Abstract
Marine-derived bacteria and fungi are promising sources of novel bioactive compounds that are important for drug discovery programs. However, as encountered in terrestrial microorganisms there is a high rate of redundancy that results in the frequent re-discovery of known compounds. Apparently only a part of the biosynthetic genes that are harbored by fungi and bacteria are transcribed under routine laboratory conditions which involve cultivation of axenic microbial strains. Many biosynthetic genes remain silent and are not expressed in vitro thereby seriously limiting the chemical diversity of microbial compounds that can be obtained through fermentation. In contrast to this, co-cultivation (also called mixed fermentation) of two or more different microorganisms tries to mimic the ecological situation where microorganisms always co-exist within complex microbial communities. The competition or antagonism experienced during co-cultivation is shown to lead to a significantly enhanced production of constitutively present compounds and/or to an accumulation of cryptic compounds that are not detected in axenic cultures of the producing strain. This review highlights the power of co-cultivation for increasing the chemical diversity of bacteria and fungi drawing on published studies from the marine and from the terrestrial habitat alike.
1. Introduction
Marine macroorganisms are already firmly established as sources of new drugs for the treatment of cancer but also for other malignancies such as chronic pain. So far, seven marine natural products or drugs derived from marine leads have entered the drug market. These compounds include among others Yondelis® (approved for use in Europe, Russia and in South Korea for the treatment of soft tissue sarcoma), Adcetris® (approved in the US and in the EU for the treatment of non-Hodgkin lymphoma or following an autologous stem cell transplantation), Halaven® (approved for use in the US, Canada and in the EU for the treatment of advanced mamma carcinoma) and Prialt® (approved in the US and in the EU for treatment of severe and chronic pain) [1,2,3,4,5,6,7,8,9]. In addition, the nucleoside analogues Cytosar-U® and Vira-A® may be mentioned that were modeled based on compounds isolated from marine sponges [7,10,11]. Many other compounds that are mainly derived from marine invertebrates are currently in clinical phases I or II, primarily for the treatment of cancer [12,13,14,15].
Research on marine-derived microorganisms is far more recent in comparison and was only seriously initiated in the nineties of the last century. As for marine macroorganisms, marine-derived bacteria and fungi quickly proved to be prolific sources of novel bioactive compounds that are of considerable interest as new drug leads. The most famous example of a marine microbial compound is salinosporamide A (marizomib), which is a fermentation product of marine Salinispora bacteria and a proteasome inhibitor that is currently undergoing clinical studies as a potential new anti-cancer drug [16,17,18,19]. It may be expected that further drug candidates obtained from marine-derived microorganisms will follow in the future.
However, as for natural products from terrestrial sources, the re-discovery of already known compounds is high and poses serious problems for marine bioprospecting from macroorganisms and microorganisms alike. New insights into the molecular biology of bacteria and fungi have demonstrated that the genetic potential of these microbes, in terms of producing a far greater chemical diversity of compounds than is currently known, has been vastly underestimated in the past [20,21,22]. Many microbial biosynthetic genes are apparently not transcribed under standard laboratory conditions but remain silent. As a consequence, only a fraction of the real biosynthetic diversity of microbes is obtained in terms of produced compounds, which leads to the currently experienced bottle neck in drug discovery from microbial sources. Several strategies exist that try to overcome these limitations during fermentation of microbes. These include among others the OSMAC approach where promising strains are cultured in a variety of media and under different culture regimes in order to maximize the diversity of compounds produced [23], in addition to epigenetic modifications. In the latter case microorganisms are treated with epigenetic modifiers such as histone deacetylase inhibitors or DNA methyl transferase inhibitors aiming at a modulation of histones or of the DNA thereby initiating the transcription of silent genes which in turn may lead to the accumulation of new compounds [24,25,26,27,28,29].
A third option tries to mimic the natural ecological situation, where microbes always co-exist within complex microbial communities. Competition for limited resources and antagonism are characteristics of these micro-habitats, which favor various defense mechanisms that rely mainly on the production of bioactive secondary metabolites [30]. It is often assumed that antibiotic production by bacteria and fungi can be interpreted within these ecological frames that will select for chemically defended microbes [31,32,33,34,35] even though other authors question this hypothesis [36,37,38]. Co-cultivation of two or more different microbes tries to mimic this setting in a laboratory scale. Competition among these microbes is deliberately provoked in the hope that biosynthetic genes that remain silent under luxurious culture conditions are activated and transcribed under stress conditions.
As shown in this review, several co-cultivation studies that have been conducted in recent years prove the feasibility of this strategy and suggest that co-cultivation is a viable experimental approach for enhancing the chemical diversity of microorganisms when grown in vitro. When preparing this review we have deliberately decided to extend our scope beyond co-cultivation studies reported from the marine field and to include those that have been carried out with microorganisms derived from the terrestrial habitat. Several reasons were decisive for this decision: first of all, the definition of a “marine” microorganism is often vague. Many microorganisms isolated from marine sources are in fact already known from the terrestrial habitat and can be cultured without the presence of sea salt in the medium. This is especially common for marine-derived fungi and has been noted by many authors [39,40,41]. It is in fact often assumed that a large part of the microbes found in the sea are of terrestrial origin and may survive under marine conditions but are by no means obligatory marine [42,43,44]. Furthermore, recent studies conducted with the fungus Aspergillus nidulans when challenged by Streptomyces rapamycinicus shed for the first time light on the molecular basis of induction of silent fungal biosynthetic gene clusters during co-cultivation [45,46,47]. These studies will certainly prove helpful for natural product chemists and molecular microbiologists who are trying to unravel the interspecies cross talk of marine-derived microorganisms in the future.
2. Results
2.1. Co-Cultivation Studies of Marine-Derived Microorganisms with Influence on Natural Product Accumulation
During the ongoing search for novel bioactive metabolites from microorganisms, several studies which were conducted mainly within the last few years proved the power of co-cultivation (also called “mixed fermentation”) as an experimental tool for either enhancing the production of constitutively present compounds and/or for inducing silent gene clusters. In general, the performed studies followed three major strategies which include (a) co-cultivation of different fungi; (b) co-cultivation of fungi with bacteria and (c) co-cultivation of different bacteria. With regard to marine-derived microorganisms (Table 1, Figure 1 and Figure 2 for structures) thirteen different co-cultivation studies have been published in recent years [34,48,49,50,51,52,53,54,55,56,57,58,59] proving that this experimental approach for increasing the chemical productivity of microbes is a highly promising strategy whereas on the other side it is still an emerging field that will require broader attention of marine natural product chemists and microbiologists alike in the future.

Table 1.
Co-cultures of marine-derived microorganisms and secondary metabolites reported.
| Co-Cultivated Microorganisms | Secondary Metabolites Reported | Reported Activity | Reference |
|---|---|---|---|
| Aspergillus sp. Aspergillus sp. | Aspergicin (n, 1) Neoaspergillic acid (k) Ergosterol (k) | Antibiotic Antibiotic - | [48] |
| Unidentified fungus Unidentified fungus | 8-Hydroxy-3-methyl-9-oxo-9H-xanthene-1-carboxylic acid methylether (n, 2) | Antifungal | [49] |
| Unidentified fungus Unidentified fungus | Marinamide (n, 3) Marinamide methylether (n, 4) | Cytotoxic Cytotoxic | [60] |
| Pestalotia sp. Unidentified bacterium | Pestalone (n, 5) | - | [52] |
| Libertella sp. Thalassopia sp. | Libertellenones A–D (n, 6–9) | Cytotoxic | [53] |
| Emericella ap. Salinospora arenicola | Emericellamide A (n, 10) Emericellamide B (n, 11) | Antibiotic Cytotoxic | [54] |
| Aspergillus fumigatus Sphingomonas sp. | Glionitrin A (n, 12) | Cytotoxic Antibiotic | [55] |
| B. thuringensis B. Megaterium S. sciuri | Indole (k) Phe-Pro diketopiperazine (k) | Antibiotic (on extract level) | [59] |
| Streptomyces tenjimariensis 12 unidentified bacteria | Istamycin (k) | Antibiotic | [34] |
n = new; k = known.
In an attempt to induce the accumulation of novel compounds through fungal-fungal co-cultivation, Zhu et al. [48] demonstrated that the mixed cultivation of two different mangrove-derived epiphytic fungi, both belonging to the genus Aspergillus, leads to the production of the new alkaloid aspergicin (1) (Figure 1) and the known compounds neoaspergillic acid and ergosterol, respectively. Aspergicin (1) and neoaspergillic acid were evaluated for their antibiotic potential towards the Gram-positive bacteria Staphylococcus aureus, Staphylococcus epidermidis, Bacillus subtilis, Bacillus dysenteriae, Bacillus proteus and against the Gram-negative Escherichia coli. Aspergicin (1) exhibited a MIC of 15.62 µg/mL against B. subtilis, whereas the MIC of neoaspergillic acid was in the range of 0.49–15.62 µg/mL against all tested bacteria [48].
Li et al. [49] showed that the co-cultivation of two epiphytic, unidentified mangrove-associated fungi resulted in the production of a novel xanthone derivative (2). Using the agar plate diffusion assay and a fixed compound concentration of 100 µg/mL, a mild antifungal activity of 2 against Gloeosporium musae and Peronophthora cichoralearum was detected [49].
