Investigation of Interspecies Interactions within Marine Micromonosporaceae Using an Improved Co-Culture Approach
Abstract
:1. Introduction

2. Results and Discussion
2.1. Microscale Fermentation of Monocultures and Co-Cultures

2.2. Evaluation of Induced Antibiotic Production in Micromonosporaceae via Co-Culture with Mycolic Acid-Containing Bacteria
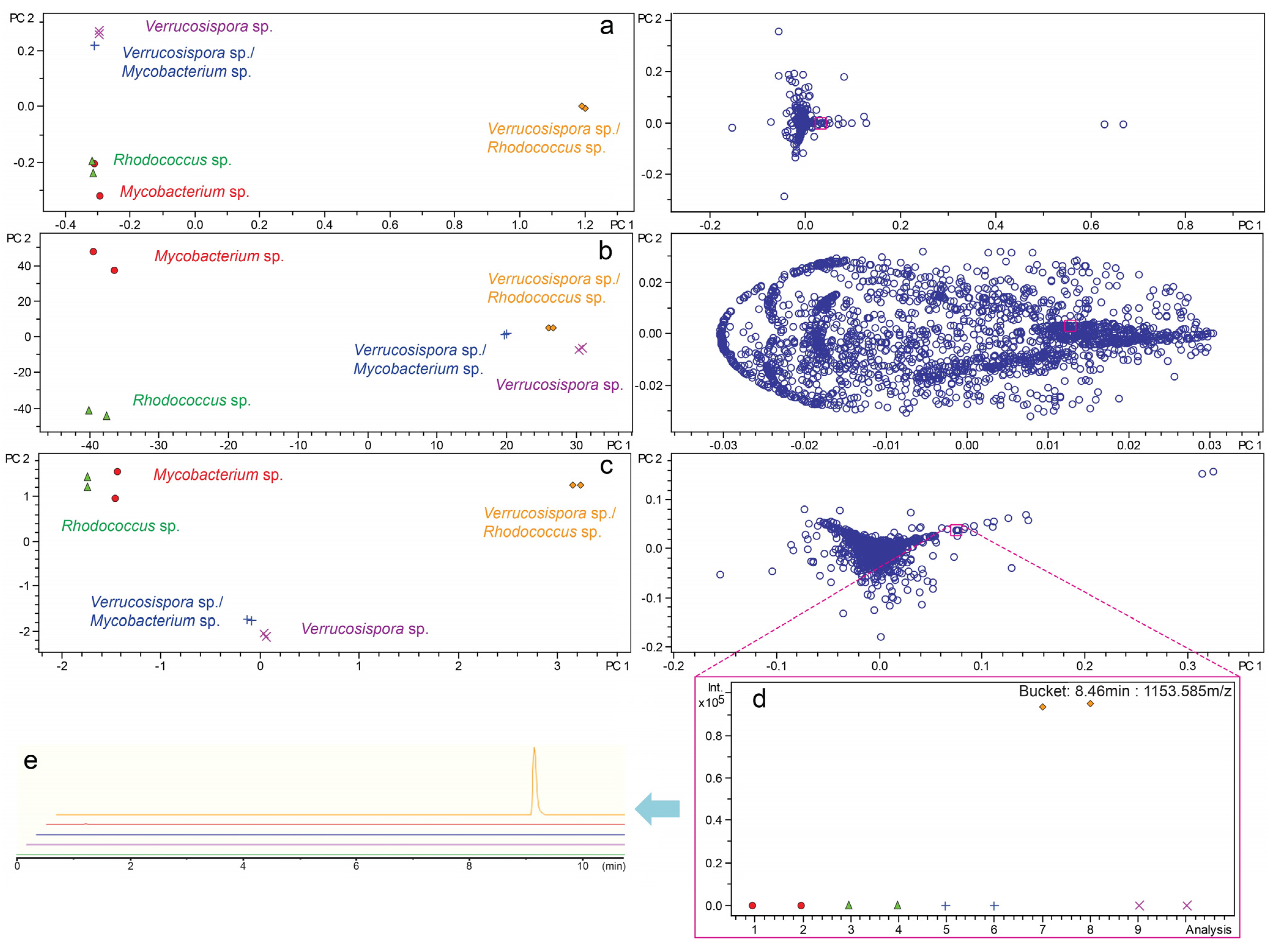
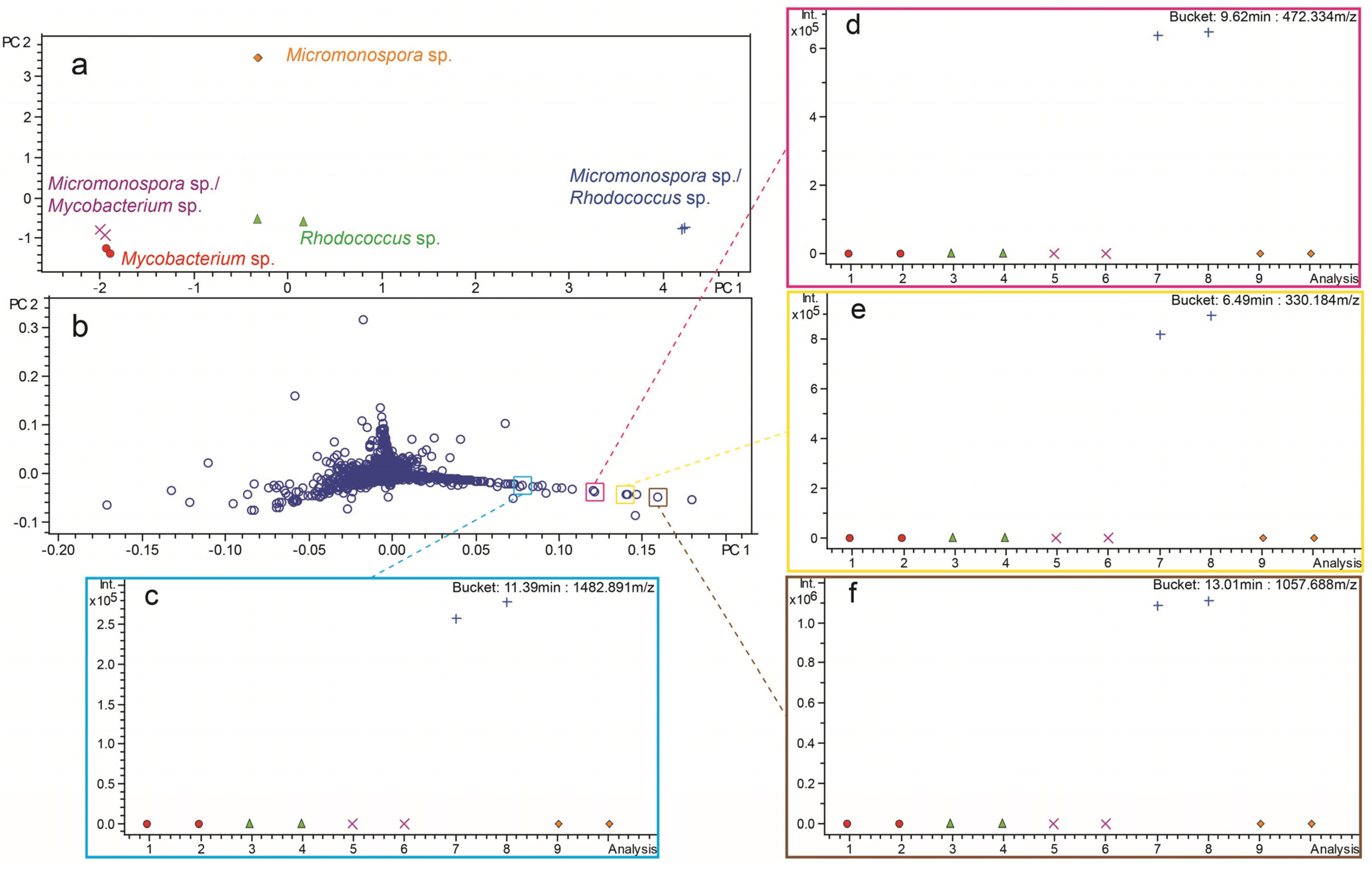
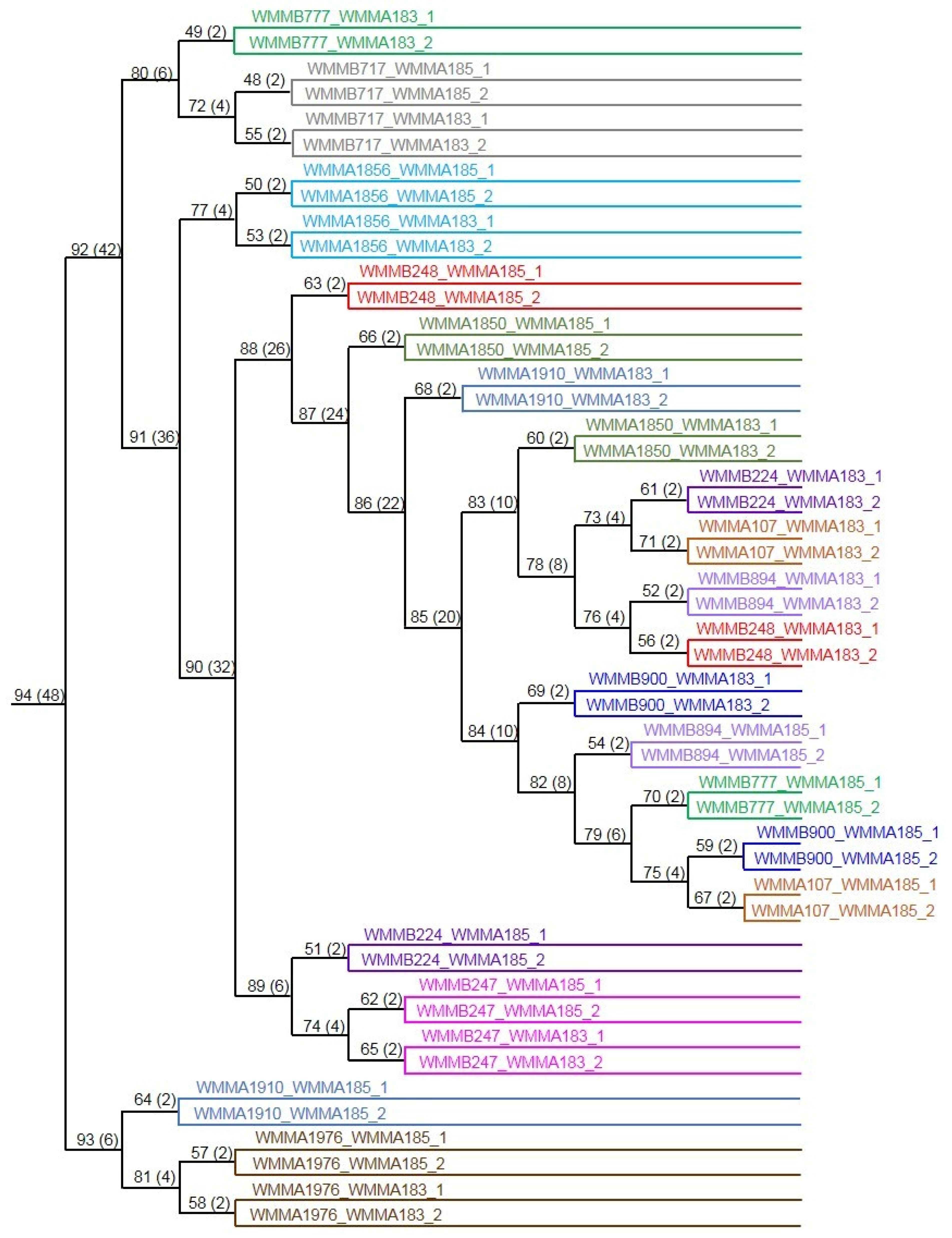
2.3. Investigation of Co-Culture by Metabolomics
| Strain | Organism | Mycobacterium sp. | Rhodococcus sp. |
|---|---|---|---|
| WMMA-1850 | Solwaraspora sp. | ■ | |
| WMMA-1856 | Solwaraspora sp. | ■ | ■ |
| WMMA-1910 | Micromonospora sp. | ■ | |
| WMMA-1949 | Micromonospora sp. | □ | □ |
| WMMA-1976 | Micromonospora sp. | ■ | ■ |
| WMMA-107 | Verrucosispora sp. | ■ | |
| WMMB-224 | Verrucosispora sp. | ■ | |
| WMMB-247 | Micromonospora sp. | ■ | ■ |
| WMMB-248 | Micromonospora sp. | ■ | |
| WMMB-717 | Micromonospora sp. | ■ | ■ |
| WMMB-777 | Micromonospora sp. | ■ | ■ |
| WMMB-894 | Micromonospora sp. | □ ■ | |
| WMMB-900 | Micromonospora sp. | ■ | ■ |
3. Experimental Section
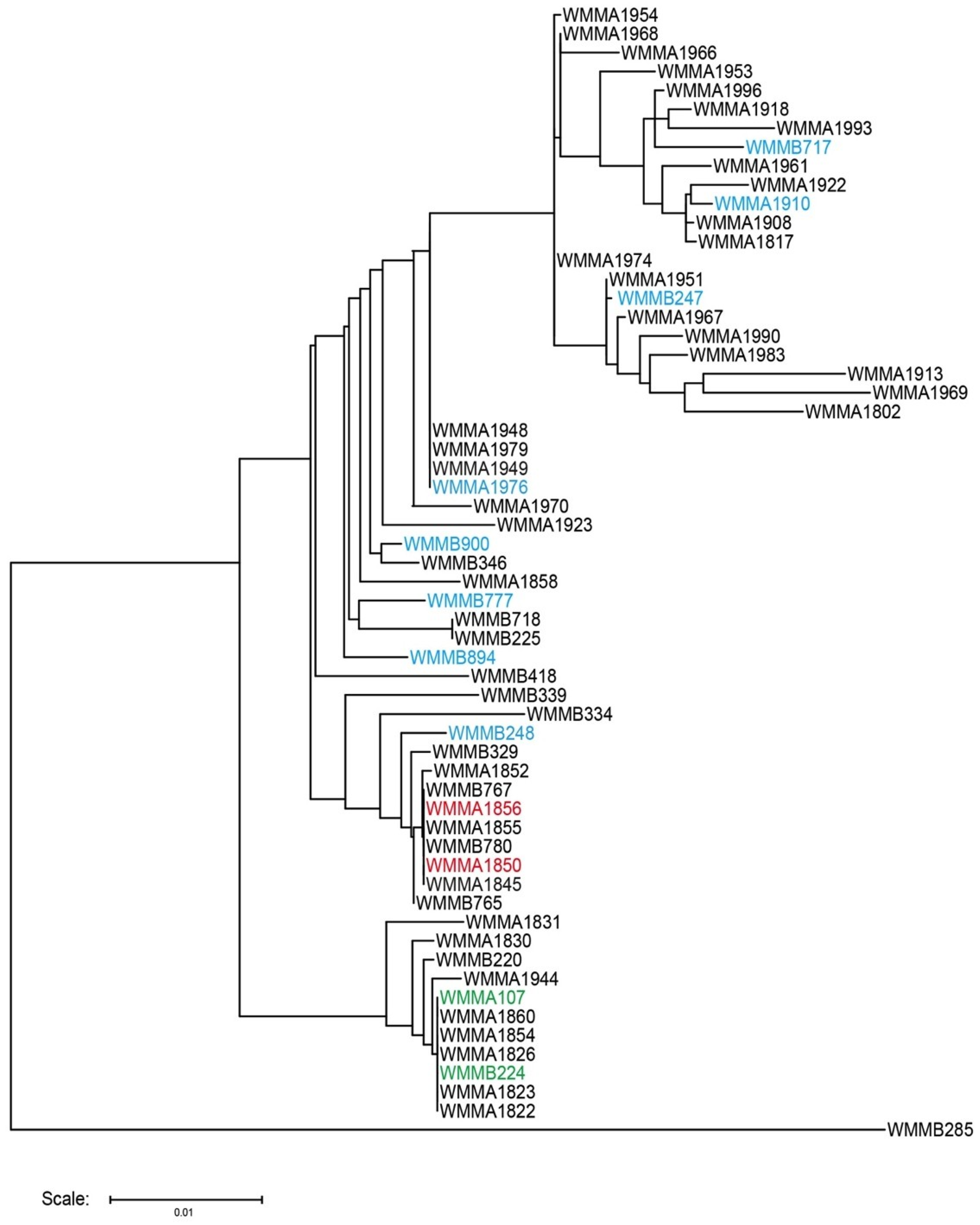
3.1. Strain Collection and Selection
3.2. Microscale Cultivation
3.3. Microscale Extraction
3.4. Antibiotic Activity Screening
3.5. Sample Processing of Microscale Cultures for UHPLC/HRESI-TOF-MS Analysis
3.6. Sample Analysis of Microscale Cultures via UHPLC/HRESI-TOF-MS
3.7. Bucketing and PCA of LC/MS Data
4. Conclusions
Supplementary Information
Acknowledgments
Author Contributions
Conflict of Interest
References
