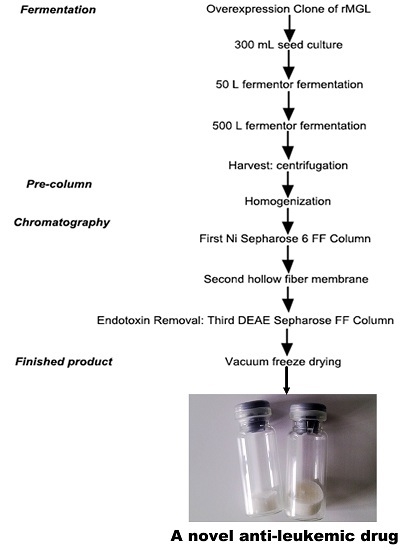
High-Level Expression, Purification and Large-Scale Production of l-Methionine γ-Lyase from Idiomarina as a Novel Anti-Leukemic Drug
Abstract
:1. Introduction
2. Results
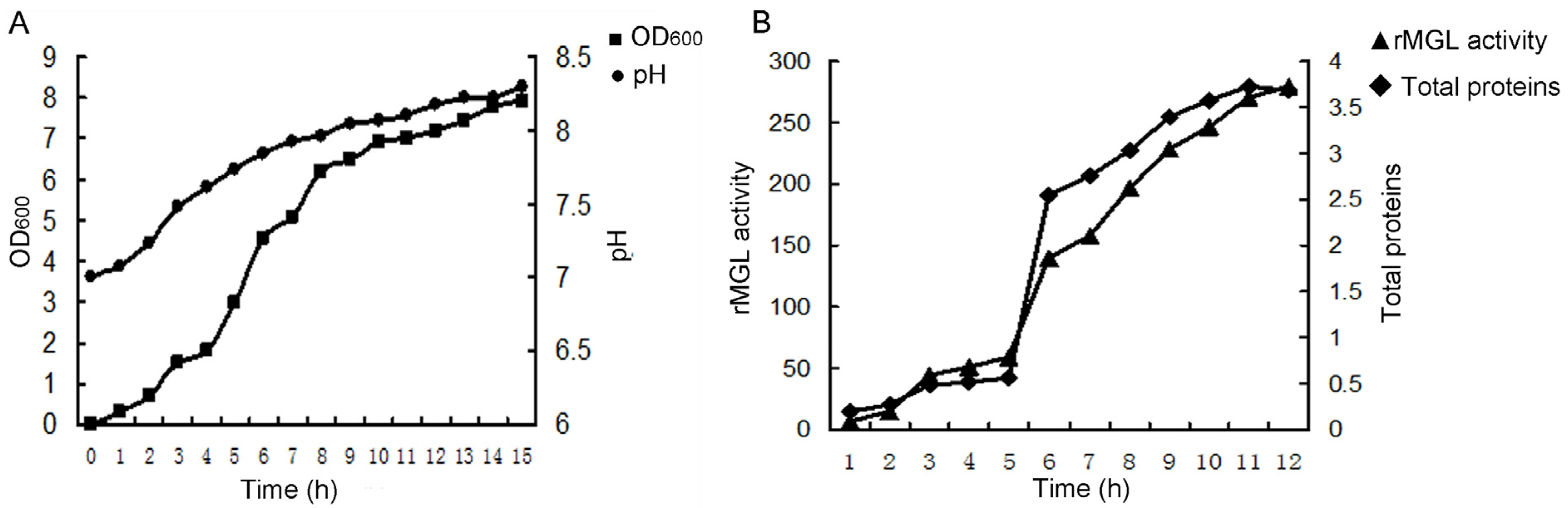
2.1. Large-Scale Fermentation for rMGL Production
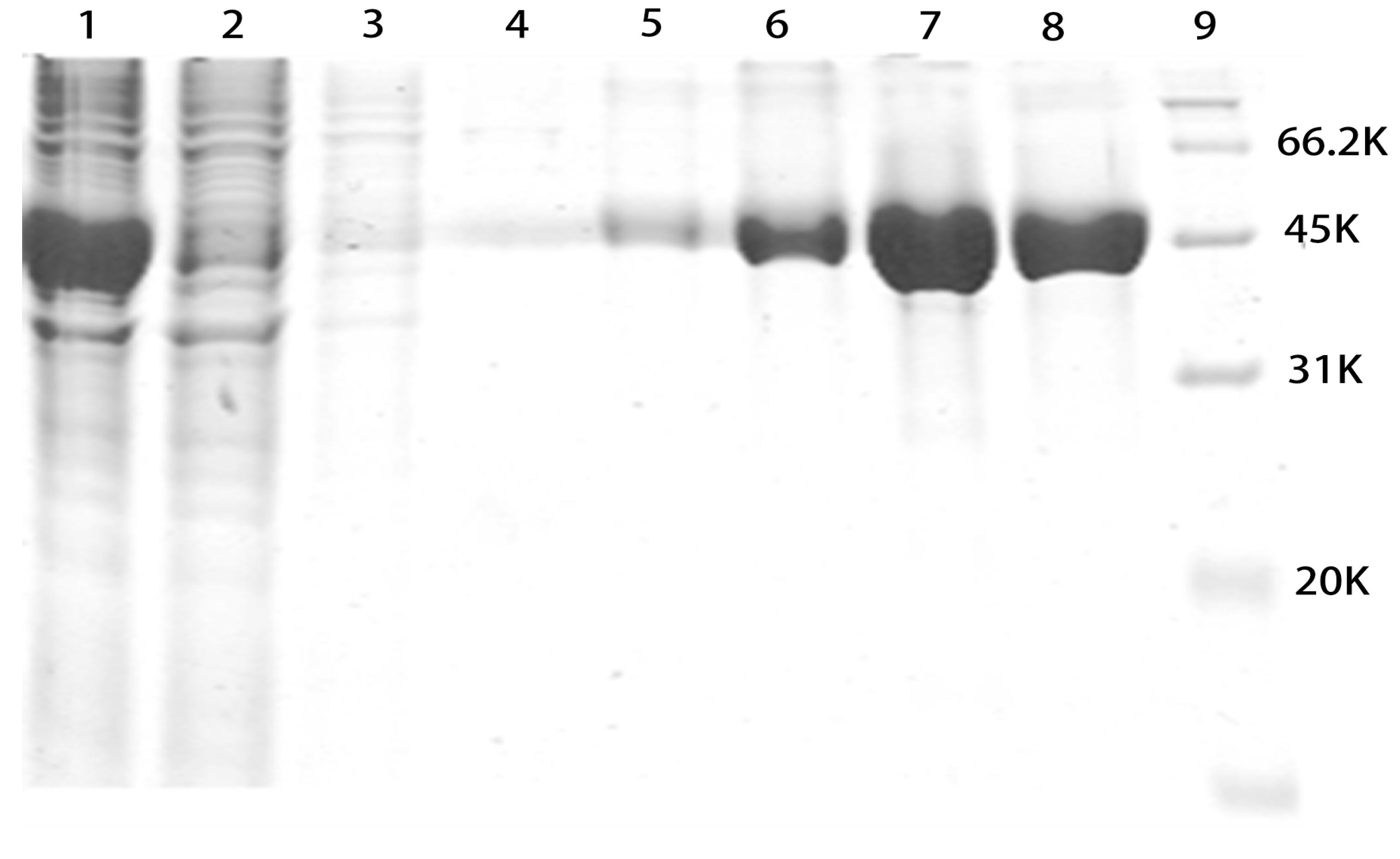
2.2. SDS-PAGE Analysis of Purified rMGL
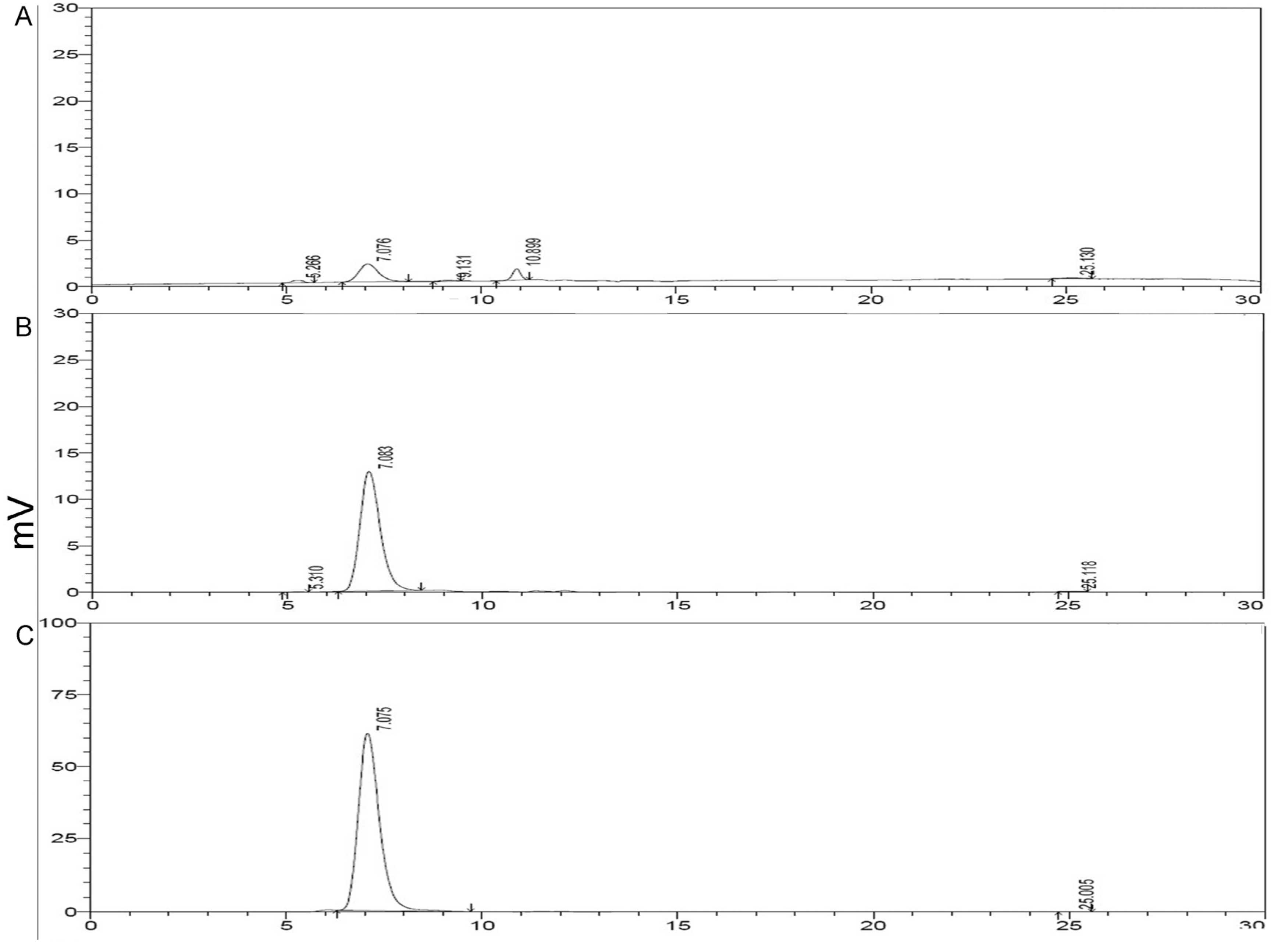
2.3. HPLC Analysis of Purified rMGL
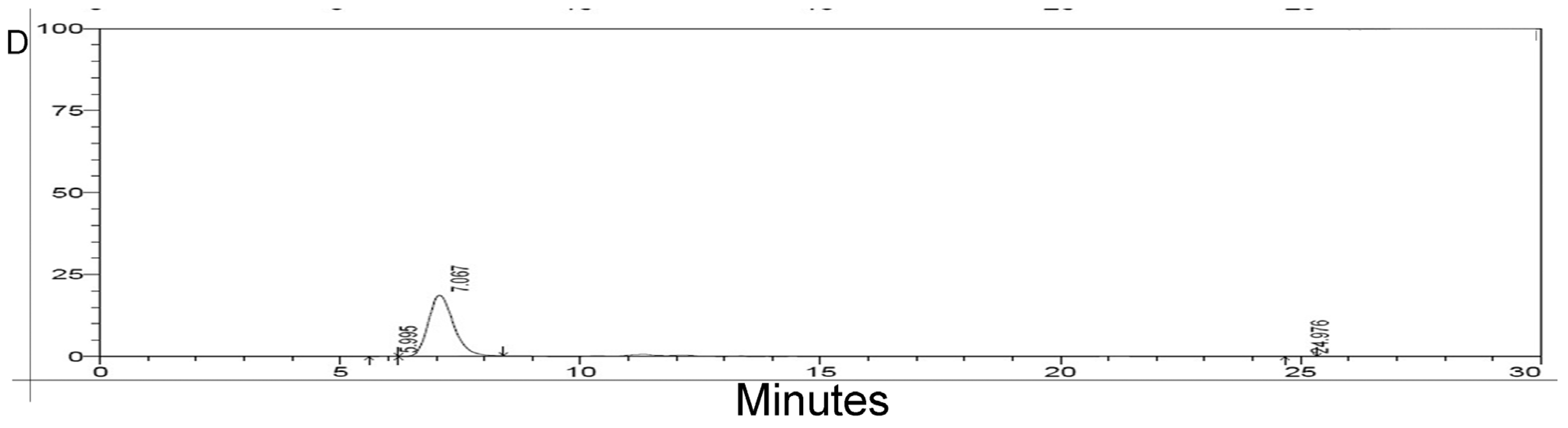
2.4. Lyophilization of rMGL
| Step | Volume (L) | Activity (U) | Protein (g) | SA * (U/mg) | Yield (%) |
|---|---|---|---|---|---|
| HG * | 30 | 75,600,000 | 1104 | 68.47 | 90 |
| Ni-FF * | 10 | 71,820,000 | 567.57 | 126.54 | 85.50 |
| DS * and CO * | 2 | 66,792,600 | 544.86 | 122.59 | 79.52 |
| EN * | 2 | 60,113,340 | 506.72 | 118.63 | 71.56 |
| Drying * | -- | 54,703,139 | 506.72 | 107.95 | 57.25 |
2.5. Comparison of rMGL Production from Various Strains
2.6. rMGL Inhibits the Proliferation of Leukemia Cell Lines

2.7. rMGL Induces Apoptosis of Leukemia Cell Lines
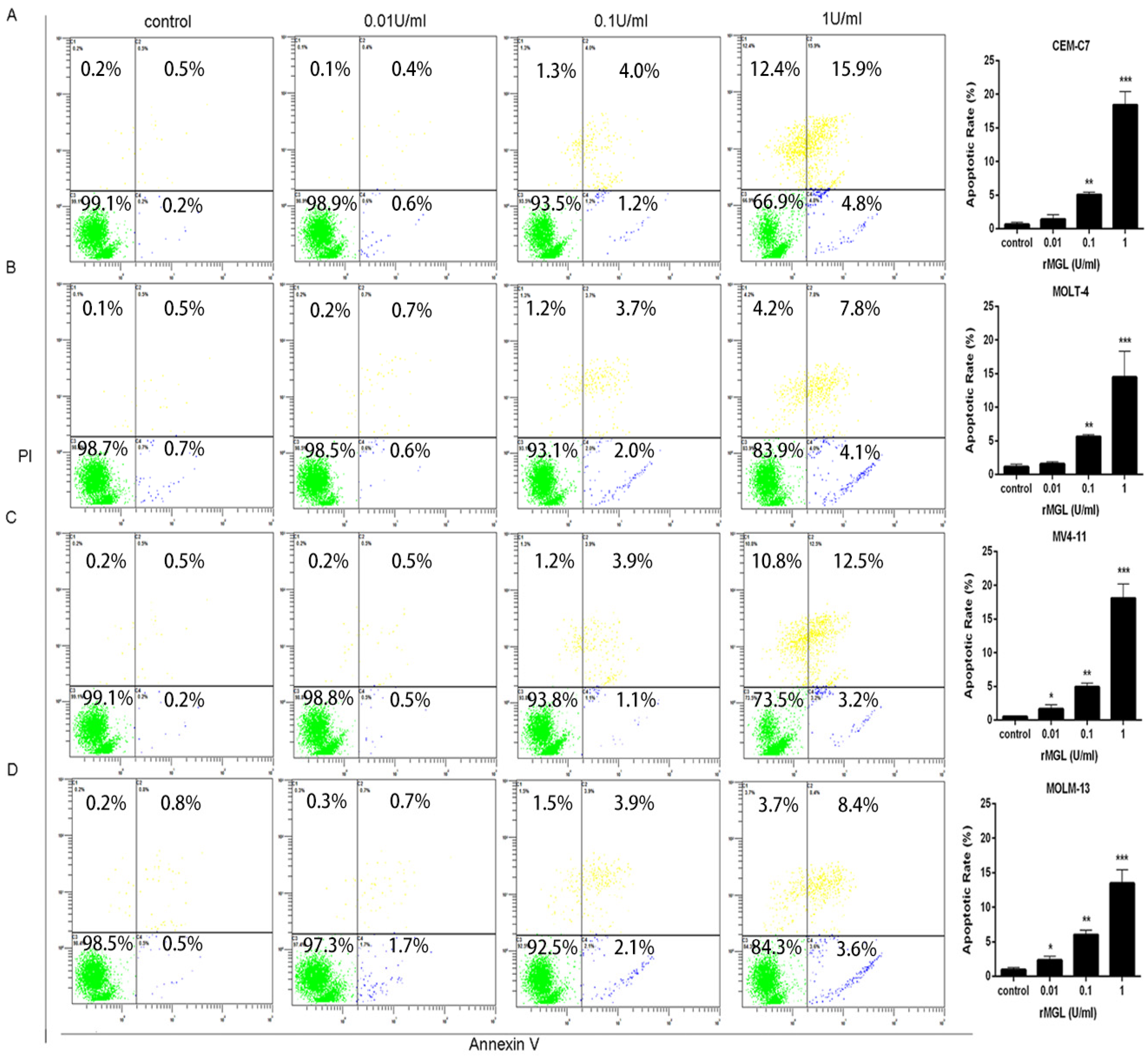
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Bacteria Strains and Media
4.3. Large-Scale Production of rMGL
4.3.1. Fermentation
4.3.2. Preparation of the Crude Enzyme
4.3.3. Chromatographic Conditions
4.3.4. Finished Product
4.4. Analysis of rMGL
4.4.1. Biomass
4.4.2. Protein Concentration
4.4.3. Activity Assay
4.4.4. HPLC
4.5. Cell Lines and Cell Culture
| Cell Line | ||||
|---|---|---|---|---|
| Characteristics | CEM-C7 | MOLT-4 | MV4-11 | MOLM-13 |
