Mycemycins A–E, New Dibenzoxazepinones Isolated from Two Different Streptomycetes
Abstract
:1. Introduction
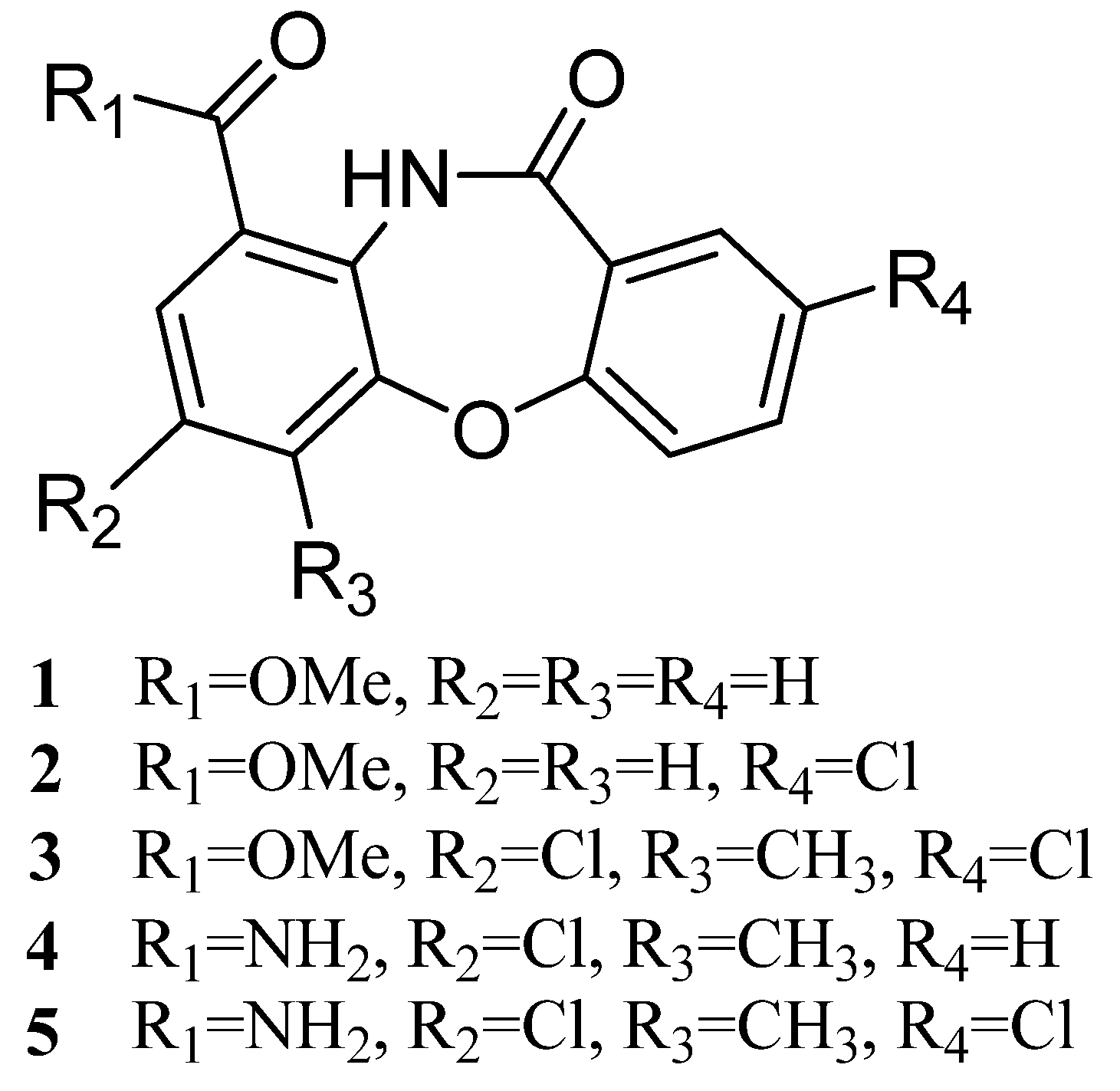
2. Results and Discussion
2.1. Genetic Manipulation of S. olivaceus FXJ8.012 and Isolation of Compounds 1–5
2.2. Structural Identification of Mycemycins A–E
| Position | 1 | 2 | 3 | 4 | ||||
|---|---|---|---|---|---|---|---|---|
| δH | δC | δH | δC | δH | δC | δH | δC | |
| 1 | ― | 164.4, C | ― | 163.1, C | ― | 163.3, C | ― | 163.9, C |
| 2 | ― | 110, C | ― | 111, C | ― | 110.8, C | ― | 109.8, C |
| 3 | 8.04 (dd, 1.5, 8.0) | 127.2, CH | 8.01 (d, 2.5) | 126.4, CH | 8.00 (d, 2.5) | 128.4, CH | 8.10 (d, 7.5) | 127.83, CH |
| 4 | 7.03 (t, 7.5) | 119.6, CH | ― | 124.5, C | ― | 124.5, C | 7.10 (t, 7.5) | 120.4, CH |
| 5 | 7.48 (dt, 1.5, 7.8) | 134.3, CH | 7.42 (dd, 2.5, 8.5–9.0) | 134.1, CH | 7.43 (dd, 2.5, 8.5–9.0) | 134.2, CH | 7.53 (t, 7.5) | 134.9, CH |
| 6 | 7.15 (d, 8.5) | 117.7, CH | 7.10 (d, 9.0) | 119.3, CH | 7.10 (d, 9.0) | 119.3, CH | 7.15 (d, 8.0) | 117.7, CH |
| 7 | ― | 159.4, C | ― | 157.9, C | ― | 157.9, C | ― | 158.4, C |
| 8 | ― | 149.8, C | ― | 149.8, C | ― | 149.3, C | ― | 148.7, C |
| 9 | 7.81 (d, 8.5) | 115, CH | 7.82 (d, 8.0) | 115.1, CH | ― | 125.4, C | ― | 124.3, C |
| 10 | 7.46 (t, 8.0) | 124.8, CH | 7.49 (t, 8.0) | 125.2, CH | ― | 131.4, C | ― | 131.9, C |
| 11 | 8.08 (d, 7.5) | 127.5, CH | 8.10 (d, 8.0) | 127.7, CH | 8.10 (s) | 126.3, CH | 8.23 (s) | 127.8, CH |