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012, 75, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Nett, M.; Ikeda, H.; Moore, B.S. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 2009, 26, 1362–1384. [Google Scholar] [CrossRef] [PubMed]
- Shank, E.A.; Kolter, R. New developments in microbial interspecies signaling. Curr. Opin. Microbiol. 2009, 12, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Degenkolb, T.; Heinze, S.; Schlegel, B.; Strobel, G.; Grafe, U. Formation of new lipoaminopeptides, acremostatins A, B, and C, by co-cultivation of Acremonium sp. Tbp-5 and Mycogone rosea DSM 12973. Biosci. Biotechnol. Biochem. 2002, 66, 883–886. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, J.; Shao, C.; Ding, W.; She, Z.; Lin, Y. A new xanthone derivative from the co-culture broth of two marine fungi (strain no. E33 and k38). Chem. Nat. Compd. 2011, 47, 382–384. [Google Scholar] [CrossRef]
- Sonnenbichler, J.; Dietrich, J.; Peipp, H. Secondary fungal metabolites and their biological activities, V. Investigations concerning the induction of the biosynthesis of toxic secondary metabolites in basidiomycetes. Biol. Chem. 1994, 375, 71–80. [Google Scholar] [CrossRef]
- Zhu, F.; Chen, G.; Chen, X.; Huang, M.; Wan, X. Aspergicin, a new antibacterial alkaloid produced by mixed fermentation of two marine-derived mangrove epiphytic fungi. Chem. Nat. Compd. 2011, 47, 767–769. [Google Scholar] [CrossRef]
- Zhu, F.; Lin, Y. Marinamide, a novel alkaloid and its methyl ester produced by the application of mixed fermentation technique to two mangrove endophytic fungi from the South China Sea. Chin. Sci. Bull. 2006, 51, 1426–1430. [Google Scholar] [CrossRef]