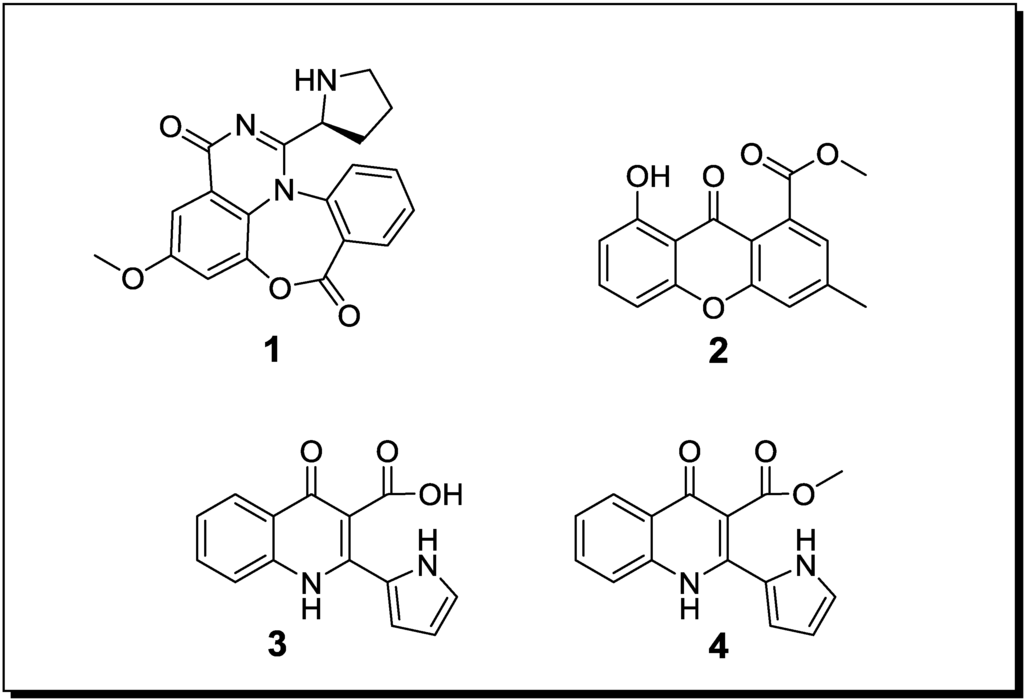
Figure 1.
New natural products reported from co-cultures of marine-derived fungi.
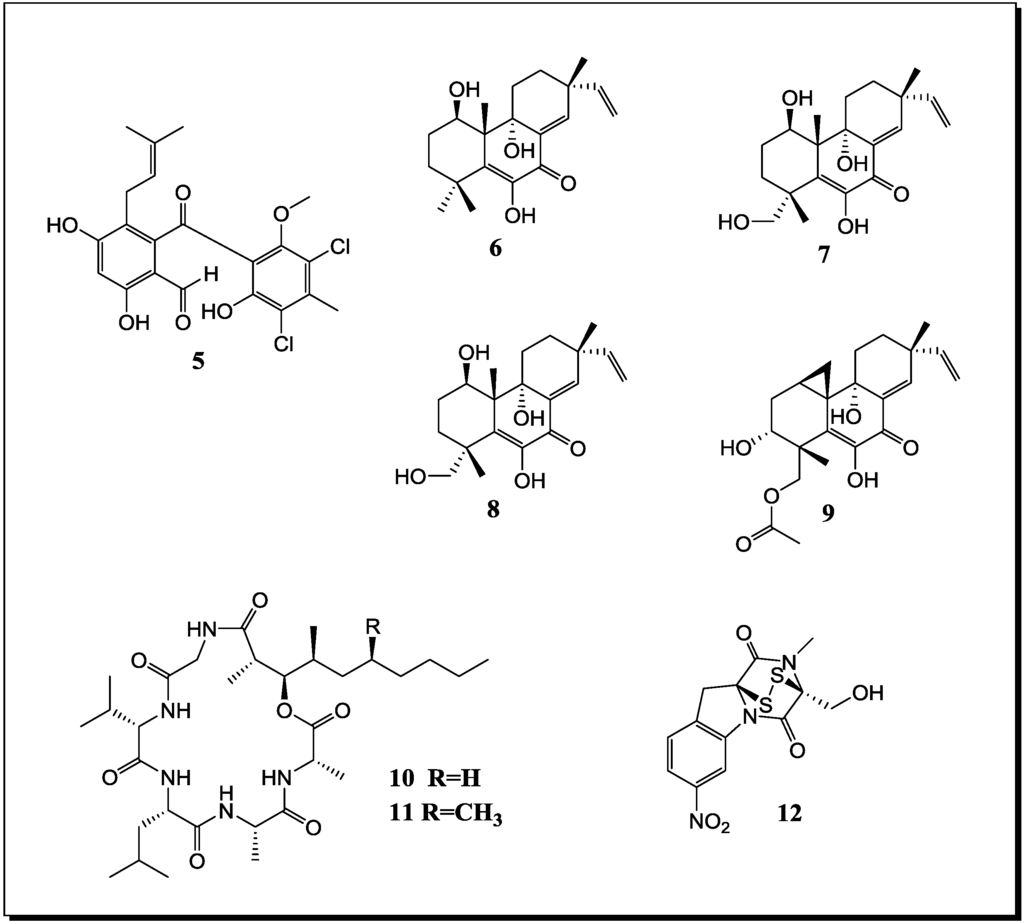
Figure 2.
New natural products reported from co-cultures of marine-derived fungi and bacteria.
Co-cultivation of two unidentified mangrove-derived endophytic fungi, resulted in the production of the new alkaloids marinamide (3) and marinamide methylether (4) [50]. When studied for their cytotoxic effects on HepG2, 95-D, MGC832 and HeLa cells, IC50 values for marinamide (3) were in the nanomolar range whereas those of marinamide methylether (4) were in the low micromolar range [60].
Until now five studies were published on the co-cultivation of marine-derived fungi and bacteria. Miao et al. [51] investigated the influence of bacterial cells and cell free bacterial broth on the fungus Arthrinium c.f. saccharicola. In total the fungus was co-cultivated with 14 different fouling bacterial taxa. Six of them, including Pseudoalteromonas spongiae, Shewanella algae, Vibrio vulnificus, Vibrio halioticoli, Pseudoalteromonas piscicida and Loktanella hongkongensis reduced the growth of the fungus during co-cultivation. This effect was especially pronounced for P. piscicida. Addition of the cell free culture broth of this bacterium significantly increased the antimicrobial activity of the fungus when measured against V. vulnificus and P. spongiae. Apparently the induction of antibiotic activity by the fungus was caused by so far unknown bacterial compounds that had been secreted into the culture broth and served as chemical signals for elicitation of the fungal secondary metabolism [51].
In 2001 Cueto et al. [52] reported the chlorinated prenylsecoanthraquinone, pestalone (5) (Figure 2), from a co-culture of the marine-derived fungus Pestalotia sp. with a likewise marine-derived unidentified Gram-negative bacterium of the genus Thalassopia (CNJ-328). Since the methyl analogue of 5 was already known from another fungus (Chrysosporium sp.), pestalone (5) was suspected to be of fungal rather than of bacterial origin. When evaluating the antibacterial activity of 5 against MRSA and against vancomycin-resistent Enterococcus faecium, MIC values of 37 ng/mL and 78 ng/mL were obtained whereas the GI50 of pestalone when measured in the NIH human tumor cell line screen amounted to 6.0 µM [52].
Co-cultivation of a marine-derived Libertella sp. fungus and the marine-derived bacterium Thalassopia sp. (CNJ-328) yielded the novel diterpenoids libertellenones A–D (6–9) which are presumably of fungal origin since diterpenes are far more common fungal rather than bacterial secondary products. The cell free bacterial culture broth as well as autoclaved cultures of CNJ-328 or an EtOAc extract of the bacterium failed to induce the accumulation of 6–9 by the fungus Libertella sp. Therefore a direct physical contact of fungus and bacterium is assumed to be necessary for induction of the libertellenones [53]. In spite of being induced by a bacterium, the libertellenones failed to show antibiotic properties against CNJ-328, MRSA or against a vancomycin-resistent E. faecium strain whereas they exhibited cytotoxic activity against HCT-116 cells with IC50 values between 0.76 and 53 µM, dependent on the derivative investigated [53]. Interestingly, in the last two studies the bacterial strain CNJ-328 was found to induce different biosynthetic pathways in Libertella sp. and in Pestalotia sp., respectively. Whereas the core structure of pestalone (5) is a polyketide which features a prenyl substituent, the libertellenones (6–9) are terpenoids.
Through co-cultivation of the fungus Emericella sp., that had been isolated from the surface of the marine green alga Halimeda sp. and the marine sediment-derived actinomyete Salinispora arenicola Oh et al. (2007) [54] were able to demonstrate a 100 fold increased production of the two fungal depsipetides emericellamide A (10) and B (11). Both compounds when evaluated for their biological activities displayed a moderate antibiotic activity against a methicillin-resistant S. aureus strain with MICs of 3.0 µM and 6.0 µM, as well as weak cytotoxic effects towards HCT-116 cells with IC50 values of 23 µM and 40 µM, respectively [54].
A novel diketopiperazine disulfide, glionitrin A (12), was obtained from a mixed fermentation of the marine-derived fungus Aspergillus fumigatus and the marine-derived bacterium Sphingomonas sp.. The compound showed strong cytotoxic activity against HCT-116, A549, AGS and DU145 cells with IC50 values of 0.82, 0.55, 0.45 and 0.24 µM, respectively, whereas the MIC against MRSA amounted to 0.78 µg/mL [55]. Interestingly, glionitrin A (12) exhibited also weak antifungal activity against A. fumigatus, which was used for the co-culture study. Presumably the core structure of glionitrin A (12) is derived from the fungus whereas the biogenetic origin of the nitro group is unknown [55].
Dusane et al. [56] isolated four surface associated bacteria from the green mussel Perna viridis and from the coral Symphyllia sp., which were identified as Bacillus sp., Bacillus licheniformis, Bacillus pumilus and as Serratia marcescens by analysis of their 16S rDNA sequences. Each bacterial strain was co-cultured with Pseudomonas aeruginosa, Bacillus pumilus and with the yeasts Candida albicans and Yarrowia lipolytica. The extracts resulting from co-cultivation were screened for increased antimicrobial activity against the respective competitors that had been used during mixed fermentation. Antibacterial activity against P. aeruginosa or against B. pumilus was enhanced for most of the studied epibiotic bacteria during co-cultivation. Biosurfactant production was increased when B. pumilus was co-cultivated with S. marcescens or with B. lichenifomis, whereas quorum-sensing inhibition was found for Bacillus sp. when co-cultured with P. aeruginosa [56].
In a similar approach Burgess et al. [57] isolated several unidentified surface associated bacteria form the marine algae Fucus vesiculosus, F. serratus, Corallina officinalis, Ophiothrix fragilis and Asterias rubens. In total 78 bacterial strains were isolated, cultured and systematically investigated for their antibiotic potential. Nine extracts showed antibiotic activity in the agar plate diffusion assay. Two unidentified strains, MH46 and SSE20, were chosen for a cultivation study employing cell free culture broths of E. coli, B. subtilis or of P. aeruginosa as well as of the unidentified surface associated bacterial strains MH1, MH2 and MH3. All resulting extracts were tested for their antibiotic properties against B. subtilis. Two major effects were noted: the antibiotically inactive bacterium MH46 suddenly showed antibiotic activity when exposed to the culture broth of P. aeruginosa. In addition the bacteria MH1, MH2 and MH3 and SSE20 showed an increased antibiotic activity when cultured in the presence of culture broths of MH1, MH2 and MH3 or of B. subtilis [57].
