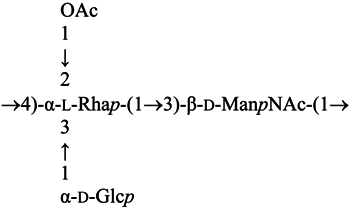

2.1. LPS O-Antigen Biosynthesis Gene Cluster (wbsalmo)
An A. salmonicida A450 cosmid-based genomic library was constructed and introduced into E. coli DH5α as previously described for other Aeromonas strains [15]. As we and others reported, A. salmonicida LPS O-antigen contains rhamnose. Thus, we used previously constructed DNA probes from A. hydrophila strain AH-3 rmlA and B genes (two biosynthetic rhamnose genes, [16]) due to their high DNA sequence conservation among Aeromonas strains, and screened the A. salmonicida A450 genomic library by colony Southern blot. Several tetracycline-resistant clones able to cross react with both probes were isolated and sequences flanking the DNA inserted were determined by using oligonucleotides complementary to the pLA2917 [15] cosmid. To complete the nucleotide sequence (GenBank KR704893) other sequence-derived oligonucleotides were designed by us, purchased (Sigma-Aldrich) and used. Analysis of the sequenced regions showed 26 complete putative open reading frames (ORFs) transcribed in the same direction, being 13 of them (ORF1 to 5 and ORF18 to 25) genes involved in the LPS O-antigen biosynthesis (wbsalmo cluster) as indicated in Figure 1 (in yellow).
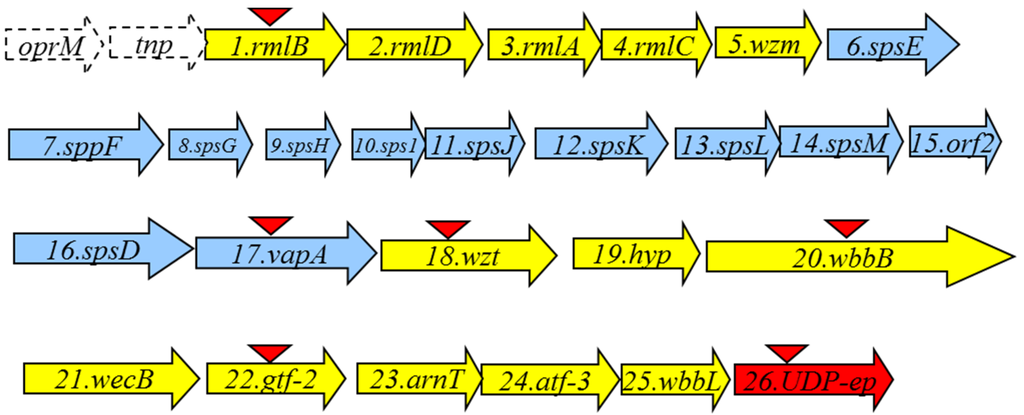
Figure 1.
The A. salmonicida A450 LPS O-antigen (wbsalmo) in yellow and S-layer cluster in blue open reading frames (ORFs) detected as complete genes. The genes, numbered according to the ORF number, were named according to the similarity found by their encoding products with proteins of well characterized functions. The UDP-ep gene (in red) does not belong to any of both clusters. The orf named tnp is encoding a putative transposase protein with a DDE domain from superfamily endonuclease. The oprM gene is incompletely sequenced and it should be noticed that it is also adjacent to the wb O34-antigen LPS from strain AH-3 previously characterized [16]. Triangles indicate the mutants obtained.
The putative transposase protein showed a DDE domain from a superfamily of endonucleases. This domain contains three carboxylate residues that are believed to be responsible for coordinating metal ions needed for catalysis. The catalytic activity of this enzyme involves DNA cleavage at a specific site followed by a strand transfer reaction.
We observed several ORFs (6 to 17), indicated in blue (Figure 1), between the genes involved in the LPS biosynthesis that belong to a type II secretion system (T2SS) plus the structural gene vapA (ORF17) which encodes the surface A-layer protein. It seems logical that these ORFs are genes that encode for the production and export/assembly of the A. salmonicida A-layer characteristic from these strains. Interestingly, the insertion point of the A-layer genes is immediately downstream of a gene encoding for a Wzm putative protein. Downstream of the A-layer genes, a complete ORF encoding a Wzt putative protein was observed. Wzm and Wzt proteins are characteristics of an ABC-2 type transporter. Finally, we found a gene (ORF26), labeled in red (Figure 1), that encodes for a NAD-dependent dehydratase or UDP-sugar epimerase (named accordingly UDP-ep). This gene does not initially belong to the wbsalmo cluster, because its mutation does not abolish the LPS O-antigen (see next section). Sequence analysis of the wbsalmo gene cluster revealed a conserved JUMPstart sequence with the 8 bp ops (operon polarity suppressor) sequence (GGCGGTAG) 119 bp upstream of the ORF1. The ops sequence is recognized by the bacterial antiterminator RfaH, which can be recruited by the transcription elongation complex to reduce pausing and termination at intergenic sites of polycistronic operons, allowing RNA polymerase to finalise transcription of the distal genes in large operons [17,18].
Analysis of ORFs from wbsalmo and S-layer clusters with their predicted function based on their homology to proteins of known function is shown in Table 2.

Table 2.
Characteristics of the A. salmonicida A450 O-antigen LPS (wbsalmo) and A-layer cluster ORFs.
| ORF | Protein Name | Protein Size (in Amino Acid Residues) | Predicted Function | Homologous Protein with Known Function | Percentage in Amino Acid Identity/Similarity |
|---|---|---|---|---|---|
| Inserted S-layer protein cluster | |||||
| 18 | Wzt | 438 | ABC transporter ATP binding protein | Wzt multispecies Aeromonas | 100/100 |
| 19 | Hyp | 216 | Hypothetical protein with domain Sulfotransferase | Sulfotransferase Vibrio cholerae | 38/54 |
| 20 | WbbB | 1122 | N-acetyl glucosaminyl transferase | WbbB Klebsiella pneumonaie | 63/77 |
| 21 | WecB | 370 | UDP-N-acetyl glucosamine 2-epimerase | WecB Serratia marcescens | 100/100 |
| 22 | Gtf-2 | 355 | Glycosyl transferase | Glycosyl transferase family group 2 Vibrio choleare | 78/91 |
| 23 | ArnT | 457 | Hypothetical protein with ArnT (4-amino-4-deoxy-l-arabinose transferase) domain | Hypothetical protein multispecies Aeromonas | 100/100 |
| 24 | Atf-3 | 348 | Acetyl transferase family 3 | Acetyltransferase Serratia marcescens | 45/65 |
| 25 | WbbL | 288 | Rhamnosyl transferase | -Glucosyl transferase family 2 A. veronii -Rhamnosyl transferase E. coli | -100/100 -43/67 |
| 26 | UDP-ep | 318 | NAD-dependent dehydratase or UDP-sugar epimerase | NAD-dependent dehydratase or UDP-sugar epimerase multispecies Aeromonas | 100/100 |
| A-layer protein cluster | |||||
| 6 | SpsE | 552 | S-layer secretion system protein E | Type II secretion system (T2SS) protein E A. salmonicida | 100/100 |
| 7 | SpsF | 395 | S-layer secretion system protein F | Type II secretion system(T2SS) protein F A. salmonicida | 100/100 |
| 8 | SpsG | 143 | S-layer secretion system protein G | Type II secretion system (T2SS) protein G A. salmonicida | 97/99 |
| 9 | SpsH | 131 | S-layer secretion system protein H | Type II secretion system (T2SS) protein H A. salmonicida | 96/98 |
| 10 | SpsI | 132 | S-layer secretion system protein I | Type II secretion system (T2SS )protein I A. salmonicida | 99/100 |
| 11 | SpsJ | 235 | S-layer secretion system protein J | Type II secretion system (T2SS) protein J A. salmonicida | 94/98 |
| 12 | SpsK | 288 | S-layer secretion system protein K | Type II secretion system (T2SS) protein K A. salmonicida | 100/100 |
| 13 | SpsL | 371 | S-layer secretion system protein L | Type II secretion system (T2SS) protein L A. salmonicida | 94/95 |
2.2. Mutant Isolation and Characterization
As described in Materials and Methods section, we obtained in-frame mutants in ORFs 1, 17, 18, 20, 22 and 26 named A450ΔrmlB, A450ΔvapA, A450Δwzt, A450ΔwbbB, A450Δgtf-2 and A450ΔUDP-ep, respectively. As shown in Figure 2, mutants A450ΔrmlB, A450Δwzt, A450ΔwbbB and A450Δgtf-2 were unable to produce LPS O-antigen when analyzed in a SDS-PAGE gel. However, A450ΔvapA and A450ΔUDP-ep mutants showed in the same gels an identical LPS profile as the wild-type strain with O-antigen ladder repetitions.
