Antifungal Compounds from Cyanobacteria
Abstract
:1. Introduction
2. Results and Discussion
2.1. Cyanobacteria Producing Antifungal Compounds
| Cyanobacteria | 16S rRNA Gene | Origin | Inhibited Organism(s) | Antifungal Compound |
|---|---|---|---|---|
| Nostoc sp. CENA 219 | KP701037 | Benthic freshwater, Brazil | Ca/Af | hassallidin |
| Anabaena sp. BIR JV1 | KP701036 | The Gulf of Finland | Ca | hassallidin |
| Anabaena sp. HAN7/1 | KP701033 | Epilithic, Finland | Ca | hassallidin |
| Nostoc calcicula 6 sf Calc | KP701034 | Dobre Pole, Czech Republic | Ca/Af | hassallidin |
| Anabaena sp. HAN21/1 | KP701032 | Gastropod, Finland | Ca/Af | scytophycin |
| Anabaena cf. cylindrica PH133 | AJ293110 | Lake Arresø, Denmark | Ca/Af | scytophycin |
| Scytonema sp. HAN3/2 | KP701039 | Green biofilm in the pond, Finland | Ca/Af | scytophycin |
| Nostoc sp. HAN11/1 | KP701035 | Small pond on a rock, Finland | Ca/Af | scytophycin |
| Fischerella sp. CENA 298 | KP701038 | Soil, Brazil | Ca/Af | unidentified |
| Scytonema hofmanni PCC 7110 | NR112180 | Limestone, Bermuda | Ca/Af | unidentified |
| Nostoc sp. N107.3 | KP701040 | Lichen, Finland | Af | unidentified |

| Strains | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Scytophycin (Sc) variant | Rt (min) | [M + Na]+ (m/z) | HAN3/2 | HAN11/1 | HAN21/1 | PH133 | ||||
| No. | Chemical variant | Exp | Cal | ∆ (ppm) | Formula | |||||
| 10 N | 6-OH-7-OMe-15-O-deMe-Sc-B | 19.9 | 858.5 | ++ | ||||||
| 15 | 6-OH-7-OMe-Sc-D/E | 20.1 | 874.5 | ++ | ||||||
| 16 N | X-OH-Sc-D/E (not 6-OH) | 20.3 | 860.5116 | 860.5131 | −1.70 | C45H75NO13 | ++ | + | ++ | |