In a further study, sixteen strains of marine-derived epibiotic bacteria isolated from the algae F. vesiculosus and the nudibranch Archidoris pseudoargus were subjected to co-cultivation with the human pathogens S. aureus, P. aeruginosa and E. coli [58]. The marine-derived bacteria were cultivated either with autoclaved cultures of S. aureus or in the same culture vessels together with live cultures of the pathogens. In the latter case the pathogens were kept within dialysis tubes with a pore size of 8000 Da thereby allowing diffusion of secreted low to medium weight compounds but avoiding physical cell-cell contact of pathogens and marine-derived bacteria. As a read out, the antibiotic activity of the resulting extracts was measured using MSSA, MRSA, E. coli and P. aeruginosa as indicator strains. Both S. aureus and P. aeruginosa induced antibiotic activity in several of the marine-derived bacteria. This induced antibiotic activity was similar for live and for autoclaved bacteria [58].
When two different Bacillus spp. (related to B. thuringiensis and B. megaterium, respectively based on 16S rDNA sequence similarity) that had been isolated from the surface of the marine alga Ulva californica were co-cultured, the production of indole and (Phe-Pro) diketopiperazines was found to increase by 450% and 320%, respectively compared to an axenic culture of the producing strain (“B. thuringiensis”) [59]. This induced accumulation of diketopiperazines coincided with an increase of antibiotic activity against the second Bacillus strain (“B. megaterium”). When “B. thuringiensis” was co-cultured with another bacterial strain isolated from U. californica (Staphylococcus sciuri), no induction of diketopiperazine accumulation was detected. At the same time the analyzed indole and (Phe-Pro) diketopiperazines failed to exert antibiotic activity against S. sciuri [59].
In a further study Slattery et al. [34] investigated the interaction between the marine-derived Streptomyces tenjimariensis and 53 further unknown marine bacteria, with regard to induction of istamycin A and B accumulation by S. tenjimariensis. Twelve out of 53 co-cultures showed at least a twofold higher production of istamycins compared to an axenic culture of S. tenjimariensis. This effect was only found when S. tenjimariensis was inoculated 24 h prior to the other bacteria. When the bacterial competitors had been inoculated 24 h before addition of S. tenjimariensis, or when both cultures were inoculated simultaneously a reduced istamycin production was observed [34].
2.2. Co-Cultivation Studies of Terrestrial Microorganisms with Influence on Natural Product Accumulation
Reported co-cultivation studies of terrestrial microorganisms follow similar research strategies as described before for marine-derived microbes and focus on interactions of fungi with fungi, fungi with bacteria or bacteria with bacteria. To the best of our knowledge seven studies investigating the effects of fungal-fungal interaction have been reported to date from the terrestrial habitat (Table 2, Figure 3 and Figure 4 for structures).

Table 2.
Co-cultures of terrestrial microorganisms and secondary metabolites reported.
| Co-Cultivated Microorganisms | Secondary Metabolites Reported | Reported Activity | Reference |
|---|---|---|---|
| Penicillium pinophilum Trichoderma harzianum | Secopenicillide C (n, 13) | n.t. | [61] |
| Penicillide (k) | n.t. | ||
| MC-141 (k) | n.t. | ||
| Pestalasin A (k) | n.t. | ||
| Stromemycin (k) | n.t. | ||
| Fusarium tricinctum Fusarium begoniae | Subenniatin A (n, 14) | - | [62] |
| Subenniatin B (n, 15) | - | ||
| Enniatin A (k) | - | ||
| Enniatin A1 (k) | - | ||
| Enniatin B (k) | - | ||
| Enniatin B1 (k) | - | ||
| Acremonium sp. Mycogone rosea | Acremostatin A (n, 16) | n.t. | [63] |
| Acremostatin B (n, 17) | n.t. | ||
| Acremostatin C (n, 18) | n.t. | ||
| Gloeophyllum abietinum Heterobasidion annosum Armillaria ostoyae | Oospoglycol (k) | n.t. | [64] |
| Oopsonol (k) | n.t. | ||
| Fomannoxin (k) | n.t. | ||
| Fomannoxinalcohol (k) | n.t. | ||
| Fomannosin (k) | n.t. | ||
| Melledonal (k) | n.t. | ||
| Melledonal C (k) | n.t. | ||
| Melleolide D (k) | n.t. | ||
| Oyadendron sulphureoochraceum Ascochyta pisi Emercillopsis minima Cylindrocarpon destructans Fusarium oxysporum | Lateritin (k) | Cytotoxic Antifungal Antibiotic | [65] |
| Paraconiothyrium sp. Alternaria sp. Phomopsis sp. | Paclitaxel (k) | n.t. | [66] |
| Streptomyces bullii Aspergillus fumigatus | Brevianamide F (k) | -/-/C | [67] |
| Spirotryprostatin A (k) | T/L/C | ||
| 6-Methoxy spirotrypostatin B (k) | -/L/C | ||
| Fumitremorgin C (k) | T/L/C | ||
| 12,13-Dihydroxy Fumitremorgin C (k) | T/L/C | ||
| Fumitremorgin B (k) | T/L/C | ||
| Verruculogen (k) | T/L/C | ||
| 11-O-Methylpseurotin A (k) | -/-/C | ||
| 11-O-Methylpseurotin A2 (n, 19) | -/L/C | ||
| Ergosterol (k) | -/-/- | ||
| Emestrin A (k) | n.t. | ||
| Emestrin B (k) | n.t. | ||
| Aspergillus fumigatus Streptomyces rapamycinicus | Fumicycline A (n, 20) | Antibiotic | [45] |
| Fumicyline B (n, 21) | Antibiotic | ||
| Aspergillus fumigatus Streptomyces peucetius | Fumiformamide (n, 22) NN′-((1Z,3Z)-1,4-bis (4-Methoxyphenyl)buta-1,3-diene-2,3diyl)Di-formamide (n, 23) | Cytotoxic | [68] |
| Fusarium tricinctum Bacillus subtilis | Macrocarpon C (n, 24) | - | [30] |
| 2-(Carboxymethylamino)benzoic acid (n, 25) | - | ||
| (−)-Citreoisocoumarinol (n, 26) | - | ||
| Lateropyrone (k) | Antibiotic | ||
| Enniatin A1 (k) | Antibiotic | ||
| Enniatin B (k) | - | ||
| Enniatin B1 (k) | Antibiotic | ||
| (+)-Citreoisocoumarinol (k) | - | ||
| Tsukamurella pulmonis Streptomyces endus | Alchivemycin A (n, 27) | Antibiotic | [69] |
n = new; k = known; n.t. = not tested; T = trypanocidal; L = leishmanocidial; C = cytotoxic; - = inactive.
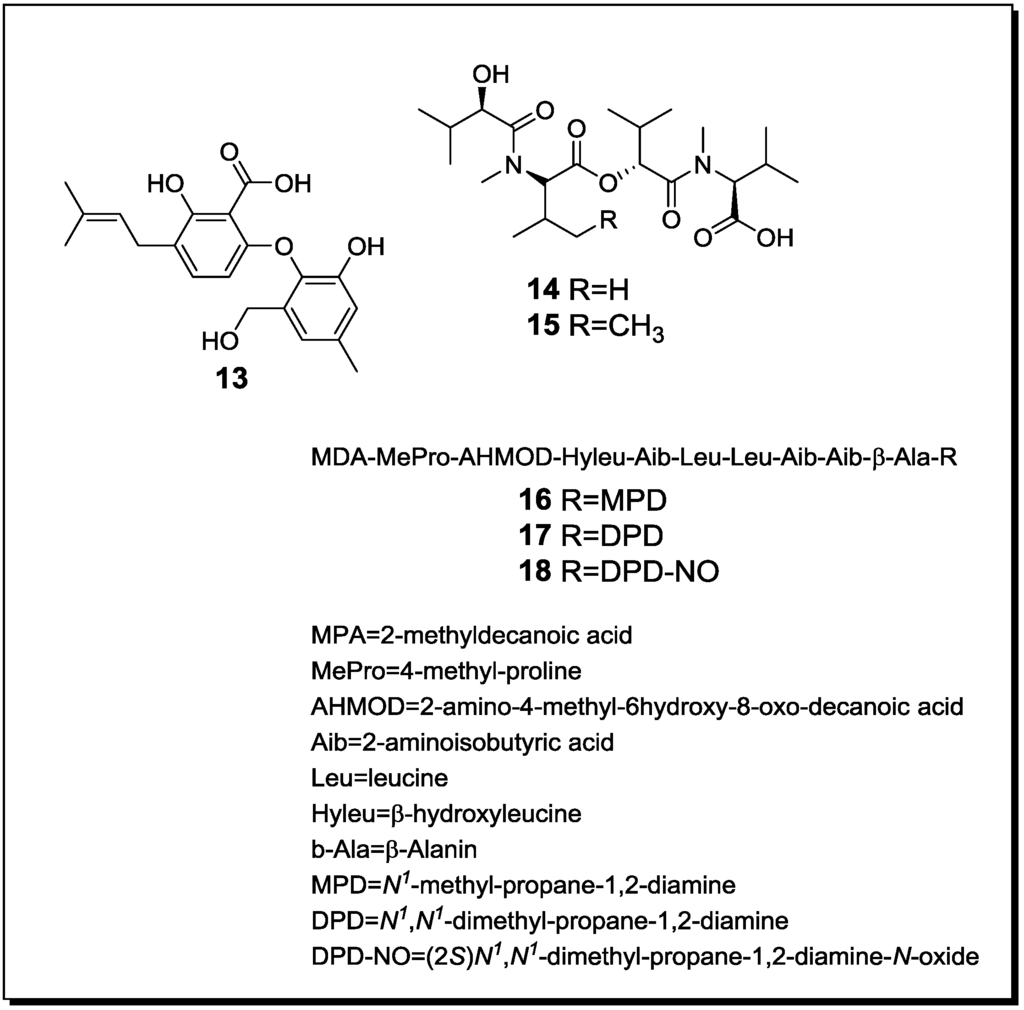
Figure 3.