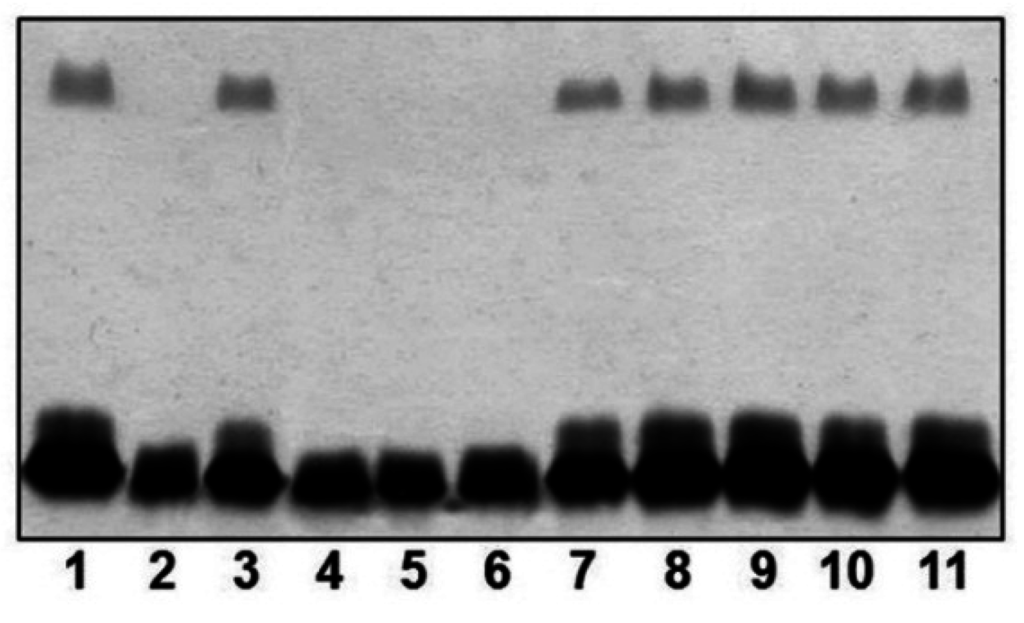
Figure 2.
LPS analysed by SDS-PAGE (12%) and silver stained from A. salmonicida A450 (wild-type, lane 1), mutants A450ΔrmlB (lane 2), A450ΔvapA (lane 3), A450Δwzt (lane 4), A450ΔwbbB (lane 5), A450Δgtf-2, (lane 6), A450ΔUDP-ep (lane 7), mutant A450ΔrmlB complemented with A450rmlB (lane 8), mutant A450Δwzt complemented with A450wzt (lane 9), mutant A450ΔwbbB complemented with A450wbbB (lane 10), and mutant A450Δgtf-2 complemented with A450gtf-2 (lane 11).
We obtained outer-membrane proteins (OMp) from A450ΔvapA mutant as described in Materials and Methods. Analysis by SDS-PAGE gels showed the lack of the major protein band of approximately 49 kDa compared with the wild type (Figure 3A). This band reacts with specific serum anti-VapA protein in Western blot analysis (Figure 3B). Mutant A450ΔUDP-ep showed no changes in OMp.
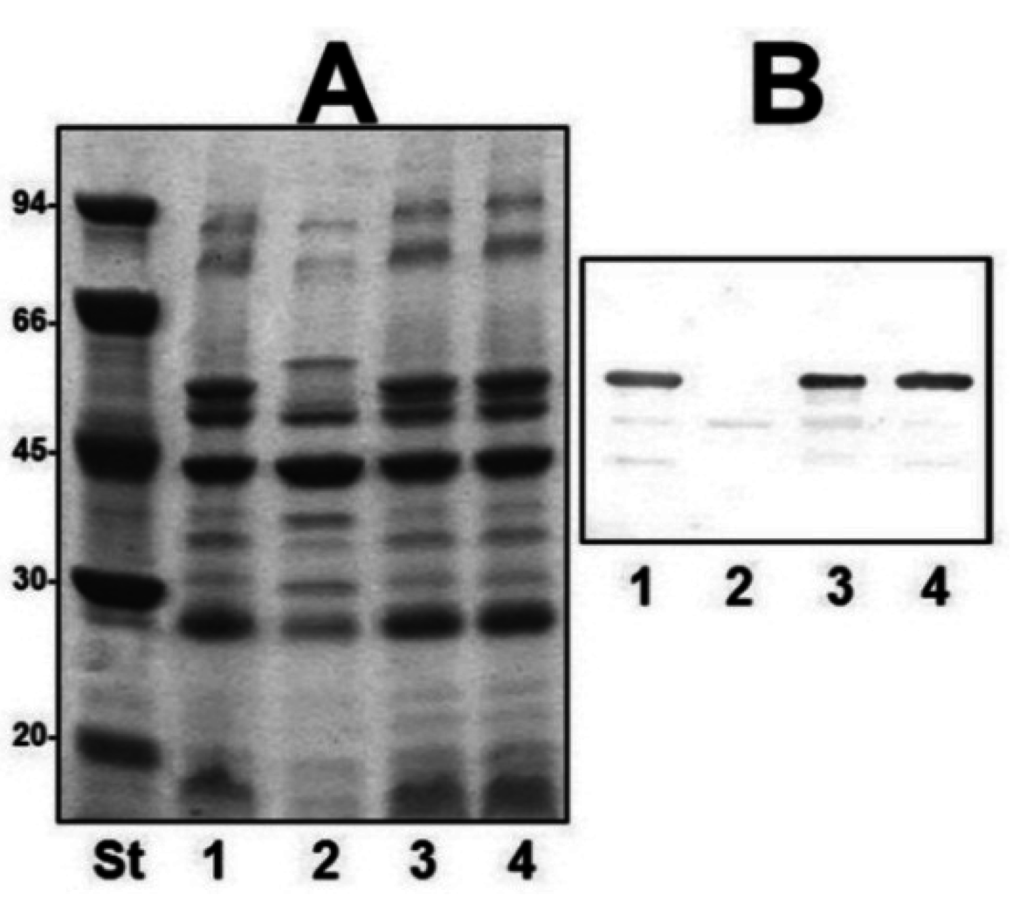
Figure 3.
