Diversity of Secondary Metabolites from Marine Bacillus Species: Chemistry and Biological Activity
Abstract
:1. Introduction
2. Bioactive Compounds
2.1. Lipopeptides
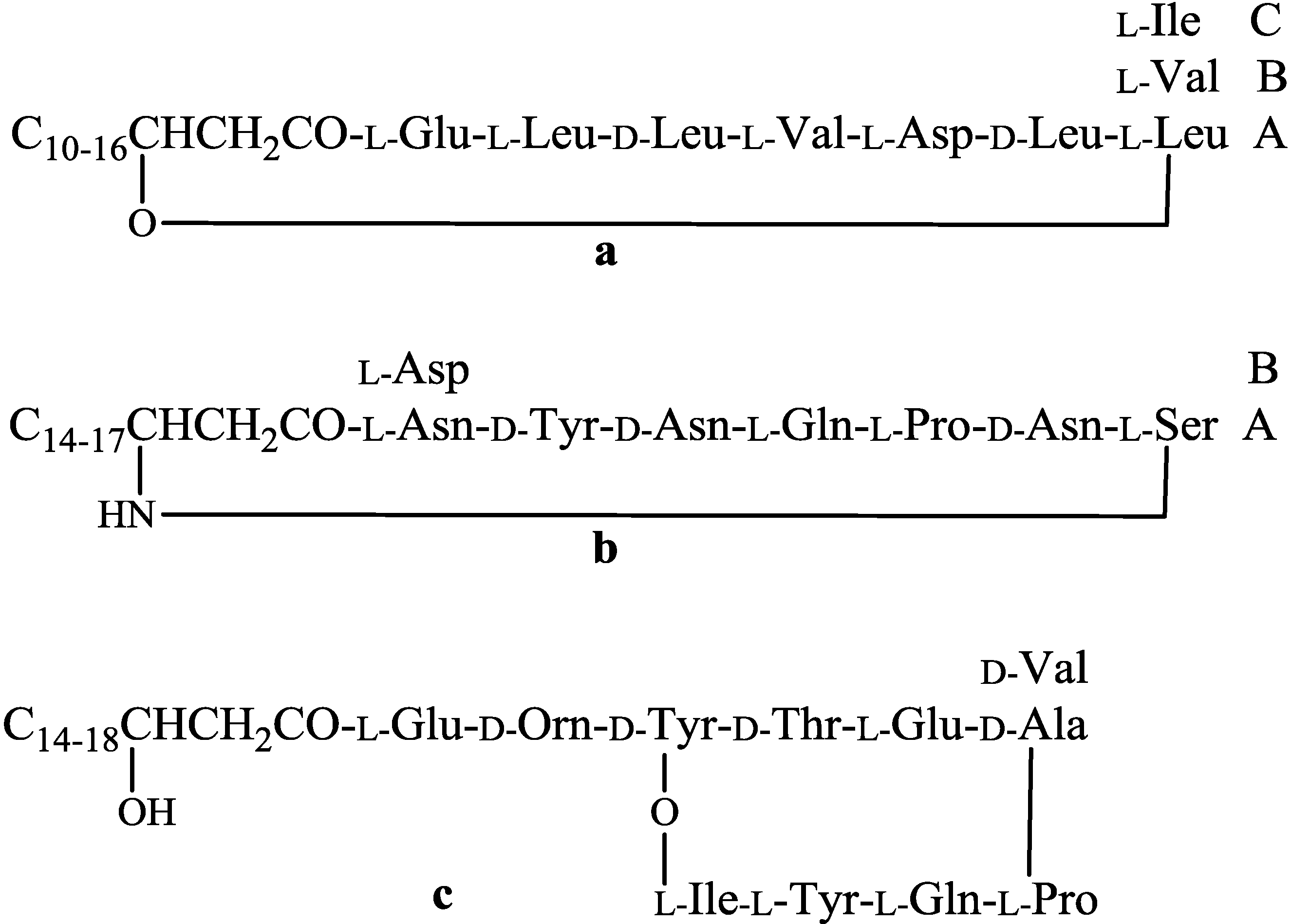
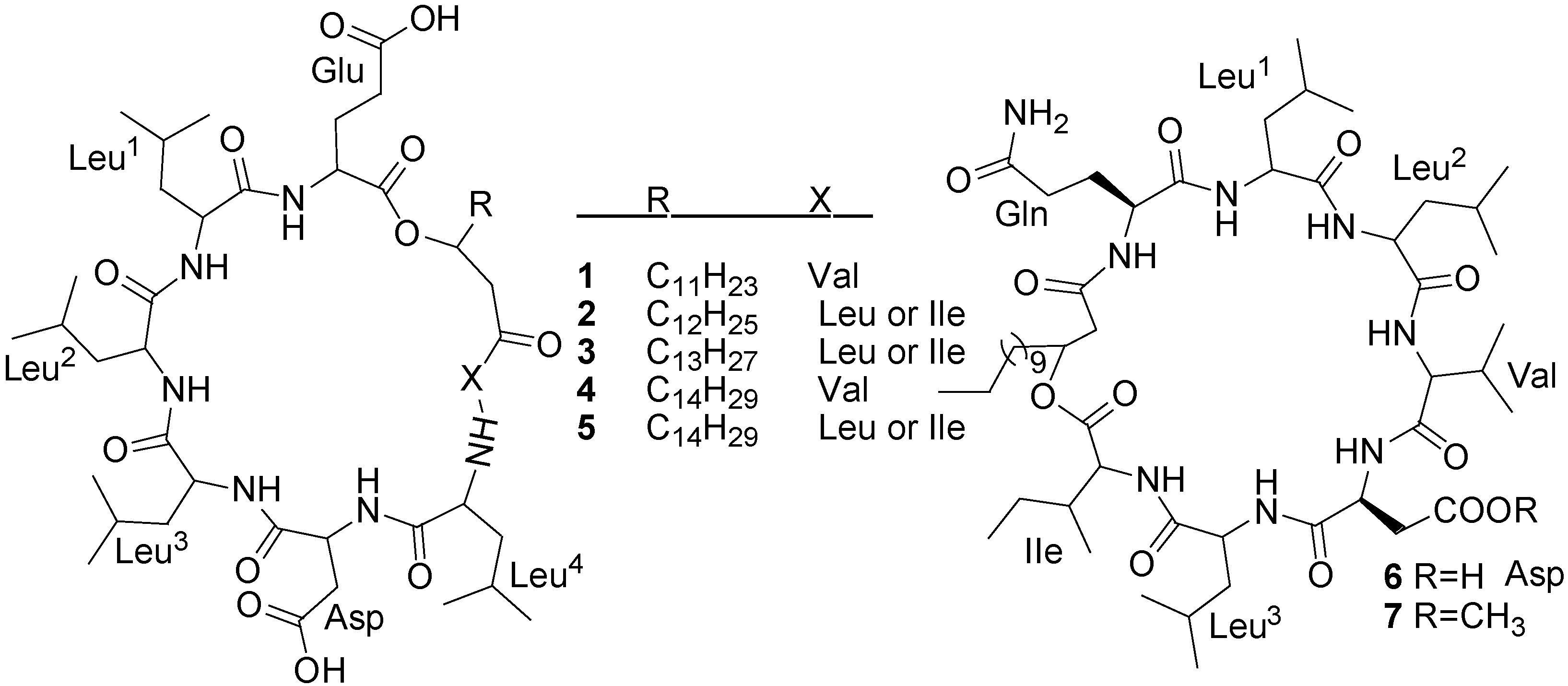