| Cell type | T-ALL | T-ALL | AML | AML |
| Primary site | PB * | PB | PB | PB |
| IC50 * (U/mL) | 1.70 | 0.30 | 0.64 | 0.03 |
4.6. Cell Proliferation Assay
4.7. Apoptosis Assay
4.8. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix


References
- Tanaka, H.; Esaki, N.; Soda, K. Bacterial l-Methionine gamma-lyase: Characterization and application. Prog. Clin. Biol. Res. 1983, 125, 365–377. [Google Scholar] [PubMed]
- Tanaka, H.; Esaki, N.; Yamamoto, T.; Soda, K. Purification and properties of methioninase from Pseudomonas ovalis. FEBS Lett. 1976, 66, 307–311. [Google Scholar] [CrossRef]
- Tanaka, H.; Esaki, N.; Soda, K. Properties of l-Methionine gamma-lyase from Pseudomonas ovalis. Biochemistry 1977, 16, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Inagaki, K.; Sugimoto, M.; Esaki, N.; Soda, K.; Tanaka, H. Structural analysis of the l-Methionine gamma-lyase gene from Pseudomonas putida. J. Biochem. 1995, 117, 1120–1125. [Google Scholar] [PubMed]
- Tan, Y.; Xu, M.; Tan, X.; Tan, X.; Wang, X.; Saikawa, Y.; Nagahama, T.; Sun, X.; Lenz, M.; Hoffman, R.M. Overexpression and large-scale production of recombinant l-Methionine-alpha-deamino-gamma-mercaptomethane-lyase for novel anticancer therapy. Protein Expr. Purif. 1997, 9, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Takakura, T.; Ito, T.; Yagi, S.; Notsu, Y.; Itakura, T.; Nakamura, T.; Inagaki, K.; Esaki, N.; Hoffman, R.M.; Takimoto, A. High-level expression and bulk crystallization of recombinant l-Methionine gamma-lyase, an anticancer agent. Appl. Microbiol. Biotechnol. 