(A) Outer membrane proteins, and (B) Western blot using antiserum against A-layer protein (anti-VapA protein) of strains: A450 (wild type, lane 1), mutant A450ΔvapA (lane 2), A450ΔvapA complemented with A450 vapA (lane 3), and A450ΔUDP-ep (lane 4). St, molecular weight standard.
The reintroduction of the corresponding wild-type genes in A450ΔrmlB, A450Δwzt, A450ΔwbbB, and A450Δgtf-2 fully restored the LPS profile of the wild-type strain in silver-stained SDS gels (Figure 2). A similar situation was observed when wild-type vapA was reintroduced in mutant A450ΔvapA, the presence in the OMp profile of the 49 kDa protein reacting with specific serum anti-A protein was restored (Figure 3).
2.3. Different A. salmonicida Subspecies Strains
When we analyzed the LPS by SDS-PAGE gels from different A. salmonicida subsp. salmonicida strains, we found an identical O-antigen LPS profile in all of them. However, when we analyzed the LPS profile in gels from different A. salmonicida subspecies strains, we found that the strains belonging to A. salmonicida subsp. pectinolytica lack the characteristic LPS O-antigen homogeneous band, while the rest of the subspecies showed it (Figure 4A).
The analysis of the OMp from different A. salmonicida subspecies strains by Western blot using specific anti-A protein antiserum is shown in Figure 4B. With the exception of two strains identified as A. salmonicida subsp. pectinolytica, all other strains produced a band of approximately 49 kDa which reacted with antiserum against A-protein (Figure 4B).
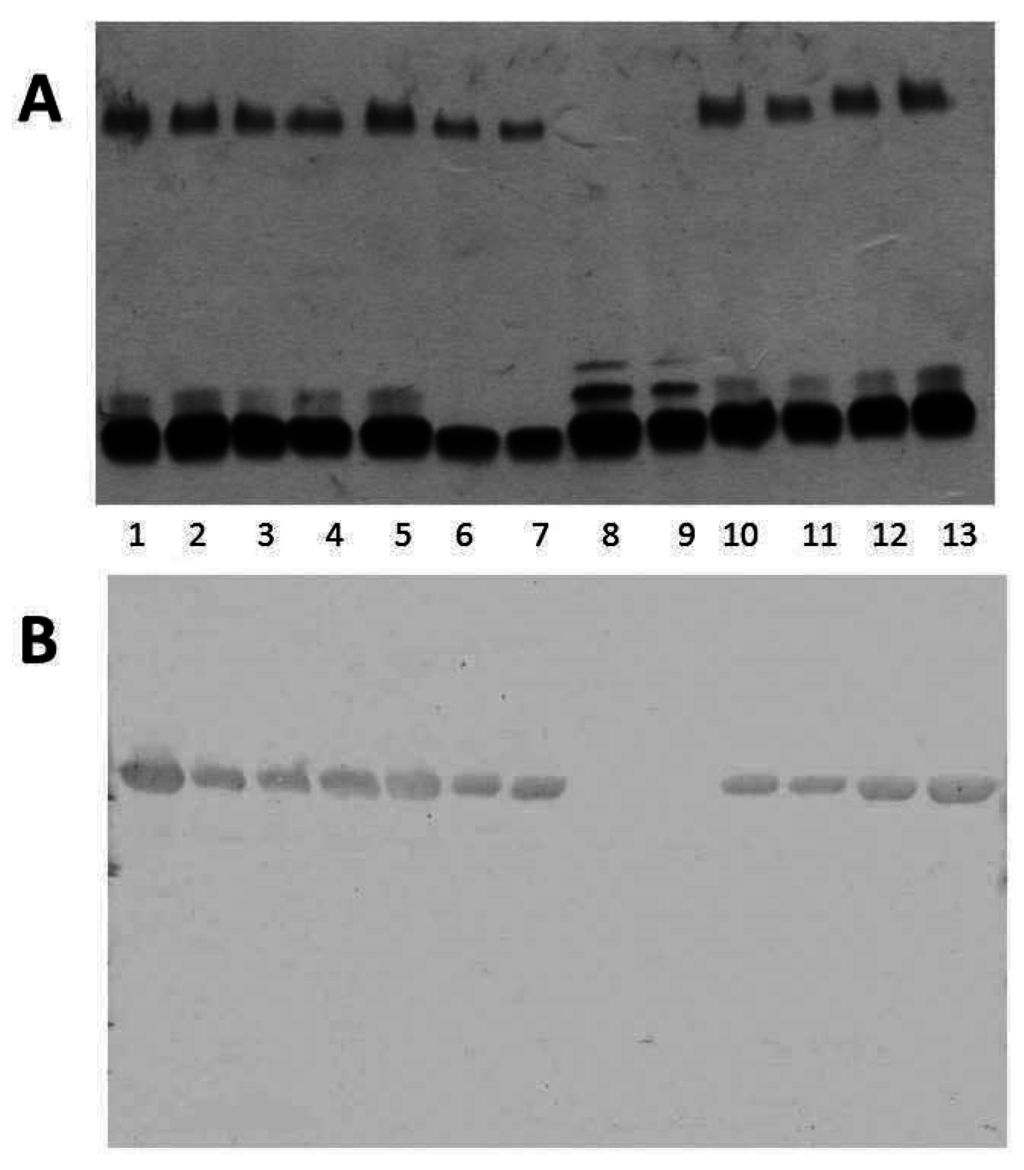
Figure 4.
(A) LPS analysed by SDS-PAGE (12%) and silver stained from different A. salmonicida subspecies strains. Lane 1, subsp. salmonicida A450; lane 2, subsp. masoucida strain CECT896T; lane 3, subsp. masoucida strain AS60; lane 4, subsp. achromogenes strain CECT4238; lane 5, subsp. achromogenes strain CECT895T; lane 6, subsp. achromogenes strain AS46; lane 7, subsp. achromogenes strain AS102; lane 8, subsp. pectinolytica strain CECT5752T; lane 9, subsp. pectinolytica strain CECT5753; lane 10, subsp. smithia strain CECT5179; lane 11, subsp. smithia strain AS74; lane 12, subsp. salmonicida strain CECT894; and lane 13, subsp. salmonicida strain CECT4235; (B) Western blot analysis using specific serum anti-A protein and OMp from different A. salmonicida subspecies strains. Lanes as in A.