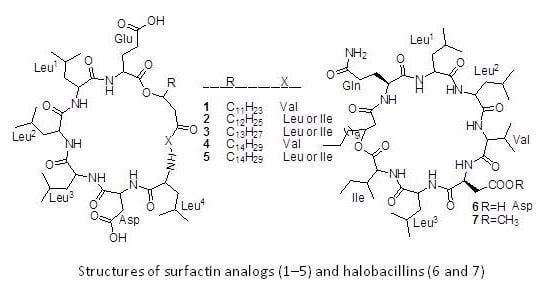
| Compounds | Producing strains | Test organisms/cell lines | Inhibitory concentrations | Nature of bioactivities | Ref. |
|---|---|---|---|---|---|
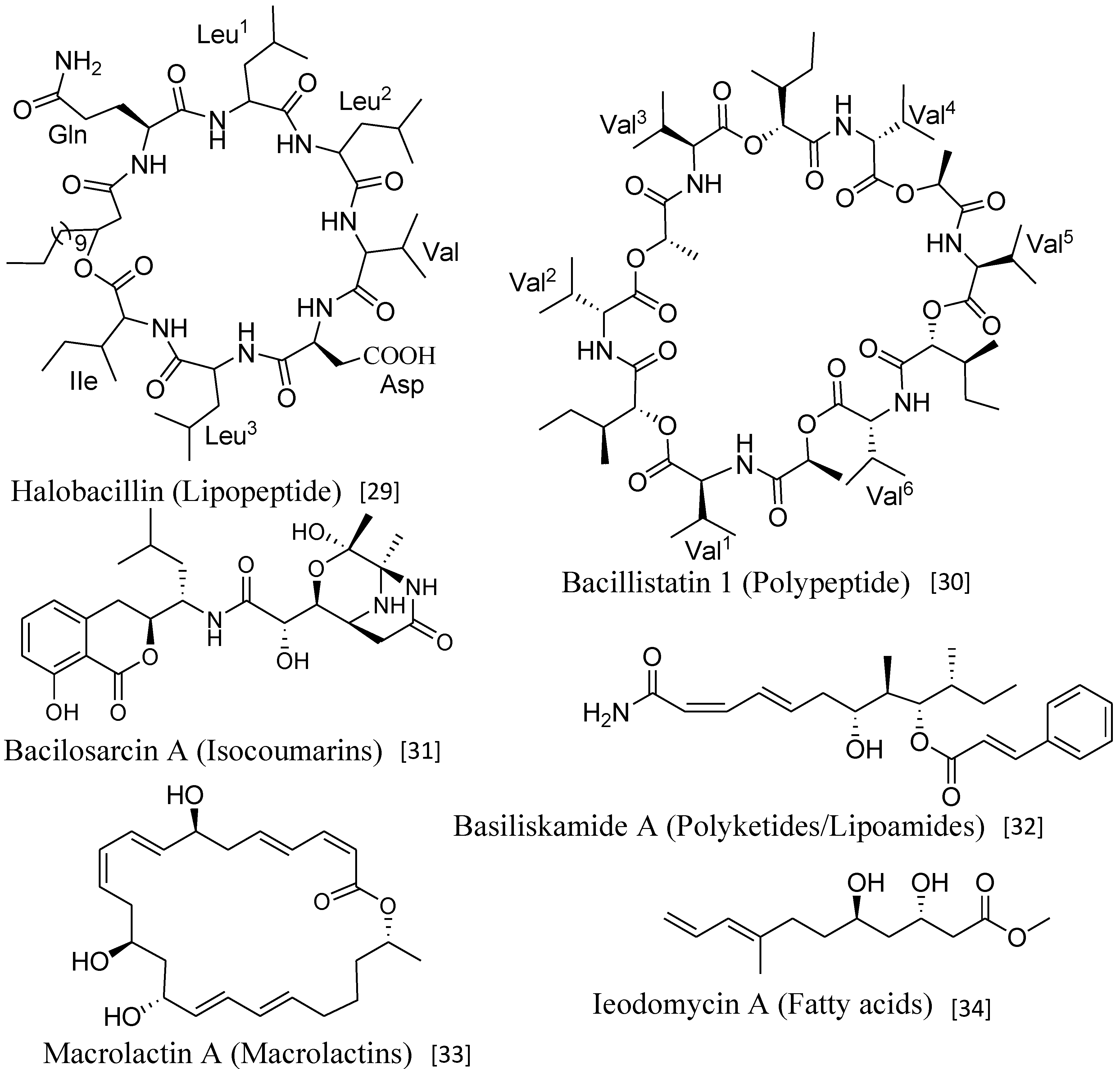