New natural products reported from co-cultures of terrestrial fungi.
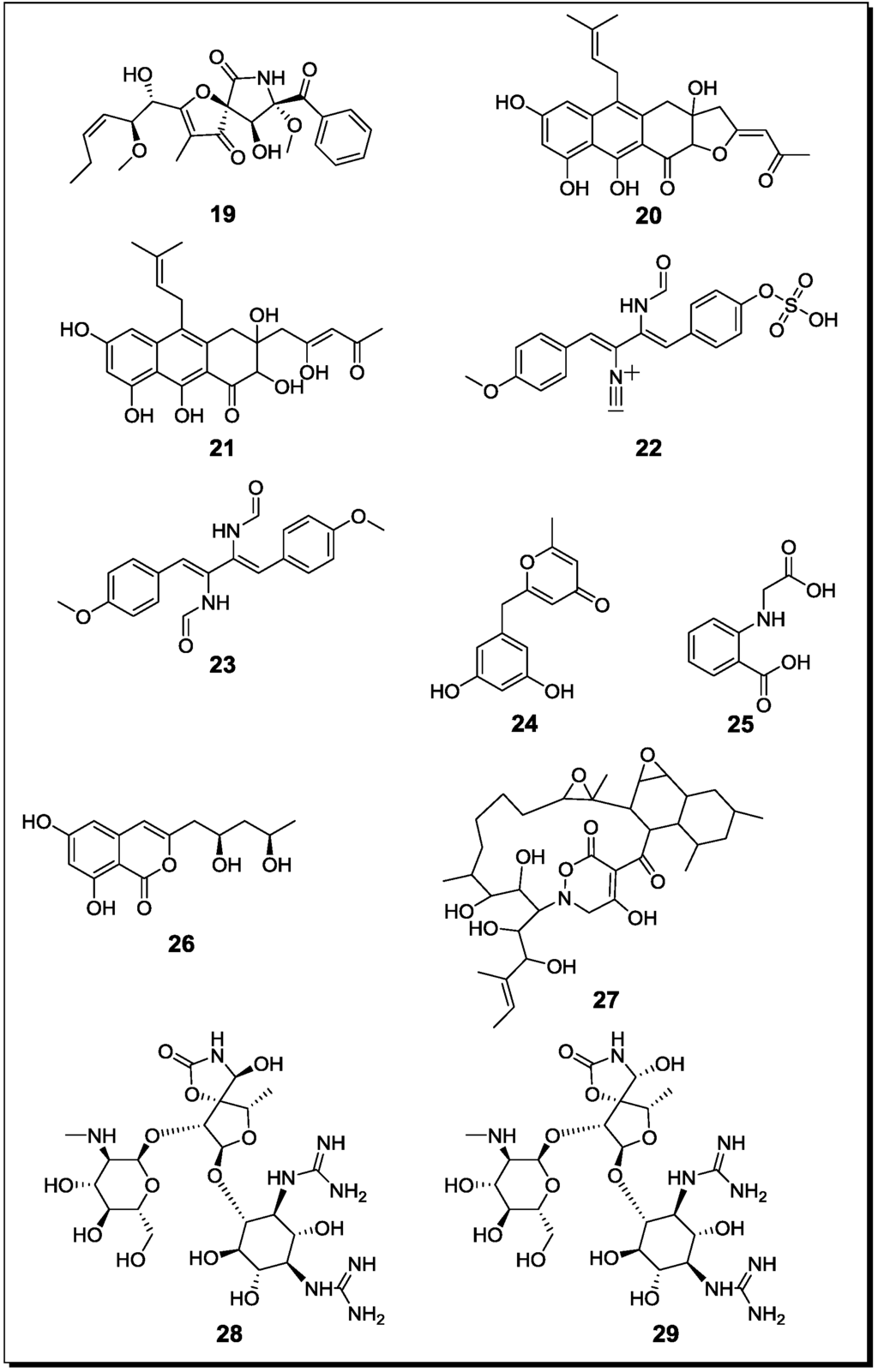
Figure 4.
New natural products reported from co-cultures of terrestrial fungi with bacteria or from bacterial co-cultures.
Co-cultivation of the soil-derived fungi Penicillium pinophilum and Trichoderma harzianum [61] resulted in an enhanced production of penicillide, MC-141, stromemycin and pestalasin A and in an induction of the new natural product secopenicillide C (13) (Figure 3). All respective compounds were produced by P. pinophilum [61].
Co-cultivation of the plant endophytic fungi Fusarium tricinctum and Fusarium begoniae yielded the new linear depsipetides subenniatin A (14) and B (15), together with the known cyclic depsipeptides enniatins A, A1, B and B1 that had been previously reported from F. tricinctum [62]. In contrast to the cyclic enniatins, the new compounds failed to show activity against the murine lymphoma cell line L5178Y or against the bacterial pathogens E. coli, S. aureus and P. aeruginosa. It may be speculated that compounds 14 and 15 are detoxification products of the cyclic enniatins that result from hydrolytic activity of F. begoniae. Interestingly, co-cultures of F. tricinctum and Fusarium equiseti failed to elicit the production of the new compounds 14 and 15 [62].
When the fungus Acremonium sp. that had been obtained as an endophyte from Taxus baccata was challenged with the mycoparasite Mycogone rosea, the novel lipoaminopeptides acremostatins A (16), B (17) and C (18) were obtained [63]. By mass spectrometry, the new metabolites were found to be structurally related to the known leucinostatins A, B and K, but differed from the latter by the presence of 2-methyldecanoic acid instead of (4S,2E)-4-methylhex-2-enoic acid as terminal substituent [63].
When pair-wise cultivating the wood-derived fungi Heterobasidion annosum, Gloeophyllum abietinum and Armillaria ostoyae, an induction of antibacterial compounds and mycotoxins was observed [64]. While co-culturing G. abietinum with H. annosum, the growth of G. abietinum was reduced, but the production of the antibiotics oospogylcol and oosponol derived from G. abietinum was increased (approximately 40 fold in liquid culture and 250 fold on agar plates). Simultaneously, H. annosum was found to react to the co-culture conditions by an approximately 430 fold increased production of the toxins fomajorin S and fomannosin. When A. ostoyae was cultivated with G. abietinum or with H. annosum, respectively, similar results with regard to an induction of secondary products were obtained. It was hypothesized, that small molecules are responsible for inducing the secondary metabolism in the investigated fungi. To prove this hypothesis cell free medium, heat-sterilized fungal cultures, or chemically and mechanically disrupted fungal cell walls, as well as either low or high molecular weight compounds from H. annosum or from G. abietinum were studied for their ability to induce the secondary metabolism of G. abietinum and of A. ostoyae, respectively. It was shown that low molecular molecules from H. annosum of less than 3000 Da are responsible for the observed increased production of secondary metabolites by G. abietinum. It was furthermore suggested that this induced accumulation of secondary products is due to a de novo biosynthesis of the respective molecules, since addition of 10 µg/mL of cycloheximide to the co-cultures stopped oospoglycol accumulation [64].
Mixed fermentation of five sediment-derived fungi, including Oyadendron sulphureoochraceum, Ascochyta pisi, Emercillopsis minima, Cylindrocarpon destructans and Fusarium oxysporum yielded lateritin as a major metabolite [65]. Lateritin showed cytotoxic effects on P388, PXPC-3, MCF-7, CNS SF268, NSC H460, KM20L2 and DU-145 tumor cells with IC50 values ranging from 1.7 to 2.0 µM. In addition, the compound showed antimicrobial activity against C. albicans, Micrococcus luteus, S. aureus, E. faecalis and against Streptococcus pneumoniae with MIC values between 2 and 16 µg/mL [65].
A systematic investigation of paclitaxel production by the endophytic fungus Paraconiothyrium sp., derived from the wood of Taxus x media, indicated that co-cultivation of Paraconiothyrium sp. with other endophytic fungi such as Alternaria sp. and Phomopsis sp. enhanced paclitaxel production between 2.7 and 3.8 fold compared to axenically grown Paraconiothyrium sp. [66]. When all three fungi were cultivated together paclitaxel production was even increased 7.8 fold, suggesting a cumulative effect.
A change of the morphology and pigmentation of the fungus Monascus sp. was observed upon co-cultivation with Saccharomyces cerevisae or with Aspergillus oryzae, while co-cultivation with Bacillus cereus did not cause such an effect [70]. It was shown that the culture broth and the ammonium sulphate precipitate of the culture broth of S. cerevisae elicited similar effects with regard to growth and pigment production by Monascus sp., while the heat sterilized culture of S. cerevisiae did not. When investigating the enzyme activities of the culture broth of S. cerevisae, chitinase and amylase activity were found. The purified chitinase from S. cerevisae was observed to cause the previously observed morphological changes of Monascus sp., while the related enzymes lysozyme, α-amylase, chitinase and protease purified from Bacillus sp., Streptomyces sp. and Staphylococcus sp. had no effect. The morphological change of Monascus sp. was linked to the hydrolytic effect of the chitinase, whereas pigment production was assumed to be a defensive reaction aimed at inhibiting chitinase activity [70].
With regard to fungal-bacterial interactions, co-cultures of Aspergillus fumigatus and different Streptomyces sp. were investigated in more detail. Addition of the soil derived Streptomyces bullii from the Atacama Desert to a culture of Aspergillus fumigatus afforded ten compounds, including the diketopiperazines brevianamide F, spirotryprostatin A, 6-methoxy spirotryprostatin B, fumitremorgin C, 12,13-dihydroxy fumitremorgin C, fumitremorgin B, verruculogen, and the pseurotins 11-O-methylpseurotin A and its new isomer 11-O-methylpseurotin A2 (19) (Figure 4), as well as ergosterol [67]. None of these compounds were detected in axenic cultures of A. fumigatus. Interestingly, when assayed for their antibiotic activity against S. aureus and E. coli, none of the induced fungal compounds was found to be active. When the fungus was treated either with the cell-free bacterial culture broth of S. bullii, a methanolic extract derived from the bacterial culture or an autoclaved bacterial culture, no induction of fungal metabolites was observed. Addition of N-butyryl-dl-homoserin, a known bacterial quorum sensing inhibitor, to the fungal culture afforded the fungal metabolites emestrin A and B. All metabolites were evaluated for their trypanocidal, leishmanicidal and cytotoxic activity (the latter using MRC5 cells). Fumitremorgin C, 12,13-dihydroxy fumitremorgin C, fumitremorgin B and verruculogen showed high activities in the used test systems with MIC values in the low to medium micromolar range [67].