| 12 | ― | 121.3, C | ― | 121.6, C | ― | 119.1, C | ― | 120.8, C |
| 13 | ― | 139.4, C | ― | 139.2, C | ― | 137.3, C | ― | 136, C |
| 14 | ― | 165.6, C | ― | 165.4, C | ― | 164.5, C | ― | 164.5, C |
| 15 | 4.06 (s) | 52.4, CH3 | 4.06 (s) | 52.5, CH3 | 4.04 (s) | 52.6, CH3 | ― | ― |
| 9-Me | ― | ― | ― | ― | 2.69 (s) | 13.7, CH3 | 2.69 (s) | 13.6, CH3 |
| 13-NH | ― | ― | ― | ― | 11.65 (s, w) | ― | 10.27 (s) | ― |
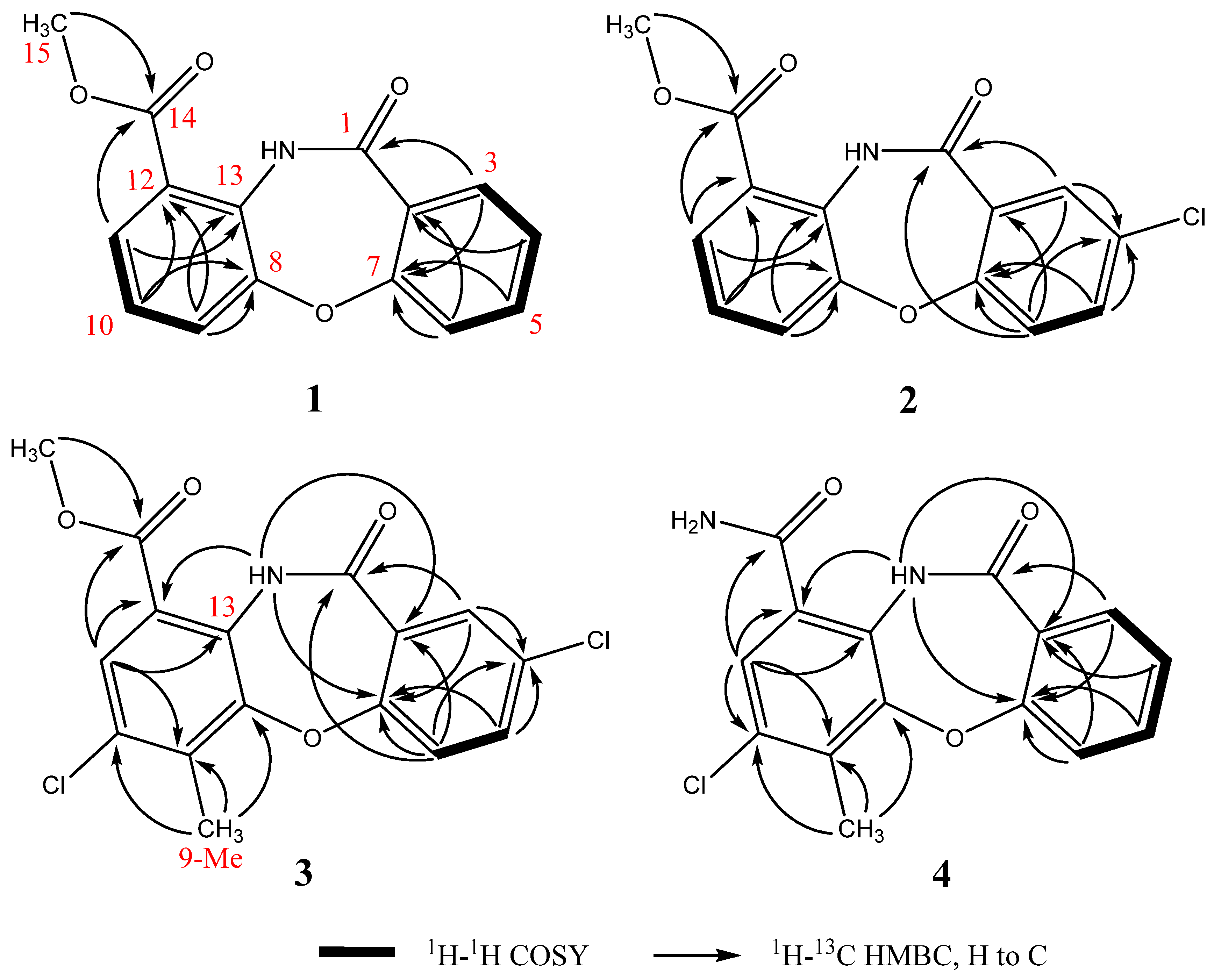
2.3. Bioactivities of Compounds 1–4
2.4. Discussion
3. Experimental Section
3.1. General
3.2. Strains, Plasmids and Media
3.3. Nucleic Acid Manipulations and Mutant Construction
3.4. Fermentation, Extraction and Isolation
3.4.1. S. sp. FXJ1.235
3.4.2. S. olivaceus FXJ8.012
3.5. Bioactivity Assay
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References and Notes
- Baltz, R.H. Antimicrobials from actinomycetes: Back to the future. Microbe 2007, 2, 125–131. [Google Scholar]
- Bérdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Bérdy, J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. 2012, 65, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Gontang, E.A.; Gaudencio, S.P.; Fenical, W.; Jensen, P.R. Sequence-based analysis of secondary-metabolite biosynthesis in marine actinobacteria. Appl. Environ. Microbiol. 2010, 76, 2487–2499. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.; Abdel-Mageed, W.M.; Ebel, R.; Bull, A.T.; Goodfellow, M.; Fiedler, H.P.; Jaspars, M. Dermacozines H–J isolated from a deep-sea strain of Dermacoccus abyssi from Mariana Trench sediments. J. Nat. Prod. 2014, 77, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Ochi, K.; Hosaka, T. New strategies for drug discovery: Activation of silent or weakly expressed microbial gene clusters. Appl. Microbiol. Biotechnol. 2013, 97, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Rebets, Y.; Brotz, E.; Tokovenko, B.; Luzhetskyy, A. Actinomycetes biosynthetic potential: How to bridge in silico and in vivo? J. Ind. Microbiol. Biotechnol. 2014, 41, 387–402. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Chater, K.F.; Chandra, G.; Niu, G.; Tan, H. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 2013, 77, 112–143. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Shang, F.; Xi, L.; Huang, Y. Tetroazolemycins A and B, two new oxazole-thiazole siderophores from deep-sea Streptomyces olivaceus FXJ8.012. Mar. Drugs 2013, 11, 1524–1533. [Google Scholar] [CrossRef]
- Hillerich, B.; Westpheling, J. A new GntR family transcriptional regulator in Streptomyces coelicolor is required for morphogenesis and antibiotic production and controls transcription of an ABC transporter in response to carbon source. J. Bacteriol. 2006, 188, 7477–7487. [Google Scholar] [CrossRef] [PubMed]