| Halobacillin (6) | Bacillus sp. CND-914 | Human HCT-116 cancer cells | 0.98 µg/mL (IC50) | Anticancer | [29] |
| Mixirin (11) | Bacillus sp. | Human HCT-116 cancer cells | 0.68 µg/mL (IC50) | Anticancer | [44] |
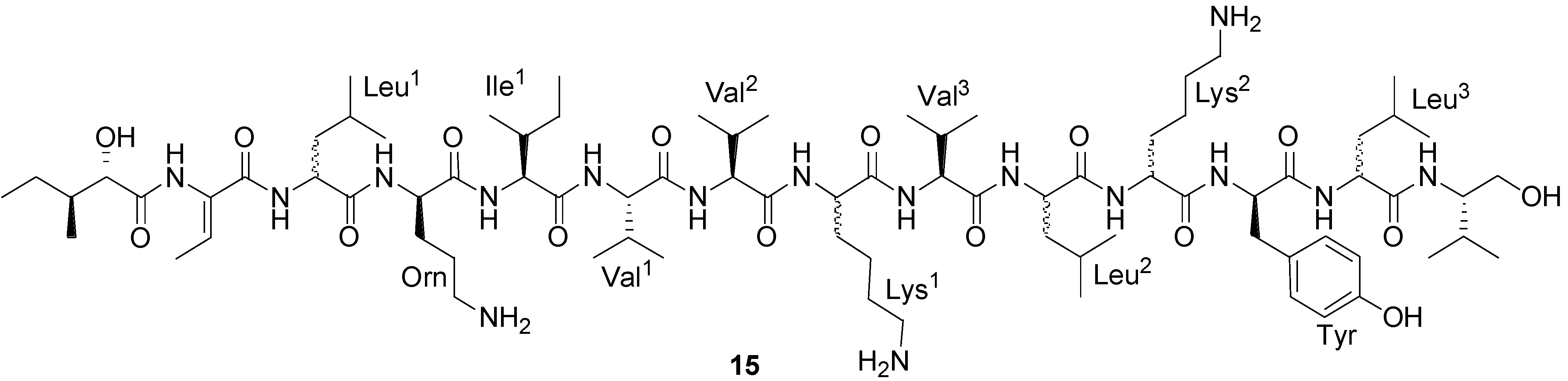