2.4. Analysis of Fully Sequenced Genomes
When we inspected all the currently available Aeromonas salmonicida genomes, we found two for A. salmonicida subsp. salmonicida strains A449 and 01-B526, one for A. salmonicida subsp. achromogenes strain AS03, one for A. salmonicida subsp. masoucida strain NBRC 13784, one for A. salmonicida subsp. pectinolytica strain 34melT, and none for A. salmonicida subsp. smithia. Despite the different genome annotations (in part because these DNA regions had not been properly studied) with a more accurate comparative genomic analysis, we can conclude that in all the A. salmonicida genomes, besides the one for A. salmonicida subsp. pectinolytica strain 34melT, the wbsalmo is nearly identical. Furthermore, the chromosomal location between the oprM and UDP-ep genes is conserved among them. However, in the A. salmonicida subsp. pectinolytica strain 34melT genome, we found a putative wb cluster completely different. Briefly, the rml genes for rhamnose biosynthesis are not together, wzm-wzt are not present, and besides finding an oprM gene upstream, downstream of the rmlC seems to be a region with partial genes and encoded bacteriophage proteins (see Figure 5 and Table 3 ).
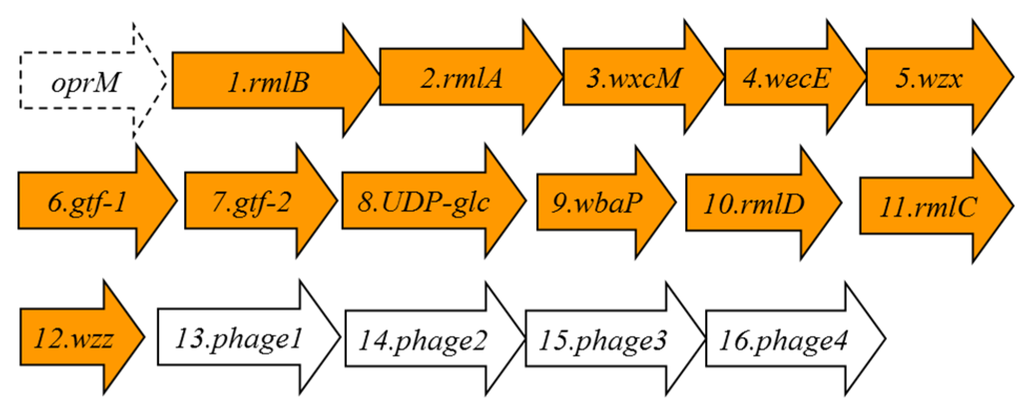
Figure 5.
The A. salmonicida subsp. pectinolytica strain 34melT putative genes for the LPS O-antigen cluster in orange. The genes, numbered according to the ORF number, were named according to their similarity, as found by their encoding proteins, with proteins of well characterized functions.

Table 3.
Characteristics of the proteins encoded by the ORFs detected in their LPS O-antigen cluster.
| ORF | Protein Name | Protein Size in Amino Acid Residues | Predicted Function |
|---|---|---|---|
| 1 | RmlB | 361 | dTDP-glucose-4-6-dehydratase RmlB |
| 2 | RmlA | 289 | Glucose-1-phosphate thymidylyl transferase RmlA |
| 3 | WxcM | 137 | dTDP-6-deoxy-3,4-keto-hexulose isomerase. |
| 4 | WecE | 367 | aminotransferase family, WecE |
| 5 | Wzx | 416 | O-antigen flippase |
| 6 | Gtf-1 | 140 | glycosyl transferase group 1 |
| 7 | Gtf-2 | 249 | glycosyl transferase group 2 |
| 8 | UDP-glc | 388 | UDP-glucose 6-dehydrogenase |
| 9 | WbaP | 423 | polyprenyl glycosyl phosphotransferase |
| 10 | RmlD | 296 | dTDP-4-dehydro rhamnose reductase |
| 11 | RmlC | 176 | dTDP-4-dehydro rhamnose 3,5-epimerase |
| 12 | Wzz | 202 | O-antigen size regulator protein |
| 13 | Phage1 | 113 | Phage terminase 1 protein |
| 14 | Phage2 | 283 | phage portal protein |
| 15 | Phage3 | 141 | phage prohead peptidase |
| 16 | Phage4 | 398 | putative phage phi-C31 gp36 major capsid-like protein |
3. Discussion
The A. salmonicida A450 LPS O-antigen gene cluster (wbsalmo) showed a G + C percentage of approximately 49.1%, which is significantly lower than the expected (59%–63%) for this species. This is characteristic of the wb clusters from different bacteria that can be 10% lower in G + C than the species average. Initially the encoded proteins showed some consistency with the chemical structure of the O-antigen LPS. ORF1 to 4 (in order RmlB, D, A and C, respectively) are the biosynthetic proteins for dTDP-rhamnose production, ORF21 (WecB) is the enzyme that converts UDP-GlcNAc into UDP-ManAc [19], and rhamnose, ManAc, and Glc are the monosaccharide components of the LPS O-antigen. There is no need for UDP-Glc specific genes. Three glycosyl transferases seem to be involved in the biosynthesis of the O-antigen repeating unit (ORF20, named WbbB; ORF22, named Gtf-2 and ORF25, named WbbL). WbbL is a presumptive rhamnosyl transferase, while the WbbB showed a HexNAc transferase domain (here probably acetyl-N-mannosamine), and Gtf-2 could be the transferase for the Glc incorporation to the LPS O-antigen repeating unit. There is also an acetyl transferase (ORF24 named Atf-3) which is in agreement with the acetylated rhamnose of the LPS O-antigen.
ORF5 and 18 were similar to an ABC-2 type transport system integral membrane and an ATP-binding protein, respectively (Table 2). The ORF5 protein hydrophobicity analysis and identification of the putative transmembrane domains [20] indicate that it is an integral membrane protein. The ORF18 protein contains the sequence GHNGAGKS (amino acid residues 57 to 64) which corresponds to the Walker box A, a motif present in ATP-binding proteins, as well as the ABC transporter family signature YSSGMYVRLAFAVQA (amino acid residues 162–175). Thus, ORF5 and 18 were named wzm and wzt respectively, despite the fact that they are usually found adjacent. Two main known pathways for O-antigen export have been established [9], the Wzy-dependent pathway for heteropolysaccharide structures and the ABC-2 transporter-dependent pathway mainly for homopolysaccharides or disaccharides. The presence of complete Wzm (ORF5) and Wzt (ORF18) showed that this LPS O-antigen belongs to the second pathway and represents the first example of an O-antigen LPS polysaccharide with three different monosaccharides in their repeating unit assembled by this pathway. Between the Wzm (ORF5) and Wzt (ORF18), we found the genes responsible for the biosynthesis and assembly of the S-layer (named A-layer), (Figure 1 and Table 2). The S-layer-encoding genes included a large group of T2SS genes plus some encoding hypothetical proteins related to this specific S-layer T2SS system (ORF6 to 16), and the last gene encoding the unique protein that forms the A-layer (vapA, ORF17) characteristic of A. salmonicida strains. The insertion point of this genomic cluster seems to be in the intergenic region between the genes encoding for a Wzm protein, located just upstream of the S-layer genes, and the gene encoding a Wzt protein (ORF18, as previously indicated). Accordingly to this, the A450ΔvapA in frame mutant showed identical LPS profile as the wild-type strain with the same O-antigen profile in SDS-PAGE gels, but lacked the VapA protein being then unable to produce the A-layer. Mutants A450ΔrmlB, A450Δwzt, A450ΔwbbB, and A450Δgtf-2, when analyzed in an SDS-PAGE gel, were unable to produce LPS O-antigen. However, the A450ΔUDP-ep mutant showed an identical LPS profile as the wild-type strain with the presence of O-antigen in the LPS. Then, we concluded that ORF22 (UDP-ep) does not belong to the wbsalmo, because it seems not to be need for LPS O-antigen biosynthesis.