| 18 | Sc-D/E | 20.6 | 844.5157 | 844.5181 | −2.90 | C45H75NO12 | ++ | + | ++ | |
| 19 N | Sc | 20.6 | 814.5a | ++ | ||||||
| 20 | 6-OH-7-OMe-Sc-B | 20.6 | 872.5 | +++ | + | |||||
| 22 N | Sc | 20.9 | 842.5 | ++ | + | |||||
| 25 | Sc-B | 21.3 | 842.5022 | 842.5025 | −0.35 | C45H73NO12 | +++ | ++ | ++ | +++ |
| 26 N | Sc, (-O from C15/16/17/19) | 21.9 | 826.5062 | 826.5076 | −1.67 | C45H73NO11 | ++ | + | + | ++ |
| 27 | Sc-C | 22.4 | 828.5221 | 828.5232 | −1.37 | C45H75NO11 | ++ | ++ | + | ++ |
| 29 N | 7-OMe-29-OAc-Sc-B | 22.6 | 884.5129 | 884.5131 | −0.18 | C47H75NO13 | ++ | |||
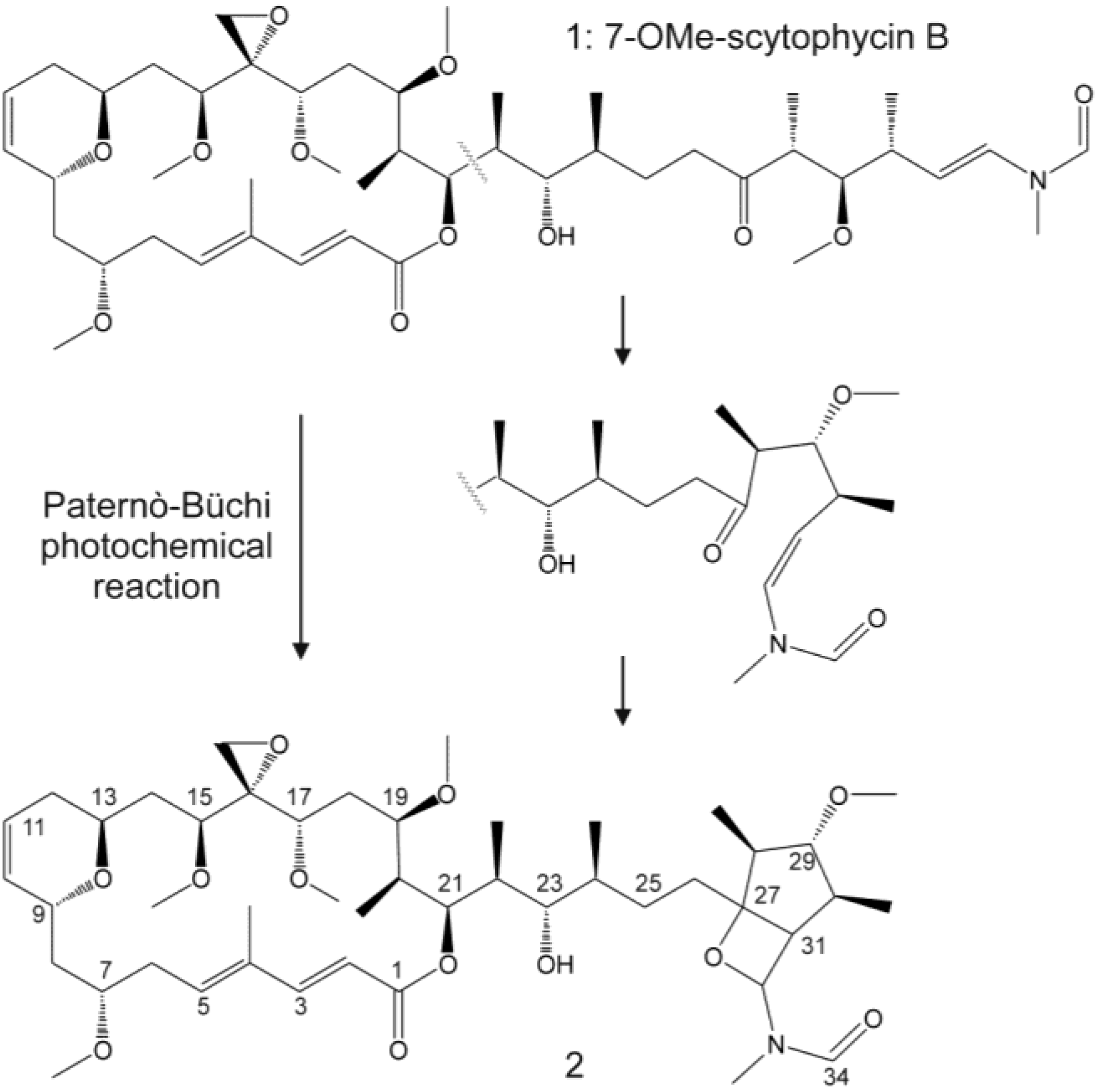
| 30 | 7-OMe-Sc-B (1) | 23.5 | 856.5 | + | + | +++ | +++ | |||
| 31 N | 7-OMe-Sc, (-O from C15/16/17) | 24.0 | 840.5 | + | ++ | + | ||||
| Strains | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Rt | Ion masses (m/z) | Monosaccharides | CENA 219 | 6sf Calc | HAN7/1 | BIR JV1 | ||||
| No | (min) | AL [M + H]+ | [M + Na]+ | M1 | M2 | M3 | ||||
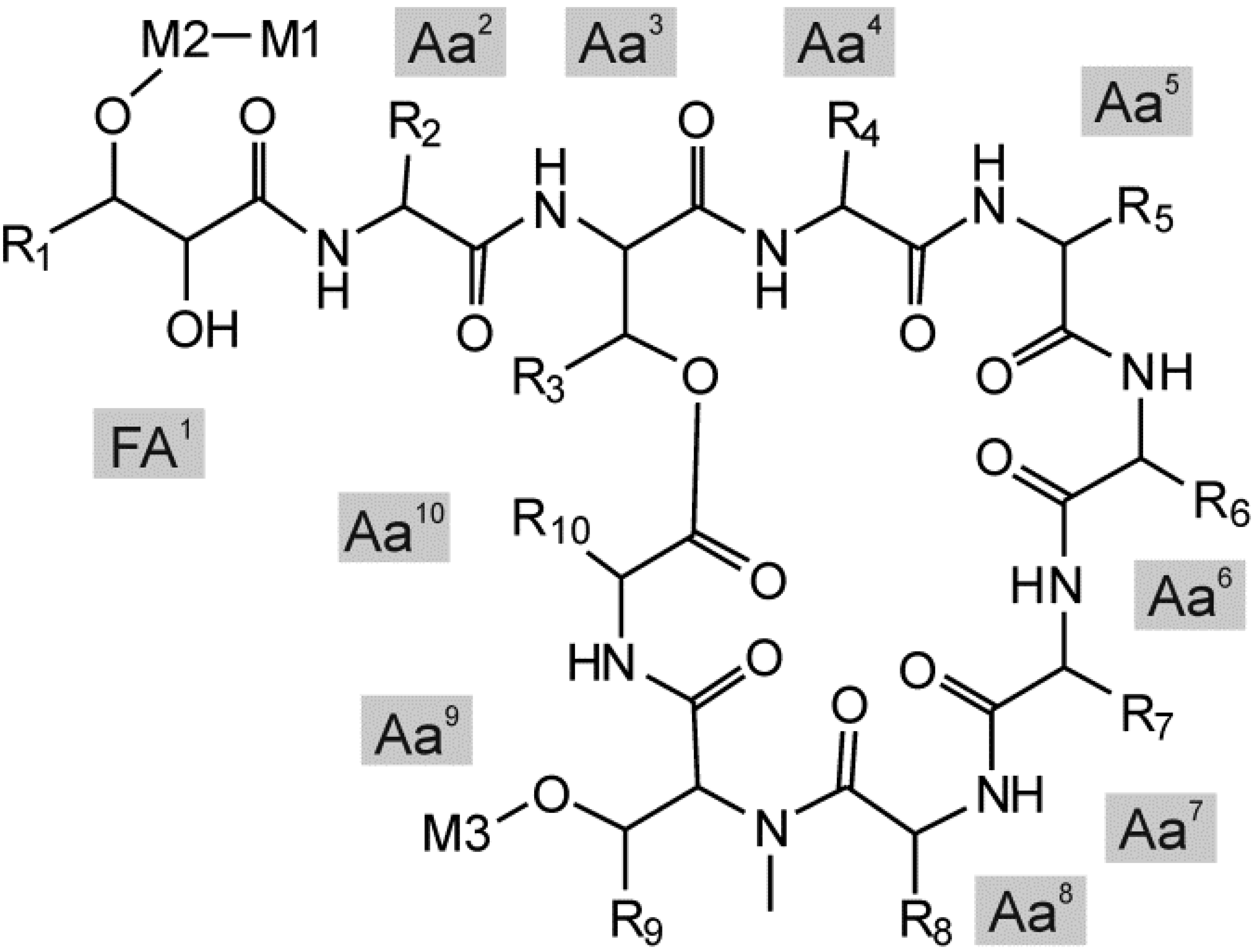
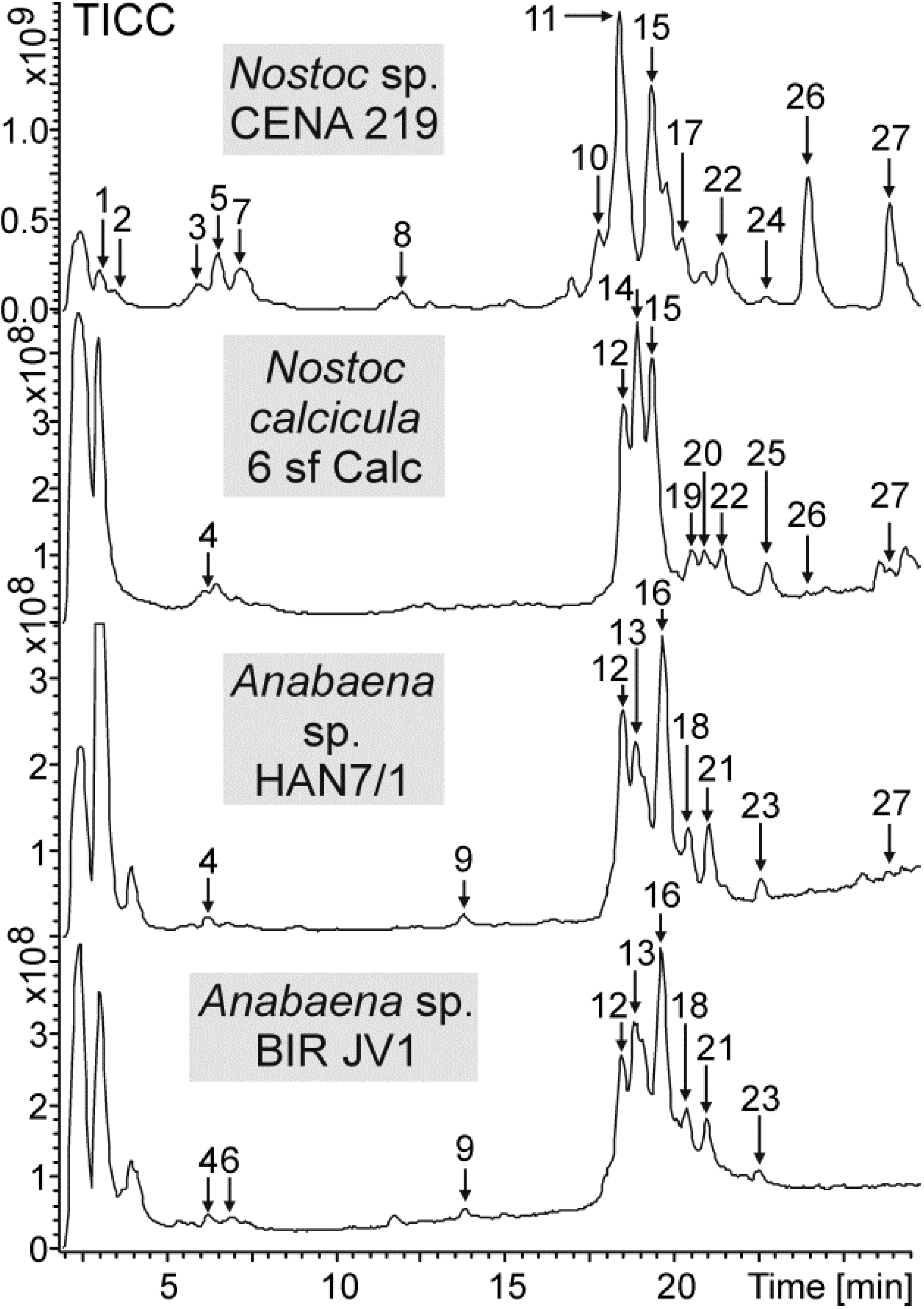
| 1 | 3.1 | 1296.6 | 1815.8 | HexNAc | Pent | Hex | x | |||
| 2 | 3.5 | 1294.8 | 1813.7 | HexNAc | Pent | Hex | x | |||
| 3 | 5.9 | 1280.7 | 1799.8 | HexNAc | Pent | Hex | x | |||
| 4 | 6.2 | 1280.7 | 1772.8 | Hex | dHex | Hex | x | x | x | |
| 5 | 6.5 | 1278.6 | 1797.8 | HexNAc | Pent | Hex | x | |||
| 6 | 6.8 | 1280.7 | 1814.9 | Hex | dHex | AcHex | x | |||
| 7 | 7.2 | 1314.7 | 1833.7 | HexNAc | Pent | Hex | x | |||
| 8 | 11.9 | 1270.7 | 1789.7 | HexNAc | Pent | Hex | x | |||
| 9 | 13.7 | 1236.7 | 1728.8 | Hex | dHex | Hex | x | x | ||