2006, 70, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Huang, Y.; Zhang, J.; Du, J.; Tan, H.; Lu, Y.; Zhou, S. Identification and characterization of a novel methionine γ-lyase gene from deep-sea sediment metagenomic library. World J. Microbiol. Biotechnol. 2011, 27, 2729–2736. [Google Scholar] [CrossRef]
- Hoffman, R.M. Methionine dependence in cancer cells—A review. in Vitro 1982, 18, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Lishko, V.K.; Herrera, H.; Groce, A.; Kubota, T.; Hoffman, R.M. Therapeutic tumor-specific cell cycle block induced by methionine starvation in vivo. Cancer Res. 1993, 53, 5676–5679. [Google Scholar] [PubMed]
- Mecham, J.O.; Rowitch, D.; Wallace, C.D.; Stern, P.H.; Hoffman, R.M. The metabolic defect of methionine dependence occurs frequently in human tumor cell lines. Biochem. Biophys. Res. Commun. 1983, 117, 429–434. [Google Scholar] [CrossRef]
- Cellarier, E.; Durando, X.; Vasson, M.P.; Farges, M.C.; Demiden, A.; Maurizis, J.C.; Madelmont, J.C.; Chollet, P. Methionine dependency and cancer treatment. Cancer Treat. Rev. 2003, 29, 489–499. [Google Scholar] [CrossRef]
- Hoffman, R.M. Development of recombinant methioninase to target the general cancer-specific metabolic defect of methionine dependence: A 40-year odyssey. Expert Opin. Biol. Ther. 2015, 15, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Singh, S.; Kanwar, S.S. l-methionase: A therapeutic enzyme to treat malignancies. Biomed Res. Int. 2014, 2014, 506287. [Google Scholar] [CrossRef] [PubMed]