- Hoskisson, P.A.; Rigali, S. Chapter 1: Variation in form and function the helix-turn-helix regulators of the GntR superfamily. Adv. Appl. Microbiol. 2009, 69, 1–22. [Google Scholar] [PubMed]
- Zhu, Q.; Li, J.; Ma, J.; Luo, M.; Wang, B.; Huang, H.; Tian, X.; Li, W.; Zhang, S.; Zhang, C.; et al. Discovery and engineered overproduction of antimicrobial nucleoside antibiotic A201A from the deep-sea marine actinomycete Marinactinospora thermotolerans SCSIO 00652. Antimicrob. Agents. Chemother. 2012, 56, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Horbal, L.; Fedorenko, V.; Bechthold, A.; Luzhetskyy, A. A transposon-based strategy to identify the regulatory gene network responsible for landomycin E biosynthesis. FEMS. Microbiol. Lett. 2013, 342, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Smanski, M.J.; Peterson, R.M.; Marchillo, K.; Andes, D.; Rajski, S.R.; Shen, B. Engineering of Streptomyces platensis MA7339 for overproduction of platencin and congeners. Org. Lett. 2010, 12, 1744–1747. [Google Scholar] [CrossRef] [PubMed]
- Modified R2 medium: sucrose (103 g), MgCl2·6H2O (10.12 g), difco casaminoacids (0.1 g), TES buffer (5.73 g), trace element solution (2 mL, containing ZnCl2 40 mg/L, FeCl3·6H2O 200 mg/L, CuCl2·2H2O 10 mg/L, MnCl2·4H2O 10 mg/L, Na2B4O7·10H2O 10 mg/L), distilled water 1000 mL, and added with KH2PO4 (0.5%) 10 mL, CaCl2·2H2O (5 M) 4 mL, NaOH (1 N) 5 mL, glucose (50%) 20 mL and l-proline (20%) 15 ml before use.