- Zhu, F.; Lin, Y.-C.; Ding, J.-H.; Wang, X.-P.; Huang, L.-S. Secondary metabolites of two marine-derived mangrove endophytic fungi (strain nos. 1924# and 3893#) by mixed fermentation. Chem. Ind. Forest Prod. 2007, 27, 8–10. [Google Scholar]
- Cueto, M.; Jensen, P.R.; Kauffman, C.; Fenical, W.; Lobkovsky, E.; Clardy, J. Pestalone, a new antibiotic produced by a marine fungus in response to bacterial challenge. J. Nat. Prod. 2001, 64, 1444–1446. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.-C.; Jensen, P.R.; Kauffman, C.A.; Fenical, W. Libertellenones A–D: Induction of cytotoxic diterpenoid biosynthesis by marine microbial competition. Bioorg. Med. Chem. 2005, 13, 5267–5273. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.-C.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J. Nat. Prod. 2007, 70, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Park, H.B.; Kwon, H.C.; Lee, C.-H.; Yang, H.O. Glionitrin A, an antibiotic-antitumor metabolite derived from competitive interaction between abandoned mine microbes. J. Nat. Prod. 2009, 72, 248–252. [Google Scholar] [CrossRef] [PubMed]
- Yakovleva, E.; Bulgakova, T. Formation of amphotericin B in mixed cultures. Pharm. Chem. J. 1978, 12, 1483–1488. [Google Scholar] [CrossRef]
- Dashti, Y.; Grkovic, T.; Abdelmohsen, U.R.; Hentschel, U.; Quinn, R.J. Production of induced secondary metabolites by a co-culture of sponge-associated actinomycetes, Actinokineospora sp. EG49 and Nocardiopsis sp. RV163. Mar. Drugs 2014, 12, 3046–3059. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, S.; Wakimoto, T.; Onaka, H.; Abe, I. Chojalactones A–C, cytotoxic butanolides isolated from Streptomyces sp. cultivated with mycolic acid containing bacterium. Org. Lett. 2015, 17, 1501–1504. [Google Scholar] [CrossRef] [PubMed]