- Yano, S.; Li, S.; Han, Q.; Tan, Y.; Bouvet, M.; Fujiwara, T.; Hoffman, R.M. Selective methioninase-induced trap of cancer cells in S/G(2) phase visualized by FUCCI imaging confers chemosensitivity. Oncotarget 2014, 5, 8729–8736. [Google Scholar] [PubMed]
- Takakura, T.; Takimoto, A.; Notsu, Y.; Yoshida, H.; Ito, T.; Nagatome, F.; Ohno, M.; Kobayashi, Y.; Yoshioka, T.; Inagaki, K.; et al. Physicochemical and pharmacokinetic characterization of highly potent recombinant l-Methionine gamma-lyase conjugated with polyethylene glycol as an antitumor agent. Cancer Res. 2006, 66, 2807–2814. [Google Scholar] [CrossRef] [PubMed]
- Bhojwani, D.; Pui, C.-H. Relapsed childhood acute lymphoblastic leukaemia. Lancet Oncol. 2013, 14, E205–E217. [Google Scholar] [CrossRef]
- Strekalova, E.; Malin, D.; Good, D.M.; Cryns, V.L. Methionine deprivation induces a targetable vulnerability in triple-negative breast cancer cells by enhancing trail receptor-2 expression. Clin. Cancer Res. 2015, 21, 2780–2791. [Google Scholar] [CrossRef] [PubMed]
- Sinha, R.; Cooper, T.K.; Rogers, C.J.; Sinha, I.; Turbitt, W.J.; Calcagnotto, A.; Perrone, C.E.; Richie, J.P., Jr. Dietary methionine restriction inhibits prostatic intraepithelial neoplasia in TRAMP mice. Prostate 2014, 74, 1663–1673. [Google Scholar] [CrossRef] [PubMed]
- Kreis, W.; Goodenow, M. Methionine requirement and replacement by homocysteine in tissue cultures of selected rodent and human malignant and normal cells. Cancer Res. 1978, 38, 2259–2262. [Google Scholar] [PubMed]
- Poirier, L.A. The role of methionine in carcinogenesis in vivo. Adv. Exp. Med. Biol. 1986, 206, 269–282. [Google Scholar] [PubMed]