It is characteristic of the LPS O-antigen disaccharides assembled and exported thought an ABC-2 type transporter to contain a large protein (WbbB), like in K. pneumoniae O12 [21,22], that shows two clear domains. The first domain, in K. pneumoniae WbbB consisted of a condensation or capsule polysaccharide biosynthesis domain from amino acid residues 125 to 300; and a second domain with glycosyltransferase activity from GT1 family (625 to 825 amino acid residues) related to HexNAc transferases [21]. The A. salmonicida WbbB (ORF20) is larger than the one in Klebsiella, and showed three domains: capsule polysaccharide biosynthesis (130 to 290 amino acid residues), a glycosyltransferase activity from GT1family 1 (500 to 825 amino acid residues), and an additional glycosyltransferase activity domain from GT2 family (850 to 1050 amino acid residues). The fact there is an additional glycosyltransferase domain in the A. salmonicida WbbB seems to be in accordance with the O-antigen being a trisaccharide instead of a disaccharide.
At this moment, not much can be said about the hypothetical protein with a sulfotransferase domain (ORF 19, named Hyp), no such residue seems to be found in the chemical structure of the LPS O-antigen in these strains [10,12], and also about the hypothetical protein (ORF23 named ArnT) with a 4-amino-4-deoxy-l-arabinose transferase domain. However, it is true that in Aeromonas has been described the presence of a 4-amino-4-deoxy-l-arabinose in the lipid A [23]. Both genes encoding these proteins are clearly conserved among the different A. salmonicida wbsalmo. The biosynthesis of O-antigens starts with the assembly of monomers on an undecaprenol phosphate (lipid carrier), before their incorporation into the LPS molecules, by enzymes that can be present or not in the wb gene cluster [9]. In wbsalmo we could not find such enzyme.
When we inspected and deeply studied all the currently available A. salmonicida genomes, we found that the wbsalmo was conserved in all of the different subspecies with identical gene location besides in A. salmonicida subsp. pectinolytica. This fact is in full agreement with the LPS profiles in SDS-PAGE obtained for the different A. salmonicida strains from several subspecies. Sequence analysis of the A. salmonicida subsp. pectinolytica wb gene cluster from strain 34melT revealed that the LPS O-antigen seems to be exported through a Wzx-Wzy pathway and not an ABC-2 transporter-dependent pathway. However, besides the presence of the typical genes encoding the Wzx and Wzz proteins, no gene encoding a Wzy protein could be found in this cluster. Wzy mutants showed only a single O-antigen repeating unit [9] and, in the case of the LPS profile of A. salmonicida subsp. pectinolytica strains, it seems that a single O-antigen unit could be observed. A. salmonicida subsp. pectinolytica strains clearly show a different O-antigen LPS compared to the other subspecies and lack the A-layer according to our results. No such A-layer protein encoded by vapA gene could be detected by blot hybridization or PCR using appropriate DNA primers (Figure 6). This gene could not be found in the complete genome of A. salmonicida subsp pectinolytica strain 34melT.
These two features, lack of wbsalmo and A-layer, are in agreement with the fact that atypical A. salmonicida subsp. pectinolytica strains are the only ones not found as pathogens in a wide variety of fish species [24]. Besides, no genome is available for A. salmonicida subsp. smithia. Our results suggest that the wbsalmo should also be conserved in these strains, according to the LPS profile in SDS-PAGE gels.
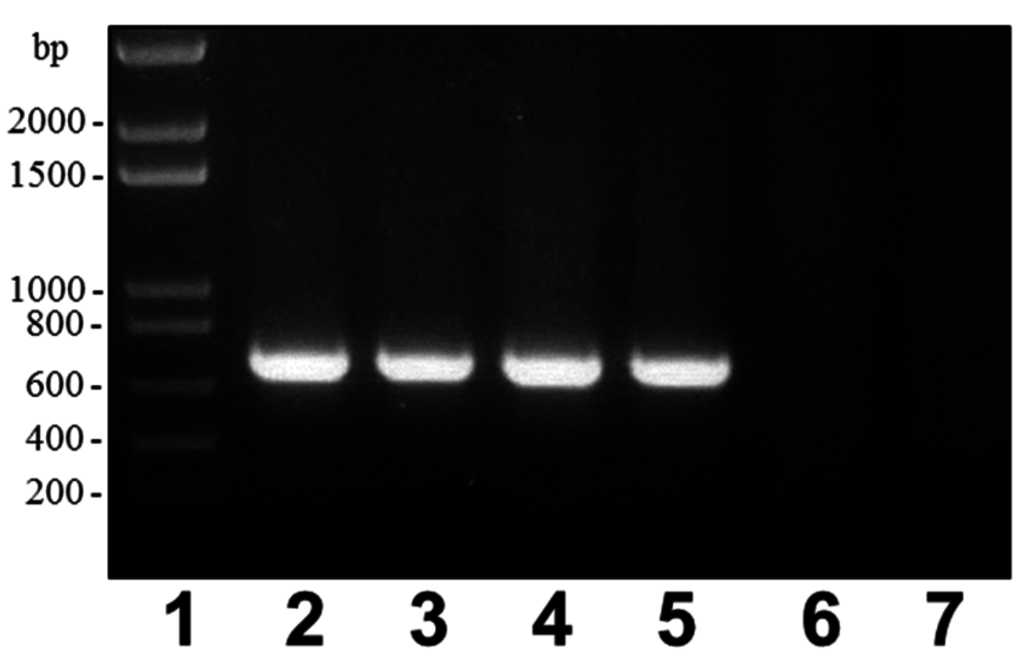
Figure 6.
PCR amplified band (696 bp) with primers VapA-for: 5′-ATCGACAGCAATGGCAAG-3′ and VapA-rev: 5′-ATCACGGGTGAGGATGAAG-3′ and A. salmonicida genomic DNAs from strains: subspecies salmonicida A450 (lane 2), subspecies masoucida CECT4235 (lane 3), subspecies achromogenes CECT4238 (lane 4), subspecies smithia CECT5179 (lane 5), subspecies pectinolytica CECT5752T (lane 6), and subspecies pectinolytica CECT5753 (lane 7). Lane 1, DNA molecular weight standard.
4. Materials and Methods
4.1. Bacterial Strains, Plasmids and Growth Conditions
The bacterial strains and plasmids used in this study are listed in Table 4. E. coli strains were grown on Luria-Bertani (LB) Miller broth and LB Miller agar at 37 °C, while Aeromonas salmonicida strains were grown either in tryptic soy broth (TSB) or agar (TSA) at 20 °C. Ampicillin (100 µg·mL−1), chloramphenicol (25 µg·mL−1), tetracycline (20 µg·mL−1), kanamycin (50 µg·mL−1), nalidixic acid (20 µg·mL−1) and gentamycin (20 µg·mL−1) were added to the different media when required. A. salmonicida AS46, AS60, AS74, and AS102 were kindly provided by Prof. Brian Austin (University of Stirling, Scotland).