- Kurosawa, K.; Ghiviriga, I.; Sambandan, T.; Lessard, P.A.; Barbara, J.E.; Rha, C.; Sinskey, A.J. Rhodostreptomycins, antibiotics biosynthesized following horizontal gene transfer from Streptomyces padanus to Rhodococcus fascians. J. Am. Chem. Soc. 2008, 130, 1126–1127. [Google Scholar] [CrossRef] [PubMed]
- Onaka, H.; Mori, Y.; Igarashi, Y.; Furumai, T. Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl. Environ. Microbiol. 2011, 77, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Slattery, M.; Rajbhandari, I.; Wesson, K. Competition-mediated antibiotic induction in the marine bacterium Streptomyces tenjimariensis. Microb. Ecol. 2001, 41, 90–96. [Google Scholar] [PubMed]
- Trischman, J.A.; Oeffner, R.E.; de Luna, M.G.; Kazaoka, M. Competitive induction and enhancement of indole and a diketopiperazine in marine bacteria. Mar. Biotechnol. 2004, 6, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, S.; Bohni, N.; Schnee, S.; Schumpp, O.; Gindro, K.; Wolfender, J.-L. Metabolite induction via microorganism co-culture: A potential way to enhance chemical diversity for drug discovery. Biotechnol. Adv. 2014, 32, 1180–1204. [Google Scholar] [CrossRef] [PubMed]
- Marmann, A.; Aly, A.H.; Lin, W.; Wang, B.; Proksch, P. Co-cultivation—A powerful emerging tool for enhancing the chemical diversity of microorganisms. Mar. Drugs 2014, 12, 1043–1065. [Google Scholar] [CrossRef] [PubMed]