- Ghoshal, A.K.; Sarma, D.S.; Farber, E. Ethionine in the analysis of the possible separate roles of methionine and choline deficiencies in carcinogenesis. Adv. Exp. Med. Biol. 1986, 206, 283–292. [Google Scholar] [PubMed]
- Howarth, G.S.; Wang, H. Role of endogenous microbiota, probiotics and their biological products in human health. Nutrients 2013, 5, 58–81. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Xu, M.; Guo, H.; Sun, X.; Kubota, T.; Hoffman, R.M. Anticancer efficacy of methioninase in vivo. Anticancer Res. 1996, 16, 3931–3936. [Google Scholar] [PubMed]
- Zhao, R.; Domann, F.E.; Zhong, W. Apoptosis induced by selenomethionine and methioninase is superoxide mediated and p53 dependent in human prostate cancer cells. Mol. Cancer Ther. 2006, 5, 3275–3284. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Zavala, J., Sr.; Han, Q.; Xu, M.; Sun, X.; Tan, X.; Tan, X.; Magana, R.; Geller, J.; Hoffman, R.M. Recombinant methioninase infusion reduces the biochemical endpoint of serum methionine with minimal toxicity in high-stage cancer patients. Anticancer Res. 1997, 17, 3857–3860. [Google Scholar] [PubMed]
- Smith, M.; Barnett, M.; Bassan, R.; Gatta, G.; Tondini, C.; Kern, W. Adult acute myeloid leukaemia. Crit. Rev. Oncol. Hematol. 2004, 50, 197–221. [Google Scholar] [CrossRef] [PubMed]
- Kindler, T.; Lipka, D.B.; Fischer, T. FLT3 as a therapeutic target in AML: Still challenging after all these years. Blood 2010, 116, 5089–5102. [Google Scholar] [CrossRef] [PubMed]