4.1.1. General DNA Methods
General DNA manipulations were done essentially as previously described [30]. DNA restriction endonucleases, T4 DNA ligase, E. coli DNA polymerase (Klenow fragment), and alkaline phosphatase were used as recommended by the Sigma-Aldrich (St Louis, MO, USA).
4.1.2. DNA Sequencing and Computer Analysis of Sequence Data
Double-stranded DNA sequencing was performed by using the dideoxy-chain termination method [31] from PCR amplified DNA fragments with the ABI Prism dye terminator cycle sequencing kit (PerkinElmer, Barcelona, Spain). Oligonucleotides used for genomic DNA amplifications and DNA sequencing were purchased from Sigma-Aldrich (St Louis, MO, USA). Deduced amino acid sequences were compared with those of DNA translated in all six frames from non-redundant GenBank and EMBL databases by using the BLAST [32] network service at the National Center for Biotechnology Information and the European Biotechnology Information. ClustalW was used for multiple-sequence alignments [33].

Table 4.
Bacterial strains and plasmids used.
| Strain or Plasmid | Relevant Characteristics | Reference or Source |
|---|---|---|
| E. coli strains | ||
| DH5α | F− end A hsdR17 (rK− mK+) supE44 thi-1 recA1 gyr-A96 _80lacZM15 | [25] |
| MC1061 | thi thr1 leu6 proA2 his4 argE2 lacY1 galK2 ara14 xyl5, supE44, λpir | [26] |
| A. salmonicida strains | ||
| A450 | Wild type, subsp. salmonicida | [27] |
| A450nal R | A450 nalidixic acid resistant | [27] |
| A450ΔrmlB | A450 rmlB in frame mutant unable to produce LPS O-antigen | This study |
| A450Δwzt | A450 wzt in frame mutant unable to produce LPS O-antigen | This study |
| A450ΔwbbB | A450 wbbB in frame mutant unable to produce LPS O-antigen | This study |
| A450Δgtf-2 | A450 gtf-2 in frame mutant unable to produce LPS O-antigen | This study |
| A450ΔvapA | A450 vapA in frame mutant, unable to produce A-layer but able to produce LPS O-antigen | This study |
| A450ΔUDP-ep | A450 UDP-ep in frame mutant, able to produce LPS O-antigen and A-layer | This study |
| CECT894 | Wild type, subsp. salmonicida | CECT |
| CECT4235 | Wild type, subsp. salmonicida | CECT |
| CECT896T | Wild type, subsp. masoucida | CECT |
| AS60 | Wild type, subsp. masoucida | [28] |
| CECT4238 | Wild type, subsp. achromogenes | CECT |
| CECT895T | Wild type, subsp. achromogenes | CECT |
| AS46 | Wild type, subsp. achromogenes | [28] |
| AS102 | Wild type, subsp. achromogenes | [28] |
| CECT5752T | Wild type, subsp. pectinolytica | CECT |
| CECT5753 | Wild type, subsp. pectinolytica | CECT |
| CECT5179 | Wild type, subsp. smithia | CECT |
| AS74 | Wild type, subsp. smithia | [28] |
| Plasmids | ||
| pGEMT easy | PCR generated DNA fragment cloning vector Amp R | Promega |
| pBAD33-Gm | Arabinose-inducible expression vector, Gm R | [27,29] |
| pDM4 | pir dependent with sacAB genes; oriR6K; Cm R | [27] |
| pLA2917 | Cosmid vector, Km R, Tc R | [15] |
R = resistant, CECT = SPANISH TYPE CULTURE COLLECTION.
4.1.3. Mutant and Plasmid Constructions, Mutant Complementation Studies
The chromosomal in-frame A450ΔrmlB, A450ΔvapA, A450Δwzt, A450ΔwbbB, A450Δgtf-2 and A450ΔUDP-ep deletion mutants were constructed by allelic exchange as described by Milton et al. [26]. Plasmids were transferred to A. salmonicida strains as previously indicated [27]. To complete the allelic exchange, the integrated suicide plasmid was forced to recombine out of the chromosome by growing on agar plates containing 10% sucrose. Mutants were selected based on their survival on plates containing 10% sucrose and loss of the chloramphenicol resistant marker of vector pDM4. The mutations were confirmed by sequencing of the whole constructs in amplified PCR products. The primers used are shown in Table 5.
Table 5.
(A) Primers used in the construction of chromosomal in-frame deletion mutants. (B) Primers used for mutant complementation using vector pBAD33-Gm.

| A | |
|---|---|
| Primers a,b | Amplified Fragment |
| rmlB | |
| A: 5′-CGCGGATCCCAAGTTCTGCCTGGTAT-3′ | AB (632 bp) |
| B: 5′-TGTTTAAGTTTAGTGGATGGGTGCACCACCAGTGACAAG-3′ | |
| C: 5′-CCCATCCACTAAACTTAAACAAGTGGTGCCTACCAATCCT-3′ | CD (704 bp) |
| D: 5′-CGCGGATCCAACATCGGGTTTGCTCT-3′ | |
| AD (1312 bp) | |
| vapA | |
| A: 5′-GAAGATCTGCCGATTCAGGTAAAACAG-3′ | AB (717 bp) |
| B: 5′-TGTTTAAGTTTAGTGGATGGGGCTAATCACGACATCAGCA-3′ | |
| C: 5′-CCCATCCACTAAACTTAAACA GAAGGCGTGGATATTCAGA-3′ | CD (670 bp) |
| D: 5′-GAAGATCTAACGATCATCCATCTCTCG-3′ | |
| AD (1366 bp) | |
| wzt | |
| A: 5′-CGCGGATCCGAGCTGGCTGATCTCTTCA-3′ | AB (721 bp) |
| B: 5′-TGTTTAAGTTTAGTGGATGGGGGAACGATAGATGGGAAATG-3′ | |
| C: 5′-CCCATCCACTAAACTTAAACAGATGTCGCCATGTTTCAAG-3′ | CD (653 bp) |
| D: 5′-CGCGGATCCTGATTGGGCGAAAATA-3′ | |
| AD (1353 bp) | |
| wbbB | |
| A: 5′-CGCGGATCCTACTTGCCCGAGATACCAG-3′ | AB (659 bp) |
| B: 5′-TGTTTAAGTTTAGTGGATGGGACCTAGCACGACCCAAAG-3′ | |
| C: 5′-CCCATCCACTAAACTTAAACAGTTAAGCAGGCGCTATTTG-3′ | CD (753 bp) |
| D: 5′-CGCGGATCCTACGATGCGATGTTACCAA-3′ | |
| AD (1391 bp) | |
| gtf-2 | |
| A: 5′-CGCGGATCCGCACCTACGCAAATTTCTC-3′ | AB (722 bp) |
| B: 5′-TGTTTAAGTTTAGTGGATGGGCACCGGTGAAAGATAAACC-3′ | |
| C: 5′-CCCATCCACTAAACTTAAACATTTCATAATAGTGGCGATGC-3′ | CD (631bp) |
| D: 5′-CGCGGATCCGACTGCCGTCTCTTTGAAC-3′ | |