| 10 | 17.7 | 1282.7 | 1801.9 | HexNAc | Pent | Hex | x | |||
| 11 | 18.3 | 1298.7 | 1817.8 | HexNAc | Pent | Hex | x | |||
| 12 | 18.4 | 1298.7 | 1790.6 | Hex | dHex | Hex | x | x | x | |
| 13 | 18.8 | 1298.7 | 1832.8 | Hex | dHex | AcHex | x | x | ||
| 14 | 18.9 | 1298.7 | 1790.8 | Hex | dHex | Hex | x | |||
| 15 | 19.3 | 1298.6 | 1614.7 | Pent | Hex | x | x | |||
| 16 | 19.6 | 1298.7 | 1874.8 | Hex | dHex | diAcHex | x | x | ||
| 17 | 20.2 | 1264.7 | 1783.9 | HexNAc | Pent | Hex | x | |||
| 18 | 20.3 | 1264.7 | 1756.6 | Hex | dHex | Hex | x | x | ||
| 19 | 20.5 | 1298.7 | 1628.7 | dHex | Hex | x | ||||
| 20 | 20.9 | 1264.7 | 1756.8 | Hex | dHex | Hex | x | |||
| 21 | 21.0 | 1298.7 | 1916.8 | Hex | dHex | triAcHex | x | x | ||
| 22 | 21.4 | 1264.7 | 1580.8 | Pent | Hex | x | x | |||
| 23 | 22.5 | 1280.6 | 1958.7 | Hex | dHex | tetraAcHex | x | x | ||
| 24 | 22.7 | 1266.7 | 1785.8 | HexNAc | Pent | Hex | x | |||
| 25 | 22.7 | 1298.7 | 1452.7 | Pent | x | |||||
| 26 | 23.9 | 1298.7 | 1482.7 | Hex | x | x | ||||
| 27 | 26.3 | 1264.7 | 1448.7 | Hex | x | x | x | |||
2.2. Evolutionary Relation of the Cyanobacterial Strains Producing Antifungal Compounds
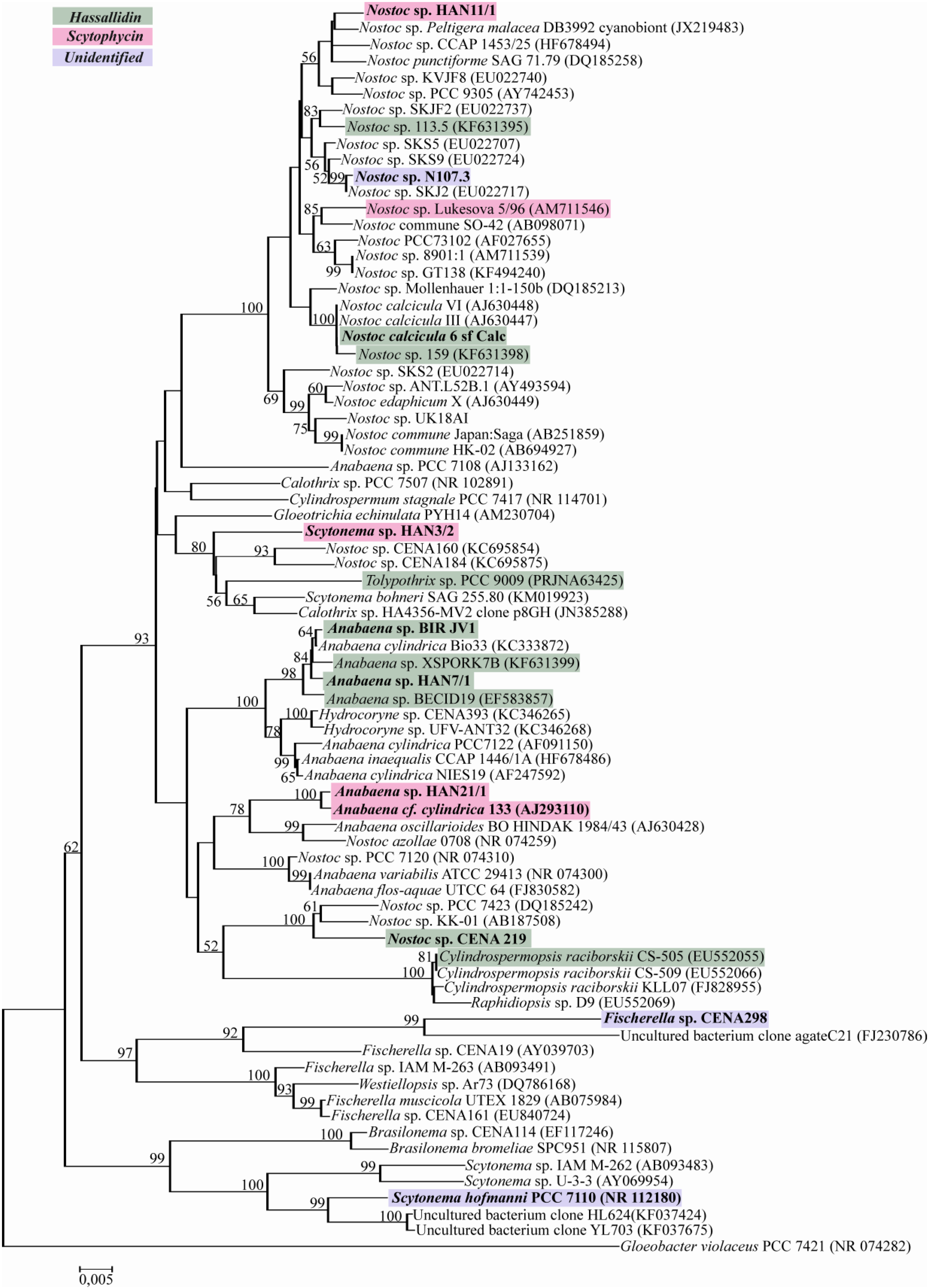
2.3. Discussion
3. Experimental Section
3.1. Cultivation of Cyanobacterial Strains
3.2. Extraction of Intracellular Cyanobacterial Compounds
3.3. Disc Diffusion Assay
3.4. Chemical Analysis
3.5. Purification of Scytophycin
3.6. NMR of Scytophycin
3.7. Microdilution Assay of Scytophycin
3.8. DNA Extraction, Sequencing and Phylogenetic Analysis