| Bogorol A (15) | Bacillus sp. | MRSA | 2 µg/mL (MIC) | Antibacterial | [45] |
| Loloatin B (18) | Bacillus sp. | MRSA, VRE | 1–2 µg/mL (MIC) | Antibacterial | [46] |
| Bacillistatins 1 (19) and 2 (20) | B. silvestris | Human cancer cell line | 10−4–10−5 µg/mL (GI50) | Anticancer | [30] |
| Bacillamide (27) | Bacillus sp. | C. polykrikoides | LC50 after 6 h: 3.2 µg/mL | Antialgal | [47] |
| Bacilosarcin A (35) | B. subtilis | Barnyard millet sprouts | 82% inhibition at 50 µM | Plant growth regulator | [31] |
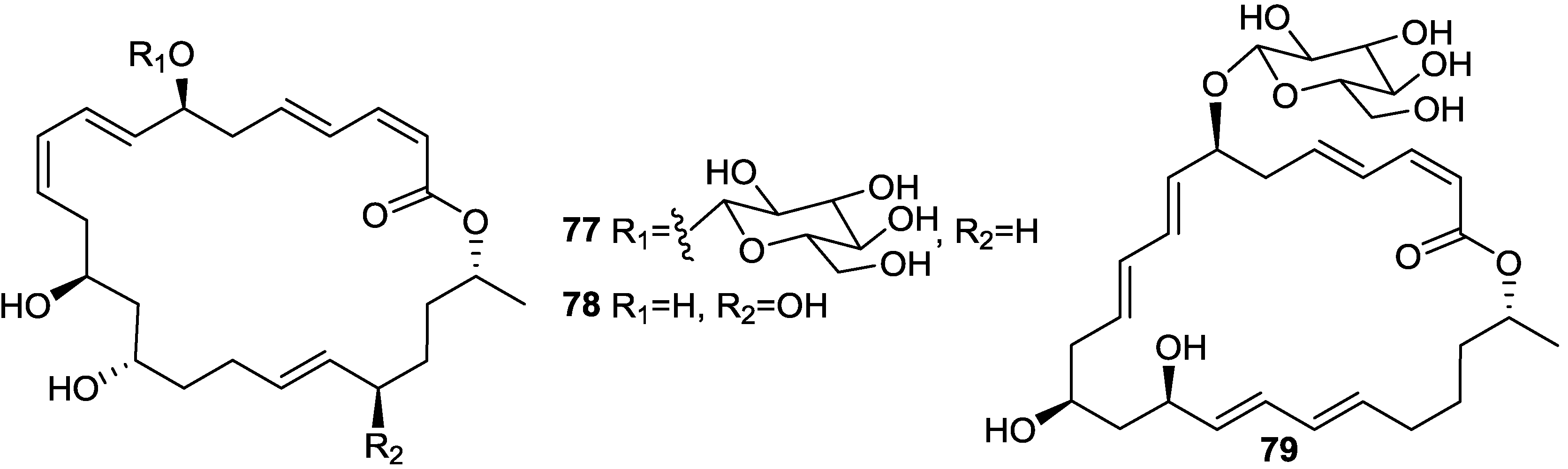
| Macrolactin S (69) | B. amyloliquefaciens | E. coli and S. aureus | 0.3 and 0.1 µg/mL (MIC) | Antibacterial | [48] |
| Macrolactin V (86) | B. amyloliquefaciens | E. coli, B. subtilis and S. aureus | 0.1 µg/mL (MIC) | Antibacterial | [48] |
| Basiliskamides A (21) and B (22) | B. laterosporus | C. albicans and A. fumigatus | 1.0 and 3.1 µg/mL 2.5 and 5.0 µg/mL | Antifungal | [32] |
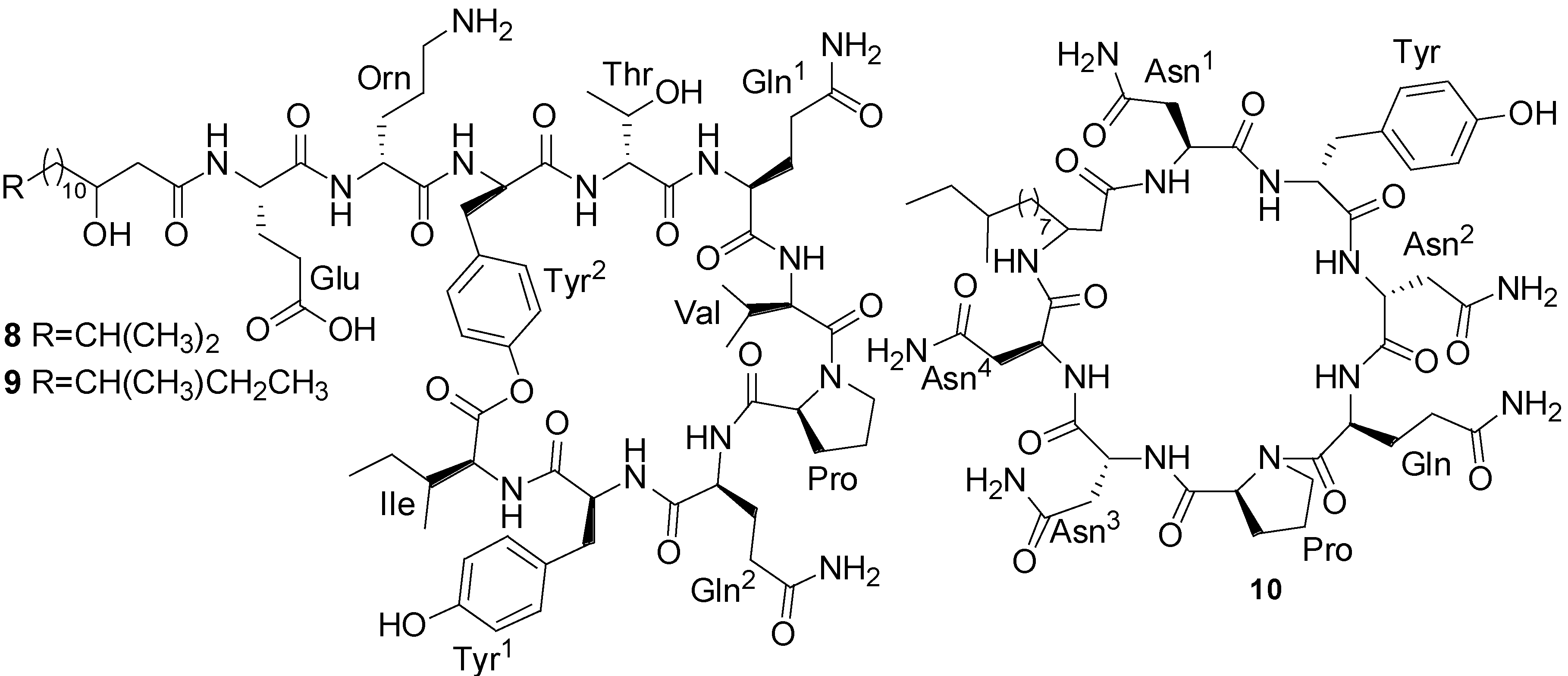
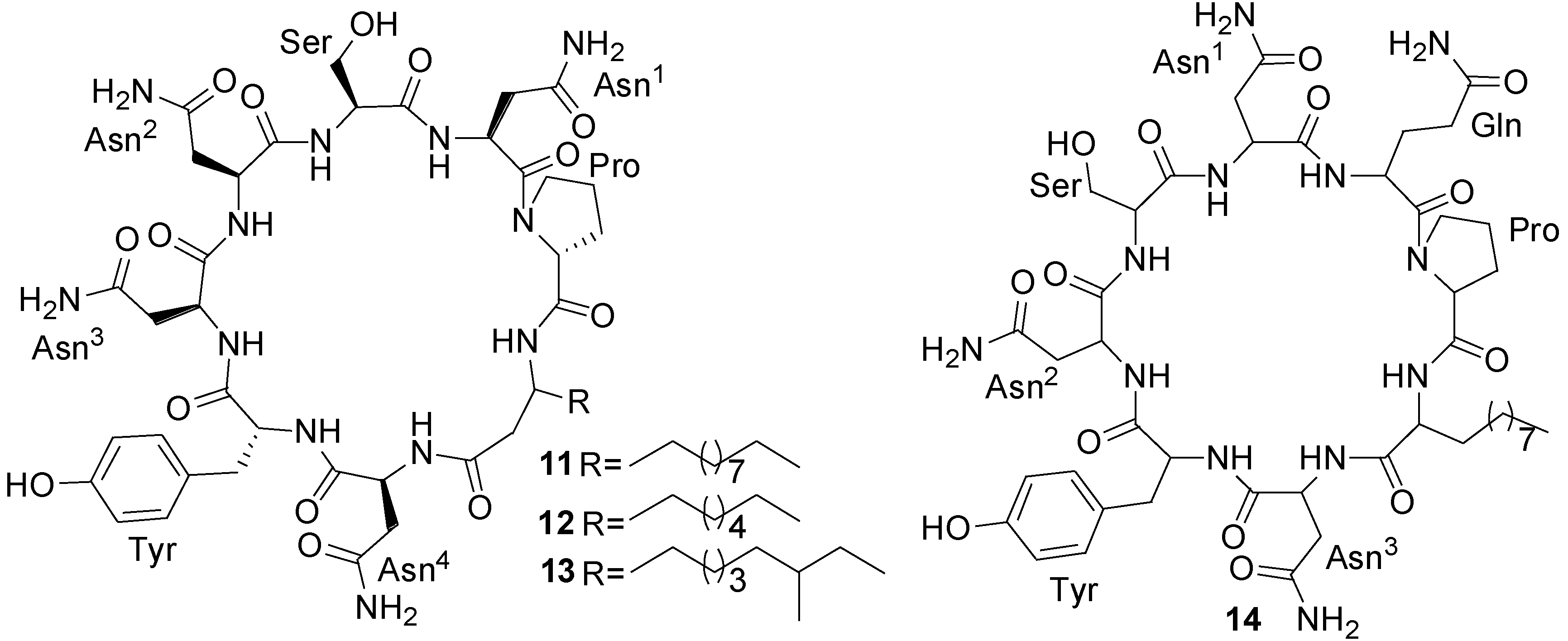