- Pettit, R.K. Mixed fermentation for natural product drug discovery. Appl. Microbiol. Biot. 2009, 83, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Vetsigian, K.; Jajoo, R.; Kishony, R. Structure and evolution of Streptomyces interaction networks in soil and in silico. PLoS Biol. 2011, 9, e1001184. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, D.J.; Haste, N.M.; Hollands, A.; Fleming, T.C.; Hamby, M.; Pogliano, K.; Nizet, V.; Dorrestein, P.C. Microbial competition between Bacillus subtilis and Staphylococcus aureus monitored by imaging mass spectrometry. Microbiology 2011, 157, 2485–2492. [Google Scholar] [CrossRef] [PubMed]
- Traxler, M.F.; Watrous, J.D.; Alexandrov, T.; Dorrestein, P.C.; Kolter, R. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. mBio 2013, 4, e00450413. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, F.; Larsen, O.; Thiel, V.; Rapp, H.T.; Pape, T.; Michaelis, W.; Reitner, J. An anaerobic world in sponges. Geomicrobiol. J. 2005, 22, 1–10. [Google Scholar] [CrossRef]
- Taylor, M.W.; Radax, R.; Steger, D.; Wagner, M. Sponge-associated microorganisms: Evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 2007, 71, 295–347. [Google Scholar] [CrossRef] [PubMed]
- Webster, N.; Hill, R. The culturable microbial community of the Great Barrier Reef sponge Rhopaloeides odorabile is dominated by an α-Proteobacterium. Mar. Biol. 2001, 138, 843–851. [Google Scholar] [CrossRef]
- Schmidt, E.W.; Obraztsova, A.Y.; Davidson, S.K.; Faulkner, D.J.; Haygood, M.G. Identification of the antifungal peptide-containing symbiont of the marine sponge Theonella swinhoei as a novel delta-Proteobacterium, “Candidatus Entotheonella palauensis”. Mar. Biol. 2000, 136, 969–977. [Google Scholar] [CrossRef]