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Whitton, B.A.; Potts, M. Introduction to cyanobacteria. In The Ecology of Cyanobacteria. Their Diversity in Time and Space; Whitton, B.A., Potts, M., Eds.; Kluwer Academic: Dodrecht, The Netherlands, 2000; pp. 61–120. [Google Scholar]
- Kaasalainen, U.; Fewer, D.P.; Jokela, J.; Wahlsten, M.; Sivonen, K.; Rikkinen, J. Cyanobacteria produce a high variety of hepatotoxic peptides in lichen symbiosis. Proc. Natl. Acad. Sci. USA. 2012, 109, 5886–5891. [Google Scholar] [CrossRef] [PubMed]
- Sønstebø, J.H.; Rohrlack, T. Possible implications of chytrid parasitism for population subdivision in freshwater cyanobacteria of the genus Planktothrix. Appl. Environ. Microbiol. 2011, 77, 1344–1351. [Google Scholar] [CrossRef] [PubMed]
- Rohrlack, T.; Christiansen, G.; Kurmayer, R. Putative antiparasite defensive system involving ribosomal and nonribosomal oligopeptides in cyanobacteria of the genus Planktothrix. Appl. Environ. Microbiol. 2013, 79, 2642–2647. [Google Scholar] [CrossRef] [PubMed]
- Kajiyama, S.; Kanzaki, H.; Kawazu, K.; Kobayashi, A. Nostofungicidine, an antifungal lipopeptide from the field-grown terrestrial blue-green alga Nostoc commune. Tetrahedron Lett. 1998, 39, 3737–3740. [Google Scholar] [CrossRef]
- Burja, A.M.; Banaigs, B.; Abou-Mansour, E.; Burgess, J.G.; Wright, P.C. Marine cyanobacteria—A prolific source of natural products. Tetrahedron 2001, 57, 9347–9377. [Google Scholar] [CrossRef]