König et al. [45] demonstrated the production of the new fungal derived polyketides fumicyclines A (20) and B (21) in a mixed fermentation of A. fumigatus and Streptomyces rapamycinicus. Both compounds showed a weak antibiotic effect in the agar plate diffusion assay against S. rapamycinicus. When incubating the fungus with the cell free bacterial culture broth or when the fungus was co-cultivated with the bacterium kept in a dialysis tube, no induction of fumicyclines was observed, indicating that a direct physical contact of fungus and bacterium is necessary for elicitation of fungal metabolites. Electron microscopy of co-cultured fungus and bacterium revealed an intimate physical contact between the fungal mycelia and the bacterial filaments. It was concluded that S. rapamycinicus is able to alter gene expression in A. fumigatus by modulating regulatory processes. Treatment of the fungus with the known histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA) caused an increased transcription level of the PKS gene that is involved in the biosynthesis of fumicyclines, while no PKS gene transcription was observed following treatment with the histone acetyltransferase inhibitor anacardic acid [45].
A further class of fungal compounds, xanthocillin analogues, was produced by A. fumigatus in the presence of Streptomyces peucetius [68]. The induced compounds included the new derivatives fumiformamide (22) and N,N′-((1Z,3Z)-1,4-bis(4-methoxyphenyl)buta-1,3-diene-2,3diyl)di-formamide (23), together with the known N-formyl derivatives and BU-4704. Compound 23 exhibited strong cytotoxic effects in the NCI-60 cell line screen with MIC values between 0.65 and 1.12 µM [68].
A co-culture of the fungal endophyte F. tricinctum and B. subtilis, resulted in an up to 78 fold enhanced production of the constitutively present fungal secondary metabolites lateropyrone, fusaristatin and enniatins A1, B and B1 compared to an axenic culture of F. tricinctum [30]. In addition, four compounds that included (+)-citreoisocoumarinol, the new macrocarpon C (24), 2-(carboxymethylamino)benzoic acid (25) and (−)-citreoisocoumarinol (26) were detected, which were absent in axenic cultures of both, the fungus and the bacterium. Interestingly, this induction of fungal metabolism was only found when B. subtilis was inoculated several days prior to the fungus to the culture medium. When inoculated simultaneously, the fungus inhibited the bacterium and no clear induction of fungal compounds was detected. When studied for their antibacterial properties, enniatins B1 and A1 showed MIC values against B. subtilis of 8 and 16 µg/mL, respectively, whereas the MIC values against the human pathogens S. aureus, S. pneumoniae and E. faecalis were between 2 and 8 µg/mL. Additionally, the constitutively present fungal metabolite lateropyrone exhibited MIC values between 2 and 8 µg/mL against all mentioned bacterial strains [30]. Interestingly, when co-cultured with two streptomycetes (S. coelicolor and S. lividans), F. tricinctum was shown to produce further, so far unknown products that differ from those detected during co-cultivation of the fungus and B. subtilis. This suggests specific reactions of the fungus in response to different bacteria.
When the yeast C. albicans was co-cultured with the bacterium P. aeruginosa, the formation of a pyocyanin related red pigment that was located intracellularly within C. albicans was observed [71]. It was suggested that a possible intermediate, 5-methyl-phenazinium-1-carboxylat (5MPCA) produced by P. aeruginosa, was transformed to the red pigment within the yeast. This hypothesis was corroborated by a feeding experiment with 5MPCA, which was shown to be transformed to the red pigment by the yeast. Co-cultivation of C. albicans with related bacterial taxa such as Pseudomonas sp., P. putida, P. fluorescens or P. chlororaphis failed to elicit the production of the red pigment, suggesting that pigment formation is a specific response of C. albicans to P. aeruginosa. Pigment formation was observed to coincide with a reduced fungal viability compared to controls [71].
Mycolic acid producing bacteria were shown to be able to elicit the accumulation of cryptic natural products in other bacteria. A mixed fermentation of S. lividans with the mycolic acid producing bacterium Tsukamurella pulmonis induced the production of a red pigment in S. lividans [69]. For pigment production, a direct cell-cell contact between S. lividans and T. pulmonis was found to be necessary. In the same study, Streptomyces endus was found to produce the new antibiotic alchivemycin A (27) when co-cultured with T. pulmonis. Alchivemycin A revealed a MIC of 0.06 µg/mL against Micrococcus luteus that was used as an indicator strain to assess antibiotic activity [69].
When investigating bacterial-bacterial interactions, the survival of Salmonella duesseldorf in soil in the presence of Streptomyces lividans or Streptomyces bikiniensis was investigated [72]. While S. bikiniensis is a known streptomycin producer, S. lividans is a non-antibiotic producer. S. duesseldorf was shown to be sensitive towards streptomycin, resulting in a reduced survival of S. duesseldorf in the presence of S. bikiniensis in non-sterile and sterile amended soil. Competition in sterile amended soil indicated a greater survival of S. duesseldorf in the presence of S. lividans compared to axenically grown S. duesseldorf. It was hypothesized that the presence of vegetative mycelia of S. lividans has a positive effect on the accessibility of nutrients by S. duesseldorf. The study provided evidence for antibiotic production in soil with regard to accumulation of streptomycin by S. bikiniensis [72].
With the purpose of investigating the role of antibiotics [31], two Streptomyces spp. were co-cultured with B. subtilis under semi-ecological conditions. The Streptomyces spp. used for this study included S. griseus and S. coelicolor. Three different experimental set ups were investigated: B. subtilis was first established in a growth medium which was then consecutively inoculated with one of the two Streptomyces spp. Alternatively, B. subtilis and a chosen Streptomyces strain were inoculated simultaneously, or as a third alternative Streptomyces sp. was inoculated prior to addition of B. subtilis. Antibiotic production by S. griseus and by S. coelicolor was found to prevent invasion by B. subtilis, whereas no advantage for the streptomycetes was seen when invading an already existing population of B. subtilis [31].
2.3. Molecular Biology and Accumulation of Cryptic Natural Products in Mixed Fermentations of the Model Organism Aspergillus Nidulans and Streptomyces Rapamycinicus
Since the genome of Aspergillus nidulans is known from sequencing, this fungal species was chosen by the groups of Hertweck and Brackhage for detailed molecular studies related to the induced accumulation of natural products in mixed fermentations with soil-derived actinomycetes [46,47]. Bioinformatic genome analysis of A. nidulans revealed 28 putative polyketide and 24 putative nonribosomal peptide biosynthetic gene loci, giving the fungus theoretically the possibility to produce at least 52 different secondary metabolites [47]. Analysis of the secondary metabolites hitherto reported from A. nidulans when grown under standard laboratory conditions, however, indicated that only a small part of the biosynthetic genes are expressed. Using the A. nidulans secondary metabolism array (ASMA) which monitors the central enzymes of each biosynthetic pathway, the fungus was co-cultured with 58 different actinomycetes [46]. Fungal mRNA and cDNA were isolated during co-cultivation and hybridized with the ASMA. Interestingly, only one actinomycete strain, Streptomyces rapamycinicus, led to an up-regulation of two putative fungal PKS and NRPS genes, AN7909 and AN7884. Using full-genome microarrays, 395 genes were found to be differentially expressed in A. nidulans when co-cultured with S. rapamycinicus, including 248 genes that were up-regulated, while 147 genes were down-regulated in the region between AN7874 and AN7914. In order to determine the gene regulating mechanism present in S. rapamycinicus, A. nidulans was monitored by qRT-PCR, while being incubated either with the cell-free bacterial broth, with (co)culture-derived extracts, with heat killed bacteria or with living bacteria that were kept in a dialysis tube and were thus physically separated from the fungus. None of these experiments led to the activation of fungal gene clusters, indicating the necessity of an intimate contact between fungus and streptomycete for fungal gene modulation. Electron microscopy showed a close physical interaction of both microorganisms. HPLC analysis of axenic fungal and bacterial cultures and of mixed-fermentations of A. nidulans and S. rapamycinicus demonstrated striking differences. Mixed fermentations of A. nidulans and S. rapamycinicus yielded four cryptic fungal compounds that included orsellinic acid, lecanoric acid, F-9775 and F-9775B. None of these metabolites were detected either in axenic cultures of the fungus or in PKS lacking mutants of A. nidulans [46].
In order to investigate the mechanism of gene regulation in A. nidulans in more detail, the fungus was treated with the histone acetyltransferase inhibitor anacardic acid or with the histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA) [47]. The histone deacetylase inhibitor SAHA caused an activation of fungal gene expression, as observed during co-cultivation of A. nidulans with S. rapamycinicus, indicating that also the streptomycete triggers modification of fungal histones. Treatment with anacardic acid inhibited fungal biosynthetic gene expression. Upon deleting 36 out of 40 fungal histone acetyltransferases, it was demonstrated that the fungal Saga/Ada complex that contains the histone acetyltransferase GcnE and the AdaB protein is required for induction of the fungal orsellinic acid gene cluster by S. rapamycinicus. A Saga/Ada dependent increase of histone 3 acetylation at the lysine residue 9 was found to take place during co-cultivation of the fungus and the streptomycete proving for the first time that fungal histone acetylation is caused by interaction with the streptomycete, and that this increased histone acetylation in turn leads to an accumulation of cryptic fungal natural products [47].
3. Discussion and Outlook
The studies reported in this review even though still limited in number clearly underline the power of co-cultivation as an emerging tool to increase the chemical diversity of secondary products that are produced by fungi or bacteria during in vitro fermentation. Co-cultivation is expected in the future to routinely complement other experimental approaches that likewise aim at diversifying secondary compound production by microorganisms such as mutagenesis [23,47], the OSMAC approach [23] or treatment of microbes with epigenetic modifiers [24] to name just a few. In contrast to the latter methods, co-cultivation is an ecologically driven approach that tries to mimic the natural situation where a given microbe is always embedded in a more or less complex microbial community and exposed to a multitude of chemical signals that are exchanged between the different taxa that compose this community. This chemical interaction will be even more complex if one considers that many marine-derived microbes are not free living in seawater or in the sediment, but are members of a microbial community that are e.g., hosted by marine invertebrates [73,74,75,76,77] or within algae [78,79,80,81] or even mangroves [82,83,84,85]. Compounds produced by the hosts are likely to have an influence on this microbial community too and may even act as stimulating clues that induce the production of certain microbial compounds that may not be accumulated in the absence of these clues.