- Stary, J.; Zimmermann, M.; Campbell, M.; Castillo, L.; Dibar, E.; Donska, S.; Gonzalez, A.; Izraeli, S.; Janic, D.; Jazbec, J.; et al. Intensive chemotherapy for childhood acute lymphoblastic leukemia: Results of the randomized intercontinental trial ALL IC-BFM 2002. J. Clin. Oncol. 2013, 31. [Google Scholar] [CrossRef] [PubMed]
- Inukai, T.; Kiyokawa, N.; Campana, D.; Coustan-Smith, E.; Kikuchi, A.; Kobayashi, M.; Takahashi, H.; Koh, K.; Manabe, A.; Kumagai, M.; et al. Clinical significance of early T-cell precursor acute lymphoblastic leukaemia: Results of the Tokyo Children’s Cancer Study Group Study L99-15. Br. J. Haematol. 2012, 156, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.-Q.; Ke, Z.-Y.; Huang, L.-B.; Guan, X.-Q.; Zhang, Y.-C.; Zhang, X.-L. High-risk childhood acute lymphoblastic leukemia in China: Factors influencing the treatment and outcome. Pediatr. Blood Cancer 2009, 52, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Zhao, X.-X.; Li, W.-J.; Cui, L.; Zhao, W.; Liu, S.-G.; Yue, Z.-X.; Jiao, Y.; Wu, M.-Y.; Li, Z.-G. Clinical features, early treatment responses, and outcomes of pediatric acute lymphoblastic leukemia in China with or without specific fusion transcripts: A single institutional study of 1004 patients. Am. J. Hematol. 2012, 87, 1022–1027. [Google Scholar] [CrossRef] [PubMed]
- Widjajanto, P.H.; Sutaryo, S.; Purwanto, I.; Ven, P.M.V.; Veerman, A.J.P. Early response to dexamethasone as prognostic factor: Result from Indonesian Childhood WK-ALL protocol in Yogyakarta. J. Oncol. 2012, 2012, 417941. [Google Scholar] [CrossRef] [PubMed]