- Abed, R.M.; Dobretsov, S.; Sudesh, K. Applications of cyanobacteria in biotechnology. J. Appl. Microbiol. 2009, 106, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cane, D.E.; Walsh, C.T.; Khosla, C. Harnessing the biosynthetic code: Combinations, permutations, and mutations. Science 1998, 282, 63. [Google Scholar] [CrossRef] [PubMed]
- Cane, D.E.; Walsh, C.T. The parallel and convergent universes of polyketide synthases and nonribosomal peptide synthetases. Chem. Biol. 1999, 6, R319–R325. [Google Scholar] [CrossRef] [PubMed]
- Vestola, J.; Shishido, T.K.; Jokela, J.; Fewer, D.P.; Aitio, O.; Permi, P.; Wahlsten, M.; Wang, H.; Rouhiainen, L.; Sivonen, K. Hassallidins, antifungal glycolipopeptides, are widespread among cyanobacteria and are the end-product of a nonribosomal pathway. Proc. Natl. Acad. Sci. USA 2014, 111, E1909–E1917. [Google Scholar] [CrossRef] [PubMed]
- Loeffler, J.; Stevens, D.A. Antifungal drug resistance. Clin. Infect. Dis. 2003, 36, S31–S41. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.A.; Ahmad, I.; Aqil, F.; Owais, M.; Shahid, M.; Musarrat, J. Virulence and pathogenicity of fungal pathogens with special reference to Candida albicans. In Combating Fungal Infections; Ahmad, I., Owais, M., Shahid, M., Aqil, F., Eds.; Springer-Verlag: Berlin, Germany, 2010; pp. 21–45. [Google Scholar]
- World Health Organization. Antifungal drug resistance: The example of invasive Candidiasis. In Antimicrobial Resistance: Global Report on Surveillance; WHO Press: Geneva, Switzerland, 2014; pp. 62–65. [Google Scholar]
- Gugger, M.; Lyra, C.; Henriksen, P.; Couté, A.; Humbert, J.F.; Sivonen, K. Phylogenetic comparison of the cyanobacterial genera Anabaena and Aphanizomenon. Int. J. Syst. Evol. Microbiol. 2002, 52, 1867–1880. [Google Scholar] [CrossRef]