| AD (1332 bp) | |
| UDP-ep | |
| A: 5′-CGCGGATCCTGGCGTTGAATAATGGAG-3′ | AB (646 bp) |
| B: 5′-TGTTTAAGTTTAGTGGATGGGCTTACCAACAAACCCGTTG-3′ | |
| C: 5′-CCCATCCACTAAACTTAAACAAAGGCTCAGAGGCGATTAC-3′ | CD (771 bp) |
| D: 5′-CGCGGATCCACCATCCCCCATAAAGAT-3′ | |
| AD (1395 bp) | |

| B | ||
|---|---|---|
| Plasmid | Primers | Amplified Fragment |
| pBADGm-rmlB c | RmlB-FOR: 5′-TCCCCCGGGTTAAAAGCAGCGAACTG-3′ | 1380 bp |
| RmlB-REV: 5′-GCTCTAGACGCTGGAGTCAAAATCAAC-3′ | ||
| pBADGm-vapA c | VapA-FOR: 5′-TCCCCCGGGTGATCAACGGATAGGTTCAA-3′ | 1666 bp |
| VapA-REV: 5′-GCTCTAGAAGGGAACAAATGAAACTGCT-3′ | ||
| pBADGm-wzt c | Wzt-FOR: 5′-TCCCCCGGGTGACCACAGCCCTTATTTC-3′ | 1473 bp |
| Wzt-REV: 5′-GCTCTAGATGCAGTAGTCCCACCTTTT-3′ | ||
| pBADGm-wbbB d | WbbB-FOR: 5′GGAATTCTAAGCTCACGGTTGCACAG-3′ | 3689 bp |
| WbbB-REV: 5′-TCCCCCGGGATAACCGGAGCCATTTTGAT-3′ | ||
| pBADGm-gtf2 c | gtf2-FOR: 5′-TCCCCCGGGATGGCTAAAGGTTCTTCACC-3′ | 1269 bp |
| gtf2-REV: 5′-GCTCTAGACATGACTGAAATACCCTGGA-3′ | ||
a Italic letters show overlapping regions. b Underlined letters show BamHI or BglII restriction site. c Primers contain SmaI(bold) and XbaI(underlined), the PCR amplified product was ligated to SmaI-XbaI digested pBAD33-Gm. d Primers contain EcoRI (doubleunderlined) and SmaI(bold), the PCR amplified product was ligated to EcoRI-SmaI digested pBAD33-Gm.
For complementation studies, the A. salmonicida A450 rmlB, vapA, wzt, wbbB, and gtf-2 genes were PCR amplified using appropriate oligonucleotides obtained from the sequenced clones and chromosomal A450 DNA as template, ligated to plasmid pGEMT (Promega), and transformed into E. coli DH5α. After checked, the corresponding genes were subcloned on plasmid pBAD33-Gm (Table 5) with an arabinose-inducible and glucose-repressible promoter [27,29]. Induction was obtained by adding l-arabinose to a final concentration of 0.2% (w/v). Plasmids were transferred to A. salmonicida strains as previously indicated [27].
4.1.4. LPS Characterization and SDS-PAGE
LPS was obtained after proteinase K digestion of whole cells [34] for screening purposes. LPS samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by silver staining as previously described [34].
4.1.5. LPS Isolation and O-Deacetylation
Cells (3 g dried weight) were digested with DNase, RNase (24 h, 3 mg each) and Proteinase K (36 h, 3 mg) in 25 mM Tris-HCl buffer pH 7.63 containing 2 mM CaCl2 (30 mL), the suspension was dialysed against distilled water and freeze-dried. Digested cells were extracted with aqueous 45% phenol at 68 °C [14], the extract was dialysed against tap water without separation of the layers, residual cells were removed by centrifugation, and the supernatant was freeze-dried to give lipopolysaccharide sample. An aliquot (150 mg) was degraded with 0.1 M sodium acetate buffer pH 4.2 for 4 h at 100 °C, the lipid precipitate was removed by centrifugation (13,000× g, 20 min), and high-molecular-mass O-polysaccharide (40 mg) was isolated from the supernatant by gel-permeation chromatography on a column (50 × 2.5 cm) of Sephadex G-50 Superfine in pyridinium acetate buffer (4 mL pyridine and 10 mL HOAc in 1 L water) using a Knauer differential refractometer (Knauer, Germany) for monitoring. The polysaccharide was deacetylated by heating with aqueous 12% ammonia (2 mL) for 3 h at 60 °C, ammonia was removed by stream of air, and the remaining solution was freeze-dried.
Sugar analysis and Electrospray liquid chromatography mass spectrometry analysis. For sugar analysis, a polysaccharide sample (1 mg) was hydrolyzed with 2 M CF3CO2H (100 °C, 4 h), the monosaccharides were conventionally converted into the alditol acetates [35] and analyzed by GLC on a Varian 3700 chromatograph (Varian Inc., Palo Alto, CA, USA) equipped with a fused-silica gel SPB-5 column using a temperature gradient from 150 °C (3 min) to 320 °C at 5° min−1. The absolute configurations of the monosaccharides were determined as described [36], using the same GLC conditions as in sugar analysis.
Mass spectrometry studies of purified O-polysaccharide were performed in the negative ion mode using a microTOF II instrument (Bruker Daltonics). A sample of the O-polysaccharide (~50 ng·µL−1) was dissolved in a 1:1 (v/v) water-acetonitrile mixture and sprayed at a flow rate of 3 µL·min−1. Capillary entrance voltage was set to 4.5 kV and exit voltage to −150 V; drying gas temperature was 180 °C.
4.1.6. OM Protein and S-Layer Isolation and Characterization
Outer membranes (OM) were obtained by incubating membrane suspensions with 3% Sarkosyl in 20 mM TrisHCl buffer (pH 8.0) for 20 min at room temperature, as previously described [37]. Protein was analysed by SDS-PAGE and separated protein bands were visualized by Coomassie Brilliant blue staining as previously described [37]. Anti-purified-A-layer antiserum was obtained and assayed as previously described [38]. After SDS-PAGE, immunoblotting was carried out as previously described [39].
Acknowledgments
This work was supported by Plan Nacional de I + D + i grants (Ministerio de Economía y Competitividad, Spain), and Generalitat de Catalunya (Centre de Referència en Biotecnologia). We also thank Maite Polo for her technical assistance.