- Obrador-Hevia, A.; Serra-Sitjar, M.; Rodriguez, J.; Villalonga, P.; Fernandez de Mattos, S. The tumour suppressor FOXO3 is a key regulator of mantle cell lymphoma proliferation and survival. Br. J. Haematol. 2012, 156, 334–345. [Google Scholar] [CrossRef] [PubMed]
- Essafi, M.; Baudot, A.D.; Mouska, X.; Cassuto, J.-P.; Ticchioni, M.; Deckert, M. Cell-penetrating TAT-FOXO3 fusion proteins induce apoptotic cell death in leukemic cells. Mol. Cancer Ther. 2011, 10, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Ausserlechner, M.J.; Salvador, C.; Deutschmann, A.; Bodner, M.; Viola, G.; Bortolozzi, R.; Basso, G.; Hagenbuchner, J.; Obexer, P. Therapy-resistant acute lymphoblastic leukemia (ALL) cells inactivate FOXO3 to escape apoptosis induction by trail and noxa. Oncotarget 2013, 4, 995–1007. [Google Scholar] [PubMed]
- Hussain, A.R.; Al-Rasheed, M.; Manogaran, P.S.; Al-Hussein, K.A.; Platanias, L.C.; Al Kuraya, K.; Uddin, S. Curcumin induces apoptosis via inhibition of PI3'-kinase/AKT pathway in Acute T cell Leukemias. Apoptosis 2006, 11, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Kudou, D.; Misaki, S.; Yamashita, M.; Tamura, T.; Takakura, T.; Yoshioka, T.; Yagi, S.; Hoffman, R.M.; Takimoto, A.; Esaki, N.; et al. Structure of the antitumour enzyme l-Methionine gamma-lyase from pseudomonas putida at 1.8 angstrom resolution. J. Biochem. 2007, 141, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Cavuoto, P.; Fenech, M.F. A review of methionine dependency and the role of methionine restriction in cancer growth control and life-span extension. Cancer Treat. Rev. 2012, 38, 726–736. [Google Scholar] [CrossRef] [PubMed]