- Shishido, T.K.; Kaasalainen, U.; Fewer, D.P.; Rouhiainen, L.; Jokela, J.; Wahlsten, M.; Fiore, M.F.; Yunes, J.S.; Rikkinen, J.; Sivonen, K. Convergent evolution of [d-Leucine(1)] microcystin-LR in taxonomically disparate cyanobacteria. BMC Evol. Biol. 2013, 13, 86. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, M.; Moore, R.E.; Patterson, G.M.L. Scytophycins, Cytotoxic and antimycotic agents from the cyanophyte Scytonema pseudohofmanni. J. Org. Chem. 1986, 51, 5300–5306. [Google Scholar] [CrossRef]
- Carmeli, S.; Moore, R.E.; Patterson, G.M.L. Tolytoxin and new scytophycins from three species of Scytonema. J. Nat. Prod. 1990, 53, 1533–1542. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Moore, R.E.; Patterson, G.M.L. Scytophycins from a blue-green alga belonging to the nostocaceae. Phytochemistry 1991, 30, 3615–3616. [Google Scholar] [CrossRef]
- Tomsickova, J.; Ondrej, M.; Cerny, J.; Hrouzek, P.; Kopecky, J. Analysis and detection of scytophycin variants by HPLC-ESI-MS. Chem. Nat. Compd. 2014, 49, 1170–1171. [Google Scholar] [CrossRef]
- Kubanek, J.; Jensen, P.R.; Keifer, P.A.; Sullards, M.C.; Collins, D.O.; Fenical, W. Seaweed resistance to microbial attack: A targeted chemical defense against marine fungi. Proc. Natl. Acad. Sci. USA 2003, 100, 6916–6921. [Google Scholar] [CrossRef]
- Yeung, K.-S.; Paterson, I. Actin-binding marine macrolides: Total synthesis and biological importance. Angew. Chem. Int. 2002, 41, 4632–4653. [Google Scholar] [CrossRef]