- Wilson, M.C.; Mori, T.; Ruckert, C.; Uria, A.R.; Helf, M.J.; Takada, K.; Gernert, C.; Steffens, U.A.E.; Heycke, N.; Schmitt, S.; et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 2014, 506, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.J.; Torres, J.P.; Ammon, M.A.; Marett, L.; Teichert, R.W.; Reilly, C.A.; Kwan, J.C.; Hughen, R.W.; Flores, M.; Tianero, M.D.; et al. A bacterial source for mollusk pyrone polyketides. Chem. Biol. 2013, 20, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.W. The secret to a successful relationship: Lasting chemistry between ascidians and their symbiotic bacteria. Invertebr. Biol. 2015, 134, 88–102. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.W.; Nelson, J.T.; Rasko, D.A.; Sudek, S.; Eisen, J.A.; Haygood, M.G.; Ravel, J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. Sci. USA 2005, 102, 7315–7320. [Google Scholar] [CrossRef] [PubMed]
- Duetz, W.A.; Witholt, B. Effectiveness of orbital shaking for the aeration of suspended bacterial cultures in square-deepwell microtiter plates. Biochem. Eng. J. 2001, 7, 113–115. [Google Scholar] [CrossRef]
- Duetz, W.A.; Witholt, B. Oxygen transfer by orbital shaking of square vessels and deepwell microtiter plates of various dimensions. Biochem. Eng. J. 2004, 17, 181–185. [Google Scholar] [CrossRef]
- Hou, Y.; Braun, D.R.; Michel, C.R.; Klassen, J.L.; Adnani, N.; Wyche, T.P.; Bugni, T.S. Microbial strain prioritization using metabolomics tools for the discovery of natural products. Anal. Chem. 2012, 84, 4277–4283. [Google Scholar] [CrossRef] [PubMed]
- Wyche, T.P.; Hou, Y.P.; Braun, D.; Cohen, H.C.; Xiong, M.P.; Bugni, T.S. First natural analogs of the cytotoxic thiodepsipeptide thiocoraline A from a marine Verrucosispora sp. J. Org. Chem. 2011, 76, 6542–6547. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal database project: Data and tools for high throughput rrna analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef] [PubMed]