- Janssens, V.; Rebollo, A. The role and therapeutic potential of Ser/Thr phosphatase PP2A in apoptotic signalling networks in human cancer cells. Curr. Mol. Med. 2012, 12, 268–287. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Yang, X.; Cao, X.; Liu, F.; Quan, M.; Cao, J. Casticin induces growth suppression and cell cycle arrest through activation of FOXO3a in hepatocellular carcinoma. Oncol. Rep. 2013, 29, 103–108. [Google Scholar] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, K.-Y.; Hu, H.-Y.; Tang, Y.-L.; Xia, F.-G.; Luo, X.-Q.; Liu, J.-Z. High-Level Expression, Purification and Large-Scale Production of l-Methionine γ-Lyase from Idiomarina as a Novel Anti-Leukemic Drug. Mar. Drugs 2015, 13, 5492-5507. https://doi.org/10.3390/md13085492
Huang K-Y, Hu H-Y, Tang Y-L, Xia F-G, Luo X-Q, Liu J-Z. High-Level Expression, Purification and Large-Scale Production of l-Methionine γ-Lyase from Idiomarina as a Novel Anti-Leukemic Drug. Marine Drugs. 2015; 13(8):5492-5507. https://doi.org/10.3390/md13085492
Chicago/Turabian StyleHuang, Kui-Ying, Hai-Yan Hu, Yan-Lai Tang, Feng-Geng Xia, Xue-Qun Luo, and Jian-Zhong Liu. 2015. "High-Level Expression, Purification and Large-Scale Production of l-Methionine γ-Lyase from Idiomarina as a Novel Anti-Leukemic Drug" Marine Drugs 13, no. 8: 5492-5507. https://doi.org/10.3390/md13085492
APA StyleHuang, K.-Y., Hu, H.-Y., Tang, Y.-L., Xia, F.-G., Luo, X.-Q., & Liu, J.-Z. (2015). High-Level Expression, Purification and Large-Scale Production of l-Methionine γ-Lyase from Idiomarina as a Novel Anti-Leukemic Drug. Marine Drugs, 13(8), 5492-5507. https://doi.org/10.3390/md13085492