Even when considering only the microbial inter-species crosstalk as discussed in this review, the scenario is complex and far from being understood. This is especially true for the mostly unknown nature of chemical triggers or signals that lead to an accumulation of cryptic natural products that are not detected in axenic microbial cultures and/or to a significantly enhanced production of constitutively present compounds. Several reports that have been highlighted in this review indicate specific reactions of fungi or bacteria during co-cultivation experiments that involve more than one microbial partner. For example, co-cultivation of two Bacillus strains that resembled B. thuringiensis and B. megaterium caused an increased production of indole and (Phe-Pro) diketopiperazines by the producing strain (“B. thuringiensis”), whereas co-cultivation of the latter with another marine-derived bacterial strain had no influence on diketopiperazine accumulation [59]. Interestingly, the induction of diketopiperazine production coincides with the susceptibility of inducing vs. non-inducing bacterial strains towards these secondary compounds [59]. Two further examples come from the terrestrial habitat and involve the cross talk between a fungus and different bacteria. Out of 58 different actinomycetes that were co-cultured with the fungus A. nidulans, only S. rapamycinicus was able to induce the accumulation of orsellinic acid and of biogenetically related compounds in the fungus [46]. When the fungal endophyte F. tricinctum was co-cultured with B. subtilis, the induced fungal natural products differed from those that were detected during co-cultivation of the fungus with either Streptomyces coelicolor or with S. lividans [30]. These different metabolic reactions of either bacteria or fungi during co-cultivation with a small set of other microbes argue for the presence of different, perhaps even specific clues or signal molecules that can apparently be perceived by the producing microbial strain. Whereas it is relatively easy to observe the metabolic reactions of fungi or bacteria when exposed to these different clues, it is far more difficult and challenging to identify the signals that are responsible for inducing the accumulation of cryptic natural products. Several studies have tried to locate these signals e.g., in the cell free culture broth or in the cellular biomass of inducing microbial strains. For example, it has been shown that an induced production of libertellenones by the marine-derived fungus Libertella sp. depends on a close physical contact between the fungus and the inducing bacterium CNJ-328 [53]. On the other side, the study by Sonnenbichler et al. [64] demonstrated that the induced accumulation of the antibiotics oospogylcol and oosponol by the fungus G. abietinum was triggered by low molecular weight compounds of less than 3000 Da that are secreted into the culture broth by the inducing fungus H. annosum. On the contrary, accumulation of fumicyclines A and B by A. nidulans as induced by S. rapamycinicus was found to require a direct physical contact between the fungus and the streptomycete [45].
The induction of cryptic natural products or the significant enhancement of constitutively present secondary compounds is usually considered to be a stress response caused by the presence of a competing microorganism. Hence, induction of metabolites as a result of stress during co-cultivation should be part of the chemical defense of the producer and aimed at inhibiting or even killing the competitor. Rather surprisingly, this hypothesis has so far rarely been proven in co-cultivation studies. One notable example is the co-cultivation between the fungal endophyte F. tricinctum and B. subtilis. The fungal derived enniatin derivatives that were greatly enhanced during co-cultivation showed a clear antibiotic activity that extended not only to clinically relevant bacterial pathogens such as MRSA but also to B. subtilis that had been used as an inducer in the respective co-cultivation study [30]. Other studies failed to demonstrate such a protective effect. For example, in spite of being induced by the marine-derived bacterium CNJ-328, the libertellenones that were produced by the marine-derived fungus Libertella sp. showed no antibiotic activity against the inducing bacterium [53].
The cross talk between inducer and producer during co-cultivation of microorganisms is a highly dynamic process that is also influenced by time. The enhanced production of istamycins by the marine-derived S. tenjimariensis during co-cultivation with several unknown likewise marine-derived bacteria was only detected when the istamycin producer was inoculated 24 h prior to the inducers into the culture broth. No enhancement of istamycin production was found when producer and inducer were inoculated at the same time [34]. The demonstration of the time dependent istamycin production [34] gives further evidence for the rat race between the microorganisms within a microbial community. Time was also shown to be an important factor during co-cultivation of the fungal endophyte F. tricinctum and B. subtilis. When inoculated at the same time, the fungus clearly out competed the bacterium and no clear changes in the fungal metabolite profile were visible. When the bacterium was given a head start during cultivation, the growth of the fungus was slowed down and a clear induction of fungal metabolites occurred [30].
One further aspect which is horizontal gene transfer and which may perhaps also influence the result of co-cultivation studies should finally not be overlooked. During co-cultivation of a multi-antibiotic resistant strain of the bacterium Rhodococcus fascians that by itself does not produce antibiotics and a strain of Streptomyces padanus that is an actinomycin producer a strain of Rhodococcus emerged that was found to contain large segments of Streptomyces DNA and produced the new aminoglycosides rhodostreptomycins A (28) and B (29) [86]. These compounds differ considerably from the actinomycins that are produced by S. padanus and have been interpreted to arise from horizontal gene transfer of the streptomycete to Rhodococcus during co-cultivation.
For the fungus A. nidulans, for which the genome is known through sequencing and which can therefore be considered to be a model organism, the molecular basis underlying the induction of orsellinic acid derived secondary metabolites and of other compounds by S. rapamycinicus could be demonstrated for the first time [46]. The interaction of fungus and streptomycete leads to an increased histone acetylation within gene clusters encoding for fungal natural products. This in turn leads to an induced accumulation of cryptic compounds that are not detected when the fungus is grown under axenic conditions. Interestingly an induced accumulation of fungal products was also observed when the fungus was treated with the HDAC inhibitor SAHA [47] that is also used as a tool in epigenetic studies that aim likewise at an induction of cryptic biogenetic pathways [24] thereby providing a link between co-cultivation and epigenetic modulation as two emerging tools that aim jointly at enhancing the chemical diversity of microorganisms.
It is clear that co-cultivation as an experimental tool, which aims at a diversified production of bioactive compounds that could be leads for biomedical research, is promising but nevertheless still in its infancy. Far more questions rather than answers or established protocols exist. Generalizations are dangerous, each case needs to be considered individually before any wider conclusions can be drawn. Nevertheless, taking this ecologically driven approach at enhancing the chemical diversity of microbes beyond the boundaries that can be reached by routine axenic cultivation is both thrilling and promising. It is hoped that far more researchers will adopt this approach for their own work in the future.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Li, J.W.-H.; Vederas, J.C. Drug discovery and natural products: End of an era or an endless frontier? Science 2009, 325, 161–165. [Google Scholar] [CrossRef]
- Enomoto, T.; Yasui, Y.; Takemoto, Y. Synthetic study toward ecteinascidin 743: Concise construction of the diazabicyclo [3.3.1] nonane skeleton and assembly of the pentacyclic core. J. Org. Chem. 2010, 75, 4876–4879. [Google Scholar] [CrossRef]
- Younes, A.; Yasothan, U.; Kirkpatrick, P. Brentuximab vedotin. Nat. Rev. Drug Discov. 2012, 11, 19–20. [Google Scholar] [CrossRef]
- Reichert, J.M. Marketed Therapeutic Antibodies Compendium; MAbs-Landes Bioscience: Austin, TX, USA, 2012; pp. 413–415. [Google Scholar]
- European Medicines Agency: Adcetris. Available online: http://www.ema.europa.eu/ema/index.jsp?curl=pages/medicines/human/medicines/002455/human_med_001588.jsp&mid=WC0b01ac058001d124 (accessed on 3 January 2014).
- Huyck, T.K.; Gradishar, W.; Manuguid, F.; Kirkpatrick, P. Eribulin mesylate. Nat. Rev. Drug Discovery 2011, 10, 173–174. [Google Scholar] [CrossRef]
- Mayer, A.; Glaser, K.B.; Cuevas, C.; Jacobs, R.S.; Kem, W.; Little, R.D.; McIntosh, J.M.; Newman, D.J.; Potts, B.C.; Shuster, D.E. The odyssey of marine pharmaceuticals: A current pipeline perspective. Trends Pharmacol. Sci. 2010, 31, 255–265. [Google Scholar] [CrossRef]
- Glaser, K.B.; Mayer, A. A renaissance in marine pharmacology: From preclinical curiosity to clinical reality. Biochem. Pharmacol. 2009, 78, 440–448. [Google Scholar] [CrossRef]
- Schmidtko, A.; Lötsch, J.; Freynhagen, R.; Geisslinger, G. Ziconotide for treatment of severe chronic pain. Lancet 2010, 375, 1569–1577. [Google Scholar] [CrossRef]
- Sagar, S.; Kaur, M.; Minneman, K.P. Antiviral lead compounds from marine sponges. Mar. Drugs 2010, 8, 2619–2638. [Google Scholar] [CrossRef]
- Yasuhara-Bell, J.; Lu, Y. Marine compounds and their antiviral activities. Antivir. Res. 2010, 86, 231–240. [Google Scholar] [CrossRef]
- Serova, M.; de Gramont, A.; Bieche, I.; Riveiro, M.E.; Galmarini, C.M.; Aracil, M.; Jimeno, J.; Faivre, S.; Raymond, E. Predictive factors of sensitivity to elisidepsin, a novel kahalalide F-derived marine compound. Mar. Drugs 2013, 11, 944–959. [Google Scholar] [CrossRef]
- Barboza, N.M.; Medina, D.J.; Budak-Alpdogan, T.; Aracil, M.; Jimeno, J.M.; Bertino, J.R.; Banerjee, D. Plitidepsin (Aplidin) is a potent inhibitor of diffuse large cell and Burkitt lymphoma and is synergistic with rituximab. Cancer Biol. Ther. 2012, 13, 114–122. [Google Scholar] [CrossRef]
- Molina-Guijarro, J.M.; Macías, Á.; García, C.; Muñoz, E.; García-Fernández, L.F.; David, M.; Núñez, L.; Martínez-Leal, J.F.; Moneo, V.; Cuevas, C. Irvalec inserts into the plasma membrane causing rapid loss of integrity and necrotic cell death in tumor cells. PLoS One 2011, 6, e19042. [Google Scholar] [CrossRef]
- Marine pharmaceuticals: The clinical pipeline. Available online: http://marinepharmacology.midwestern.edu/clinPipeline.htm (accessed on 3 January 2014).