- Gassner, N.C.; Tamble, C.M.; Bock, J.E.; Cotton, N.; White, K.N.; Tenney, K.; St Onge, R.P.; Proctor, M.J.; Giaever, G.; Nislow, C.; et al. Accelerating the discovery of biologically active small molecules using a high-throughput yeast halo assay. J. Nat. Prod. 2007, 70, 383–390. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adnani, N.; Vazquez-Rivera, E.; Adibhatla, S.N.; Ellis, G.A.; Braun, D.R.; Bugni, T.S. Investigation of Interspecies Interactions within Marine Micromonosporaceae Using an Improved Co-Culture Approach. Mar. Drugs 2015, 13, 6082-6098. https://doi.org/10.3390/md13106082
Adnani N, Vazquez-Rivera E, Adibhatla SN, Ellis GA, Braun DR, Bugni TS. Investigation of Interspecies Interactions within Marine Micromonosporaceae Using an Improved Co-Culture Approach. Marine Drugs. 2015; 13(10):6082-6098. https://doi.org/10.3390/md13106082
Chicago/Turabian StyleAdnani, Navid, Emmanuel Vazquez-Rivera, Srikar N. Adibhatla, Gregory A. Ellis, Doug R. Braun, and Tim S. Bugni. 2015. "Investigation of Interspecies Interactions within Marine Micromonosporaceae Using an Improved Co-Culture Approach" Marine Drugs 13, no. 10: 6082-6098. https://doi.org/10.3390/md13106082
APA StyleAdnani, N., Vazquez-Rivera, E., Adibhatla, S. N., Ellis, G. A., Braun, D. R., & Bugni, T. S. (2015). Investigation of Interspecies Interactions within Marine Micromonosporaceae Using an Improved Co-Culture Approach. Marine Drugs, 13(10), 6082-6098. https://doi.org/10.3390/md13106082