- Patterson, G.M.L.; Bolis, C.M. Fungal cell wall polysaccharides elicit an antifungal secondary metabolite (phytoalexin) in the cyanobacterium Scytonema ocelutum. J. Phycol. 1997, 33, 54–60. [Google Scholar] [CrossRef]
- Neuhof, T.; Schmieder, P.; Preussel, K.; Dieckmann, R.; Pham, H.; Bartl, F.; von Döhren, H. Hassallidin A, a glycosylated lipopeptide with antifungal activity from the cyanobacterium Hassallia sp. J. Nat. Prod. 2005, 68, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Neuhof, T.; Schmieder, P.; Seibold, M.; Preussel, K.; von Döhren, H. Hassallidin B—Second antifungal member of the Hassallidin family. Bioorg. Med. Chem. Lett. 2006, 16, 4220–4222. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.H.; Wray, V.; Nimtz, M.; Fossen, T.; Preisitsch, M.; Schröder, G.; Wende, K.; Heiden, S.E.; Mundt, S. Balticidins A-D, antifungal hassallidin-like lipopeptides from the Baltic Sea cyanobacterium Anabaena cylindrica Bio33. J. Nat. Prod. 2014, 77, 1287–1296. [Google Scholar] [CrossRef] [PubMed]
- Neuhof, T.; Dieckmann, R.; von Döhren, H.; Preussel, K.; Seibold, M.; Schmieder, P. Lipopeptides Having Pharmaceutical Activity. Patent Analysis 1698638 EP, 8 September 2006. [Google Scholar]
- Kótai, J. Instructions for Preparation of Modified Nutrient Solution Z8 for Algae; NIVA B-11/69; Norwegian Institute for Water Research: Blindern, Oslo, Norway, 1972. [Google Scholar]
- Allen, M.B. Simple conditions for growth of unicellular blue-green algae on plates. J. Phycol. 1968, 4, 1–4. [Google Scholar] [CrossRef]
- Allen, M.M.; Arnon, D.I. Studies on nitrogen-fixing blue-green algae. I. Growth and nitrogen fixation by Anabaena cylindrical Lemm. Plant Physiol. 1955, 30, 366–372. [Google Scholar] [CrossRef] [PubMed]