- Feling, R.H.; Buchanan, G.O.; Mincer, T.J.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Salinosporamide A: A highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew. Chem. Int. Ed. 2003, 42, 355–357. [Google Scholar] [CrossRef]
- Fenical, W.; Jensen, P.R.; Palladino, M.A.; Lam, K.S.; Lloyd, G.K.; Potts, B.C. Discovery and development of the anticancer agent salinosporamide A (NPI-0052). Bioorg. Med. Chem. 2009, 17, 2175–2180. [Google Scholar] [CrossRef]
- Chauhan, D.; Catley, L.; Li, G.; Podar, K.; Hideshima, T.; Velankar, M.; Mitsiades, C.; Mitsiades, N.; Yasui, H.; Letai, A. A novel orally active proteasome inhibitor induces apoptosis in multiple myeloma cells with mechanisms distinct from Bortezomib. Cancer Cell 2005, 8, 407–419. [Google Scholar] [CrossRef]
- Groll, M.; Huber, R.; Potts, B.C.M. Crystal structures of salinosporamide A (NPI-0052) and B (NPI-0047) in complex with the 20S proteasome reveal important consequences of β-lactone ring opening and a mechanism for irreversible binding. J. Am. Chem. Soc. 2006, 128, 5136–5141. [Google Scholar] [CrossRef]
- Knight, V.; Sanglier, J.J.; DiTullio, D.; Braccili, S.; Bonner, P.; Waters, J.; Hughes, D.; Zhang, L. Diversifying microbial natural products for drug discovery. Appl. Microbiol. Biotechnol. 2003, 62, 446–458. [Google Scholar] [CrossRef]
- Brakhage, A.A.; Schuemann, J.; Bergmann, S.; Scherlach, K.; Schroeckh, V.; Hertweck, C. Activation of Fungal Silent Gene Clusters: A New Avenue to Drug Discovery. In Natural Compounds as Drugs; Springer: Berlin/Heidelberg, Germany, 2008; pp. 1–12. [Google Scholar]
- Brakhage, A.A.; Schroeckh, V. Fungal secondary metabolites–strategies to activate silent gene clusters. Fungal Genet. Biol. 2011, 48, 15–22. [Google Scholar] [CrossRef]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Cichewicz, R.H. Epigenome manipulation as a pathway to new natural product scaffolds and their congeners. Nat. Prod. Rep. 2010, 27, 11–22. [Google Scholar] [CrossRef]
- Shwab, E.K.; Bok, J.W.; Tribus, M.; Galehr, J.; Graessle, S.; Keller, N.P. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryotic Cell 2007, 6, 1656–1664. [Google Scholar] [CrossRef]
- Henrikson, J.C.; Hoover, A.R.; Joyner, P.M.; Cichewicz, R.H. A chemical epigenetics approach for engineering the in situ biosynthesis of a cryptic natural product from Aspergillus niger. Org. Biomol. Chem. 2009, 7, 435–438. [Google Scholar] [CrossRef]
- Fisch, K.M.; Gillaspy, A.F.; Gipson, M.; Henrikson, J.C.; Hoover, A.R.; Jackson, L.; Najar, F.Z.; Wägele, H.; Cichewicz, R.H. Chemical induction of silent biosynthetic pathway transcription in Aspergillus niger. J. Ind. Microbiol. Biotechnol. 2009, 36, 1199–1213. [Google Scholar] [CrossRef]
- Asai, T.; Chung, Y.-M.; Sakurai, H.; Ozeki, T.; Chang, F.-R.; Yamashita, K.; Oshima, Y. Tenuipyrone, a novel skeletal polyketide from the entomopathogenic fungus, Isaria tenuipes, cultivated in the presence of epigenetic modifiers. Org. Lett. 2011, 14, 513–515. [Google Scholar]
- Vervoort, H.C.; Drašković, M.; Crews, P. Histone deacetylase inhibitors as a tool to up-regulate new fungal biosynthetic products: Isolation of EGM-556, a cyclodepsipeptide, from Microascus sp. Org. Lett. 2010, 13, 410–413. [Google Scholar]
- Ola, A.R.B.; Thomy, D.; Lai, D.; Brötz-Oesterhelt, H.; Proksch, P. Inducing secondary metabolite production by the endophytic fungus Fusarium tricinctum through coculture with Bacillus subtilis. J. Nat. Prod. 2013, 76, 2094–2099. [Google Scholar] [CrossRef]
- Wiener, P. Experimental studies on the ecological role of antibiotic production in bacteria. Evol. Ecol. 1996, 10, 405–421. [Google Scholar] [CrossRef]
- Anke, T. The antifungal strobilurins and their possible ecological role. Can. J. Bot. 1995, 73, 940–945. [Google Scholar] [CrossRef]
- Williams, S.T.; Vickers, J.C. The ecology of antibiotic production. Microb. Ecol. 1986, 12, 43–52. [Google Scholar] [CrossRef]
- Slattery, M.; Rajbhandari, I.; Wesson, K. Competition-mediated antibiotic induction in the marine bacterium Streptomyces tenjimariensis. Microb. Ecol. 2001, 41, 90–96. [Google Scholar]
- De Lorenzo, V.; Aguilar, A. Antibiotics from Gram-negative bacteria: Do they play a role in microbial ecology? Trends Biochem. Sci. 1984, 9, 266–269. [Google Scholar] [CrossRef]
- Linares, J.F.; Gustafsson, I.; Baquero, F.; Martinez, J.L. Antibiotics as intermicrobial signaling agents instead of weapons. Sci. Signal. 2006, 103, 19484. [Google Scholar]
- Fajardo, A.; Martínez, J.L. Antibiotics as signals that trigger specific bacterial responses. Curr. Opin. Microbiol. 2008, 11, 161–167. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, W.; Zhu, Y.; Gong, Q.; Yu, W.; Lu, X. Antibiotics at subinhibitory concentrations improve the quorum sensing behavior of Chromobacterium violaceum. FEMS Microbiol. Lett. 2013, 341, 37–44. [Google Scholar] [CrossRef]
- Shang, Z.; Li, X.-M.; Li, C.-S.; Wang, B.-G. Diverse secondary metabolites produced by marine-derived fungus Nigrospora sp. MA75 on various culture media. Chem. Biodivers. 2012, 9, 1338–1348. [Google Scholar] [CrossRef]
- Ebrahim, W.; Aly, A.H.; Mándi, A.; Totzke, F.; Kubbutat, M.H.G.; Wray, V.; Lin, W.H.; Dai, H.; Proksch, P.; Kurtán, T.; et al. Decalactone derivatives from Corynespora cassiicola, an endophytic fungus of the mangrove plant Laguncularia racemosa. Eur. J. Org. Chem. 2012, 2012, 3476–3484. [Google Scholar] [CrossRef]
- Xu, J.; Aly, A.H.; Wray, V.; Proksch, P. Polyketide derivatives of endophytic fungus Pestalotiopsis sp. isolated from the Chinese mangrove plant Rhizophora mucronata. Tetrahedron Lett. 2011, 52, 21–25. [Google Scholar] [CrossRef]
- Spatafora, J.W.; Volkmann-Kohlmeyer, B.; Kohlmeyer, J. Independent terrestrial origins of the Halosphaeriales (marine Ascomycota). Am. J. Bot. 1998, 85, 1569–1580. [Google Scholar] [CrossRef]
- Raghukumar, C. Marine fungal biotechnology: An ecological perspective. Fungal Divers. 2008, 31, 19–35. [Google Scholar]
- Kohlmeyer, J.; Kohlmeyer, E. Marine Mycology. The Higher Fungi; Academic Press, Inc.: Waltham, MA, USA, 1979. [Google Scholar]
- König, C.C.; Scherlach, K.; Schroeckh, V.; Horn, F.; Nietzsche, S.; Brakhage, A.A.; Hertweck, C. Bacterium induces cryptic meroterpenoid pathway in the pathogenic fungus Aspergillus fumigatus. ChemBioChem 2013, 14, 938–942. [Google Scholar] [CrossRef]
- Schroeckh, V.; Scherlach, K.; Nützmann, H.-W.; Shelest, E.; Schmidt-Heck, W.; Schuemann, J.; Martin, K.; Hertweck, C.; Brakhage, A.A. Intimate bacterial–fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 2009, 106, 14558–14563. [Google Scholar] [CrossRef]
- Nützmann, H.-W.; Reyes-Dominguez, Y.; Scherlach, K.; Schroeckh, V.; Horn, F.; Gacek, A.; Schümann, J.; Hertweck, C.; Strauss, J.; Brakhage, A.A. Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation. Proc. Natl. Acad. Sci. USA 2011, 108, 14282–14287. [Google Scholar] [CrossRef]
- Zhu, F.; Chen, G.; Chen, X.; Huang, M.; Wan, X. Aspergicin, a new antibacterial alkaloid produced by mixed fermentation of two marine-derived mangrove epiphytic fungi. Chem. Nat. Compd. 2011, 47, 767–769. [Google Scholar] [CrossRef]
- Li, C.; Zhang, J.; Shao, C.; Ding, W.; She, Z.; Lin, Y. A new xanthone derivative from the co-culture broth of two marine fungi (strain No. E33 and K38). Chem. Nat. Compd. 2011, 47, 382–384. [Google Scholar] [CrossRef]
- Zhu, F.; Chen, G.; Wu, J.; Pan, J. Structure revision and cytotoxic activity of marinamide and its methyl ester, novel alkaloids produced by co-cultures of two marine-derived mangrove endophytic fungi. Nat. Prod. Res. 2013, 27, 1960–1964. [Google Scholar] [CrossRef]
- Miao, L.I.; Kwong, T.F.N.; Qian, P.-Y. Effect of culture conditions on mycelial growth, antibacterial activity, and metabolite profiles of the marine-derived fungus Arthrinium c.f. saccharicola. Appl. Microbiol. Biotechnol. 2006, 72, 1063–1073. [Google Scholar] [CrossRef]
- Cueto, M.; Jensen, P.R.; Kauffman, C.; Fenical, W.; Lobkovsky, E.; Clardy, J. Pestalone, a new antibiotic produced by a marine fungus in response to bacterial challenge. J. Nat. Prod. 2001, 64, 1444–1446. [Google Scholar] [CrossRef]