- Liu, X.; Ouyang, S.; Yu, B.; Liu, Y.; Huang, K.; Gong, J.; Zheng, S.; Li, Z.; Li, H.; Jiang, H. PharmMapper server: A web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res. 2010, 38, W609–W614. [Google Scholar] [CrossRef] [PubMed]
- Fiorentino, A.; D’Abrosca, B.; Pacifico, S.; Cefarelli, G.; Uzzo, P.; Monaco, P. Natural dibenzoxazepinones from leaves of Carex distachya: Structural elucidation and radical scavenging activity. Bioorg. Med. Chem. Lett. 2007, 17, 636–639. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Farmer, P.S.; Quillam, M.A.; Howlett, S.E. Dibenzothiazepinones as potential calcium channel antagonists. I. Drug. Des. Discov. 1993, 10, 331–342. [Google Scholar] [PubMed]
- Li, R.; Farmer, P.S.; Wang, J.; Boyd, R.J.; Cameron, T.S.; Quilliam, M.A.; Walter, J.A.; Howlett, S.E. Molecular geometries of dibenzothiazepinone and dibenzoxazepinone calcium antagonists. Drug. Des. Discov. 1995, 12, 337–358. [Google Scholar] [PubMed]
- Hargrave, K.D.; Schmidt, G.; Engel, W.; Schromm, K.; Schromm, W.E.K.; Hargrave, K.; Schmidit, G.; Nehai, S. Prevention and treatment of HIV infection-by administering di:benz-(B,F)-(1–4)-oxazepine (and thiazepin)-11(hydroxy)-one derivs. and salts. EP419861-B; EP419861-A2; EP419861-A; AU9061916-A; CA2024040-A; JP3163021-A; HU57589-T; ZA9006834-A; EP419861-A3; AU639255-B; EP419861-B1; DE69023311-E; US5571806-A; NZ235082-A; HU211077-B; JP2862980-B2; KR165108-B1; CA2024040-C; EP419861-A2; EP419861-A. 03 Apr 1991 199114 English AU9061916-A 07 Mar 1991 199117 JP3163021-A 15 Jul 1991 199134 ZA9006834-A 27 May 1992 C07K 199228.
- Gribble, G.W. Biological activity of recently discovered halogenated marine natural products. Mar. Drugs. 2015, 13, 4044–4136. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Liu, N.; Li, X.; Ding, Y.; Shang, F.; Gao, Y.; Ruan, J.; Huang, Y. Red soils harbor diverse culturable actinomycetes that are promising sources of novel secondary metabolites. Appl. Environ. Microbiol. 2015, 81, 3086–3103. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Wang, H.; Liu, M.; Gu, Q.; Zheng, W.; Huang, Y. Streptomyces alni sp. nov., a daidzein-producing endophyte isolated from a root of Alnus nepalensis D. Don. Int. J. Syst. Evol. Microbiol. 2009, 59, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; The John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Pan, Y.; Liu, G.; Yang, H.; Tian, Y.; Tan, H. The pleiotropic regulator AdpA-L directly controls the pathway-specific activator of nikkomycin biosynthesis in Streptomyces ansochromogenes. Mol. Microbiol. 2009, 72, 710–723. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, N.; Song, F.; Shang, F.; Huang, Y. Mycemycins A–E, New Dibenzoxazepinones Isolated from Two Different Streptomycetes. Mar. Drugs 2015, 13, 6247-6258. https://doi.org/10.3390/md13106247
Liu N, Song F, Shang F, Huang Y. Mycemycins A–E, New Dibenzoxazepinones Isolated from Two Different Streptomycetes. Marine Drugs. 2015; 13(10):6247-6258. https://doi.org/10.3390/md13106247
Chicago/Turabian StyleLiu, Ning, Fangying Song, Fei Shang, and Ying Huang. 2015. "Mycemycins A–E, New Dibenzoxazepinones Isolated from Two Different Streptomycetes" Marine Drugs 13, no. 10: 6247-6258. https://doi.org/10.3390/md13106247
APA StyleLiu, N., Song, F., Shang, F., & Huang, Y. (2015). Mycemycins A–E, New Dibenzoxazepinones Isolated from Two Different Streptomycetes. Marine Drugs, 13(10), 6247-6258. https://doi.org/10.3390/md13106247