- Gorham, P.R.; McLachlan, J.; Hammer, U.T.; Kim, W.K. Isolation and culture of toxic strains of Anabaena flos-aquae (Lyngb.) de Bréb. Verh. Int. Verein. Theor. Angew. Limnol. 1964, 15, 796–804. [Google Scholar]
- Jokela, J.; Oftedal, L.; Herfindal, L.; Permi, P.; Wahlsten, M.; Døskeland, S.O.; Sivonen, K. Anabaenolysins, novel cytolytic lipopeptides from benthic Anabaena cyanobacteria. PLoS ONE 2012, 7, e41222. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shishido, T.K.; Humisto, A.; Jokela, J.; Liu, L.; Wahlsten, M.; Tamrakar, A.; Fewer, D.P.; Permi, P.; Andreote, A.P.D.; Fiore, M.F.; et al. Antifungal Compounds from Cyanobacteria. Mar. Drugs 2015, 13, 2124-2140. https://doi.org/10.3390/md13042124
Shishido TK, Humisto A, Jokela J, Liu L, Wahlsten M, Tamrakar A, Fewer DP, Permi P, Andreote APD, Fiore MF, et al. Antifungal Compounds from Cyanobacteria. Marine Drugs. 2015; 13(4):2124-2140. https://doi.org/10.3390/md13042124
Chicago/Turabian StyleShishido, Tânia K., Anu Humisto, Jouni Jokela, Liwei Liu, Matti Wahlsten, Anisha Tamrakar, David P. Fewer, Perttu Permi, Ana P. D. Andreote, Marli F. Fiore, and et al. 2015. "Antifungal Compounds from Cyanobacteria" Marine Drugs 13, no. 4: 2124-2140. https://doi.org/10.3390/md13042124
APA StyleShishido, T. K., Humisto, A., Jokela, J., Liu, L., Wahlsten, M., Tamrakar, A., Fewer, D. P., Permi, P., Andreote, A. P. D., Fiore, M. F., & Sivonen, K. (2015). Antifungal Compounds from Cyanobacteria. Marine Drugs, 13(4), 2124-2140. https://doi.org/10.3390/md13042124