- Oh, D.-C.; Jensen, P.R.; Kauffman, C.A.; Fenical, W. Libertellenones A–D: Induction of cytotoxic diterpenoid biosynthesis by marine microbial competition. Bioorg. Med. Chem. 2005, 13, 5267–5273. [Google Scholar] [CrossRef]
- Oh, D.-C.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J. Nat. Prod. 2007, 70, 515–520. [Google Scholar] [CrossRef]
- Park, H.B.; Kwon, H.C.; Lee, C.-H.; Yang, H.O. Glionitrin A, an antibiotic-antitumor metabolite derived from competitive interaction between abandoned mine microbes. J. Nat. Prod. 2009, 72, 248–252. [Google Scholar] [CrossRef]
- Dusane, D.H.; Matkar, P.; Venugopalan, V.P.; Kumar, A.R.; Zinjarde, S.S. Cross-species induction of antimicrobial compounds, biosurfactants and quorum-sensing inhibitors in tropical marine epibiotic bacteria by pathogens and biofouling microorganisms. Curr. Microbiol. 2011, 62, 974–980. [Google Scholar] [CrossRef]
- Burgess, J.G.; Jordan, E.M.; Bregu, M.; Mearns-Spragg, A.; Boyd, K.G. Microbial antagonism: A neglected avenue of natural products research. Prog. Ind. Microbiol. 1999, 35, 27–32. [Google Scholar] [CrossRef]
- Mearns-Spragg, A.; Bregu, M.; Boyd, K.G.; Burgess, J.G. Cross-species induction and enhancement of antimicrobial activity produced by epibiotic bacteria from marine algae and invertebrates, after exposure to terrestrial bacteria. Lett. Appl. Microbiol. 1998, 27, 142–146. [Google Scholar]
- Trischman, J.A.; Oeffner, R.E.; de Luna, M.G.; Kazaoka, M. Competitive induction and enhancement of indole and a diketopiperazine in marine bacteria. Mar. Biotechnol. 2004, 6, 215–220. [Google Scholar]
- Zhu, F.; Lin, Y. Marinamide, a novel alkaloid and its methyl ester produced by the application of mixed fermentation technique to two mangrove endophytic fungi from the South China Sea. Chin. Sci. Bull. 2006, 51, 1426–1430. [Google Scholar] [CrossRef]
- Nonaka, K.; Abe, T.; Iwatsuki, M.; Mori, M.; Yamamoto, T.; Shiomi, K.; Ômura, S.; Masuma, R. Enhancement of metabolites productivity of Penicillium pinophilum FKI-5653, by co-culture with Trichoderma harzianum FKI-5655. J. Antibiot. 2011, 64, 769–774. [Google Scholar] [CrossRef]
- Wang, J.-P.; Lin, W.; Wray, V.; Lai, D.; Proksch, P. Induced production of depsipeptides by co-culturing Fusarium tricinctum and Fusarium begoniae. Tetrahedron Lett. 2013, 54, 2492–2496. [Google Scholar] [CrossRef]
- Degenkolb, T.; Heinze, S.; Schlegel, B.; Strobel, G.; Gräfe, U. Formation of new lipoaminopeptides, acremostatins A, B, and C, by co-cultivation of Acremonium sp. Tbp-5 and Mycogone rosea DSM 12973. Biosci. Biotechnol. Biochem. 2002, 66, 883–886. [Google Scholar] [CrossRef]
- Sonnenbichler, J.; Dietrich, J.; Peipp, H. Secondary fungal metabolites and their biological activities, V. Investigations concerning the induction of the biosynthesis of toxic secondary metabolites in basidiomycetes. Bio. Chem. Hoppe-Seyler 1994, 375, 71–80. [Google Scholar] [CrossRef]
- Pettit, R.K.; Pettit, G.R.; Xu, J.P.; Weber, C.A.; Richert, L.A. Isolation of human cancer cell growth inhibitory, antimicrobial lateritin from a mixed fungal culture. Planta Med. 2010, 76, 500–501. [Google Scholar] [CrossRef]
- Soliman, S.S.M.; Raizada, M.N. Interactions between co-habitating fungi elicit synthesis of taxol from an endophytic fungus in host Taxus plants. Front. Microbiol. 2013, 4, 1–14. [Google Scholar]
- Rateb, M.E.; Hallyburton, I.; Houssen, W.; Bull, A.; Goodfellow, M.; Santhanam, R.; Jaspars, M.; Ebel, R. Induction of diverse secondary metabolites in Aspergillus fumigatus by microbial co-culture. RSC Adv. 2013, 3, 14444–14450. [Google Scholar] [CrossRef]
- Zuck, K.M.; Shipley, S.; Newman, D.J. Induced production of N-formyl alkaloids from Aspergillus fumigatus by co-culture with Streptomyces peucetius. J. Nat. Prod. 2011, 74, 1653–1657. [Google Scholar] [CrossRef]
- Onaka, H.; Mori, Y.; Igarashi, Y.; Furumai, T. Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl. Environ. Microbiol. 2011, 77, 400–406. [Google Scholar] [CrossRef]
- Shin, C.S.; Kim, H.J.; Kim, M.J.; Ju, J.Y. Morphological change and enhanced pigment production of Monascus when cocultured with Saccharomyces cerevisiae or Aspergillus oryzae. Biotechnol. Bioeng. 1998, 59, 576–581. [Google Scholar] [CrossRef]
- Gibson, J.; Sood, A.; Hogan, D.A. Pseudomonas aeruginosa-Candida albicans interactions: Localization and fungal toxicity of a phenazine derivative. Appl. Environ. Microbiol. 2009, 75, 504–513. [Google Scholar] [CrossRef]
- Turpin, P.E.; Dhir, V.K.; Maycroft, K.A.; Rowlands, C.; Wellington, E.M.H. The effect of Streptomyces species on the survival of Salmonella in soil. FEMS Microbiol. Lett. 1992, 101, 271–280. [Google Scholar]
- Zhou, Y.; Debbab, A.; Mándi, A.; Wray, V.; Schulz, B.; Müller, W.E.G.; Kassack, M.; Lin, W.; Kurtán, T.; Proksch, P. Alkaloids from the sponge-associated fungus Aspergillus sp. Eur. J. Org. Chem. 2013, 2013, 894–906. [Google Scholar]
- Brauers, G.; Edrada, R.A.; Ebel, R.; Proksch, P.; Wray, V.; Berg, A.; Gräfe, U.; Schächtele, C.; Totzke, F.; Finkenzeller, G. Anthraquinones and betaenone derivatives from the sponge-associated fungus Microsphaeropsis species: Novel inhibitors of protein kinases. J. Nat. Prod. 2000, 63, 739–745. [Google Scholar] [CrossRef]
- Yang, L.H.; Miao, L.; Lee, O.O.; Li, X.; Xiong, H.; Pang, K.-L.; Vrijmoed, L.; Qian, P.-Y. Effect of culture conditions on antifouling compound production of a sponge-associated fungus. Appl. Microbiol. Biotechnol. 2007, 74, 1221–1231. [Google Scholar] [CrossRef]
- Taylor, M.W.; Radax, R.; Steger, D.; Wagner, M. Sponge-associated microorganisms: Evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 2007, 71, 295–347. [Google Scholar] [CrossRef]
- Mitova, M.; Tommonaro, G.; Hentschel, U.; Müller, W.E.G.; de Rosa, S. Exocellular cyclic dipeptides from a Ruegeria strain associated with cell cultures of Suberites domuncula. Mar. Biotechnol. 2004, 6, 95–103. [Google Scholar] [CrossRef]
- König, G.M.; Kehraus, S.; Seibert, S.F.; Abdel-Lateff, A.; Müller, D. Natural products from marine organisms and their associated microbes. ChemBioChem 2006, 7, 229–238. [Google Scholar] [CrossRef]
- Zuccaro, A.; Schoch, C.L.; Spatafora, J.W.; Kohlmeyer, J.; Draeger, S.; Mitchell, J.I. Detection and identification of fungi intimately associated with the brown seaweed Fucus serratus. Appl. Environ. Microbiol. 2008, 74, 931–941. [Google Scholar] [CrossRef]
- Berland, B.R.; Bonin, D.J.; Maestrini, S.Y. Study of bacteria associated with marine algae in culture. Mar. Biol. 1970, 5, 68–76. [Google Scholar] [CrossRef]
- Wiese, J.; Thiel, V.; Nagel, K.; Staufenberger, T.; Imhoff, J.F. Diversity of antibiotic-active bacteria associated with the brown alga Laminaria saccharina from the Baltic Sea. Mar. Biotechnol. 2009, 11, 287–300. [Google Scholar] [CrossRef]
- Amrani, M.E.; Debbab, A.; Aly, A.H.; Wray, V.; Dobretsov, S.; Müller, W.E.G.; Lin, W.; Lai, D.; Proksch, P. Farinomalein derivatives from an unidentified endophytic fungus isolated from the mangrove plant Avicennia marina. Tetrahedron Lett. 2012, 53, 6721–6724. [Google Scholar] [CrossRef]
- Kjer, J.; Wray, V.; Edrada-Ebel, R.; Ebel, R.; Pretsch, A.; Lin, W.; Proksch, P. Xanalteric acids I and II and related phenolic compounds from an endophytic Alternaria sp. isolated from the mangrove plant Sonneratia alba. J. Nat. Prod. 2009, 72, 2053–2057. [Google Scholar] [CrossRef]
- Flores-Mireles, A.L.; Winans, S.C.; Holguin, G. Molecular characterization of diazotrophic and denitrifying bacteria associated with mangrove roots. Appl. Environ. Microbiol. 2007, 73, 7308–7321. [Google Scholar] [CrossRef]
- Díaz, M.P.; Boyd, K.G.; Grigson, S.J.W.; Burgess, J.G. Biodegradation of crude oil across a wide range of salinities by an extremely halotolerant bacterial consortium MPD-M, immobilized onto polypropylene fibers. Biotechnol. Bioeng. 2002, 79, 145–153. [Google Scholar] [CrossRef]
- Kurosawa, K.; Ghiviriga, I.; Sambandan, T.G.; Lessard, P.A.; Barbara, J.E.; Rha, C.; Sinskey, A.J. Rhodostreptomycins, antibiotics biosynthesized following horizontal gene transfer from Streptomyces padanus to Rhodococcus fascians. J. Am. Chem. Soc. 2008, 130, 1126–1127. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).