Author Contributions
S. Merino and J. M. Tomás conceived and designed the experiments; S. Merino, E. de Mendoza and R. Canals performed the experiments; all the authors analyzed the data; S. Merino and J. M. Tomás contributed reagents/materials/analysis tools and wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Austin, B.; Austin, D.A. Characteristics of the pathogens: Gram-negative bacteria. In Bacterial Fish Pathogens: Diseases of Farmed and Wild Fish; Austin, B., Austin, D.A., Eds.; Springer Praxis Publishing: Chichester, UK, 2007; pp. 81–150. [Google Scholar]
- Scott, M. The pathogenicity of Aeromonas salmonicida (Griffin) in sea and brackish waters. J. Gen. Microbiol. 1968, 50, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, E.E.; Kay, W.W.; Ainsworth, T.; Chamberlain, J.B.; Buckley, J.T.; Trust, T.J. Loss of virulence during culture of Aeromonas salmonicida at high temperature. J. Bacteriol. 1981, 148, 393–400. [Google Scholar]
- Belland, R.J.; Trust, T.J. Synthesis, export, and assembly of Aeromonas salmonicida A-layer analysed by transposon mutagenesis. J. Bacteriol. 1985, 163, 877–881. [Google Scholar] [PubMed]
- Dooley, J.S.G.; Engelhardt, H.; Baumeister, W.; Kay, W.W.; Trust, T.J. Three dimensional structure of an open form of the surface layer from the fish pathogen Aeromonas salmonicida. J. Bacteriol. 1989, 171, 190–197. [Google Scholar] [PubMed]
- Phipps, B.M.; Kay, W.W. Immunoglobulin binding by the regular surface array of Aeromonas salmonicida. J. Biol. Chem. 1988, 263, 9298–9303. [Google Scholar] [PubMed]
- Munn, C.B.; Ishiguro, E.E.; Kay, W.W.; Trust, T.J. Role of the surface components in serum resistance of virulent Aeromonas salmonicida. Infect. Immun. 1982, 36, 1069–1075. [Google Scholar] [PubMed]
- Merino, S.; Albertí, S.; Tomás, J.M. Aeromonas salmonicida resistance to complement-mediated killing. Infect. Immun. 1994, 62, 5483–5490. [Google Scholar] [PubMed]
- Aquilini, E.; Tomás, J.M. Lipopolysaccharides (Endotoxins). In Reference Module in Biomedical Sciences; Elsevier: Amsterdam, The Netherlands, 2015; in press. [Google Scholar]
- Shaw, D.H.; Lee, Y.Z.; Squires, M.J.; Lüderitz, O. Structural studies on the O-antigen of Aeromonas salmonicida. Eur. J. Biochem. 1983, 131, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Hart, M.J.; Lüderitz, O. Structure of the core oligosaccharide in the lipopolysaccharide isolated from Aeromonas salmonicida ssp. salmonicida. Carbohydr. Res. 1992, 231, 83–91. [Google Scholar]
- Wang, Z.; Vinogradov, E.; Larocque, S.; Harrison, B.A.; Li, J.; Altman, E. Structural and serological characterization of the O-chain polysaccharide of Aeromonas salmonicida strains A449, 80204 and 80204-1. Carbohydr. Res. 2005, 340, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, J.; Vinogradov, E.; Altman, E. Structural studies of the core region of Aeromonas salmonicida subsp. salmonicida lipopolysaccharide. Carbohydr. Res. 2006, 341, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Westphal, O.; Jann, K. Bacterial lipopolysaccharide extraction with phenol-water and further application of the procedure. Methods Carbohydr. Chem. 1965, 5, 83–89. [Google Scholar]
- Nogueras, M.M.; Merino, S.; Aguilar, A.; Benedí, V.J.; Tomás, J.M. Cloning, sequencing and role in serum susceptibility of porin II from mesophilic Aeromonas sp. Infect. Immun. 2000, 68, 1849–1854. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, N.; Canals, R.; Saló, M.T.; Vilches, S.; Merino, S.; Tomás, J.M. The Aeromonas hydrophila wb*O34 gene cluster: Genetics and temperature regulation. J. Bacteriol. 2008, 190, 4198–4209. [Google Scholar] [CrossRef] [PubMed]
- Artsimovitch, I.; Landick, R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell 2002, 109, 193–203. [Google Scholar] [CrossRef]
- Bailey, M.J.; Hughes, C.; Koronakis, V. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol. Microbiol. 1997, 26, 845–851. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Morales, F.; Prieto, A.I.; Beuzón, C.R.; Holden, D.W.; Casadesús, J. Role for Salmonella enterica Enterobacterial Common Antigen in Bile Resistance and Virulence. J. Bacteriol. 2003, 185, 5328–5332. [Google Scholar] [CrossRef] [PubMed]
- Klein, P.; Kanehisa, M.; DeLisi, C. The detection and classification of membrane-spanning proteins. Biochim. Biophys. Acta 1985, 815, 468–476. [Google Scholar] [CrossRef]
- Izquierdo, L.; Merino, S.; Regué, M.; Rodríguez, F.; Tomás, J.M. A Klebsiella pneumoniae O-antigen heteropolysaccharide (O12) requiring an ABC-2-trasnporter dependent pathway. J. Bacteriol. 2003, 185, 1634–1641. [Google Scholar] [CrossRef] [PubMed]
- Saigi, F.; Climent, N.; Piqué, N.; Sanchez, C.; Merino, S.; Rubirés, X.; Aguilar, A.; Tomás, J.M.; Regué, M. Genetic analysis of the Serratia marcescens N28b O4 antigen gene cluster. J. Bacteriol. 1999, 181, 1883–1891. [Google Scholar] [PubMed]
- Knirel, Y.A.; Vinogradov, E.; Jimenez, N.; Merino, S.; Tomás, J.M. Structural studies on the R-type lipopolysaccharide of Aeromonas hydrophila. Carbohydr. Res. 2004, 339, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Gudmundsdottir, B.K. Infections by atypical strains of the bacterium Aeromonas salmonicida. Iceland Agric. Sci. 1998, 12, 61–72. [Google Scholar]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Milton, D.L.; O’Toole, R.; Horstedt, P.; Wolf-Watz, H. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 1996, 178, 1310–1319. [Google Scholar]
- Jiménez, N.; Lacasta, A.; Vilches, S.; Reyes, M.; Vazquez, J.; Aquillini, E.; Merino, S.; Regué, M.; Tomás, J.M. Genetics and proteomics of Aeromonas salmonicida Lipopolysaccharide Core Biosynthesis. J. Bacteriol. 2009, 191, 2228–2236. [Google Scholar] [CrossRef] [PubMed]
- Austin, B.; Austin, D.A.; Dalsgaard, I.; Gudmundsdottir, B.K.; Høie, S.; Thornton, J.M.; Larsen, J.L.; O’Hici, B.; Powell, R. Characterization of atypical Aeromonas salmonicida by different methods. Syst. Appl. Microbiol. 1998, 21, 50–64. [Google Scholar] [CrossRef]
- Guzman, L.M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Hitchcock, P.J.; Brown, T.M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 1983, 154, 269–277. [Google Scholar] [PubMed]
- Sawardeker, J.S.; Sloneker, J.H.; Jeanes, A. Spectrophotometric determination of high molecular weight quaternary ammonium cations in polysaccharides. Anal. Chem. 1965, 37, 243–246. [Google Scholar]
- Widmalm, G.; Leontein, K. Structural studies of the Escherichia coli O127 O-antigen polysaccharide. Carbohydr. Res. 1993, 247, 87–89. [Google Scholar] [CrossRef]
- Esteve, C.; Amaro, C.; Toranzo, A.E. O-serogrouping and surface components of Aeromonas hydrophila and Aeromonas jandaei pathogenic for eels. FEMS Microbiol. Lett. 1994, 117, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Merino, S.; Aguilar, A.; Rubires, X.; Simón-Pujol, D.; Congregado, F.; Tomás, J.M. The role of the capsular polysaccharide of Aeromonas salmonicida in the adherence and invasion of fish cell lines. FEMS Microbiol. Lett. 1996, 142, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Vilches, S.; Jiménez, N.; Merino, S.; Tomás, J.M. The Aeromonas dsbA mutation decreased their virulence by triggering type III secretion system but not flagella production. Microb. Pathog. 2012, 52, 130–139. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).