2.2. Polypeptides

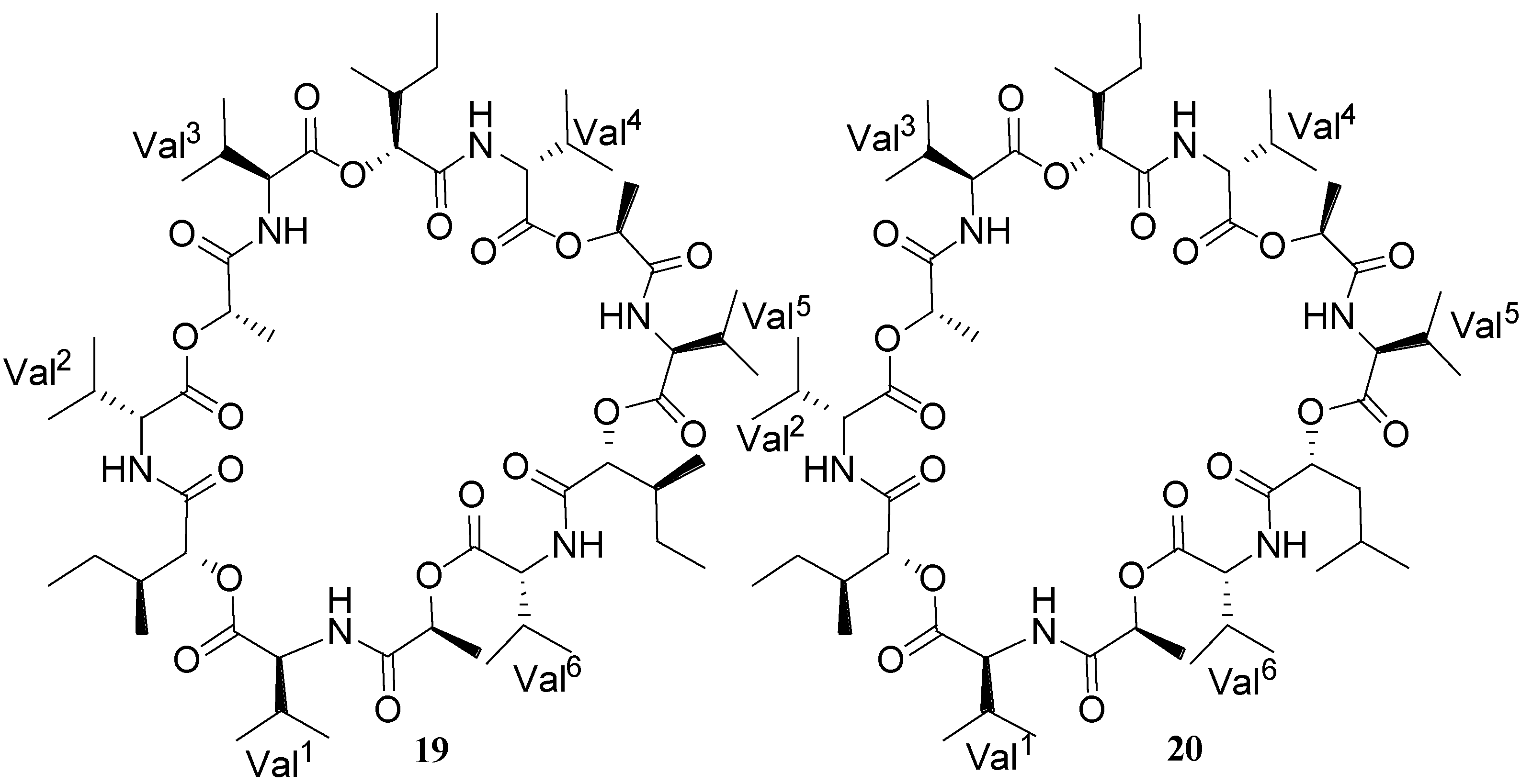
2.3. Polyketides/Lipoamides
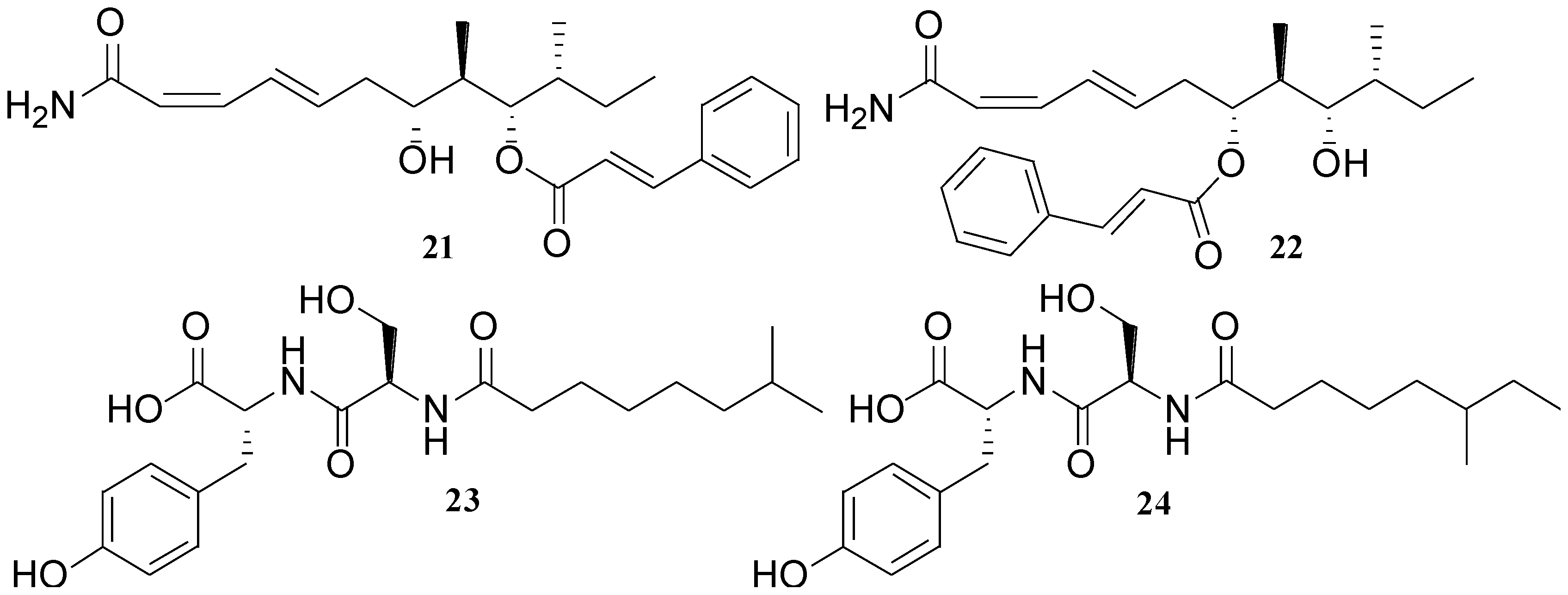
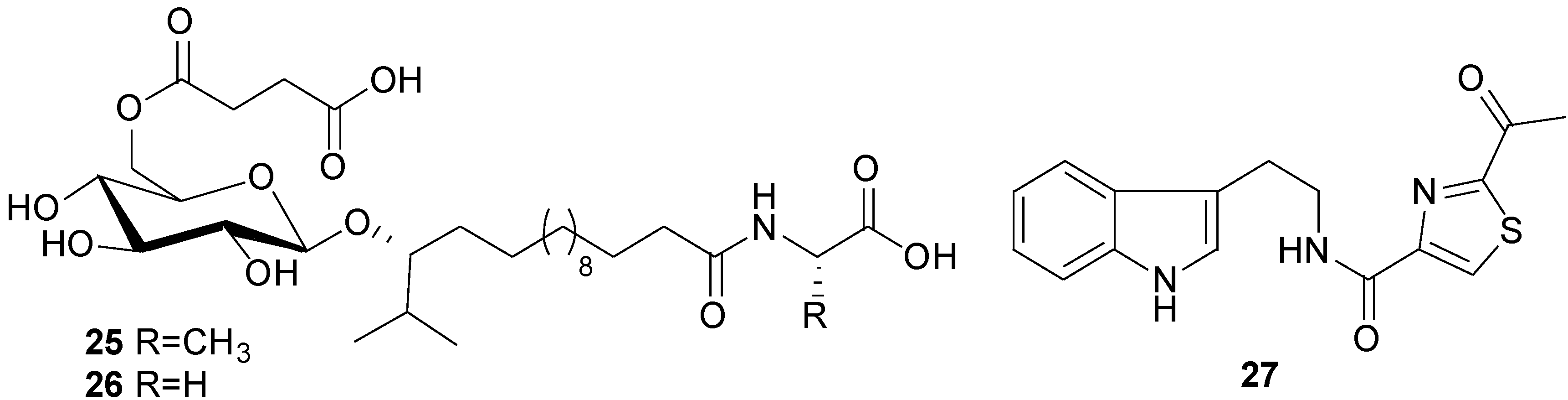
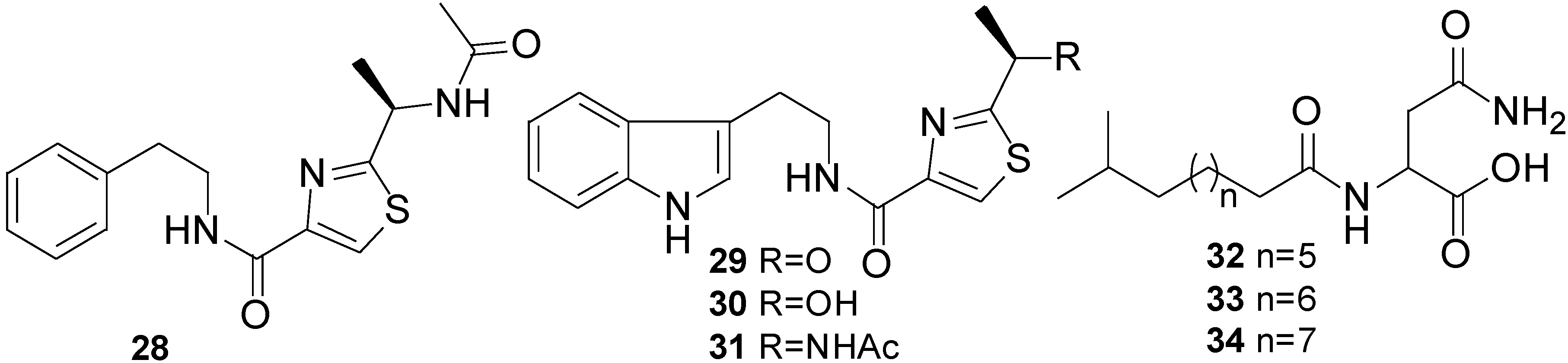
2.4. Isocoumarins
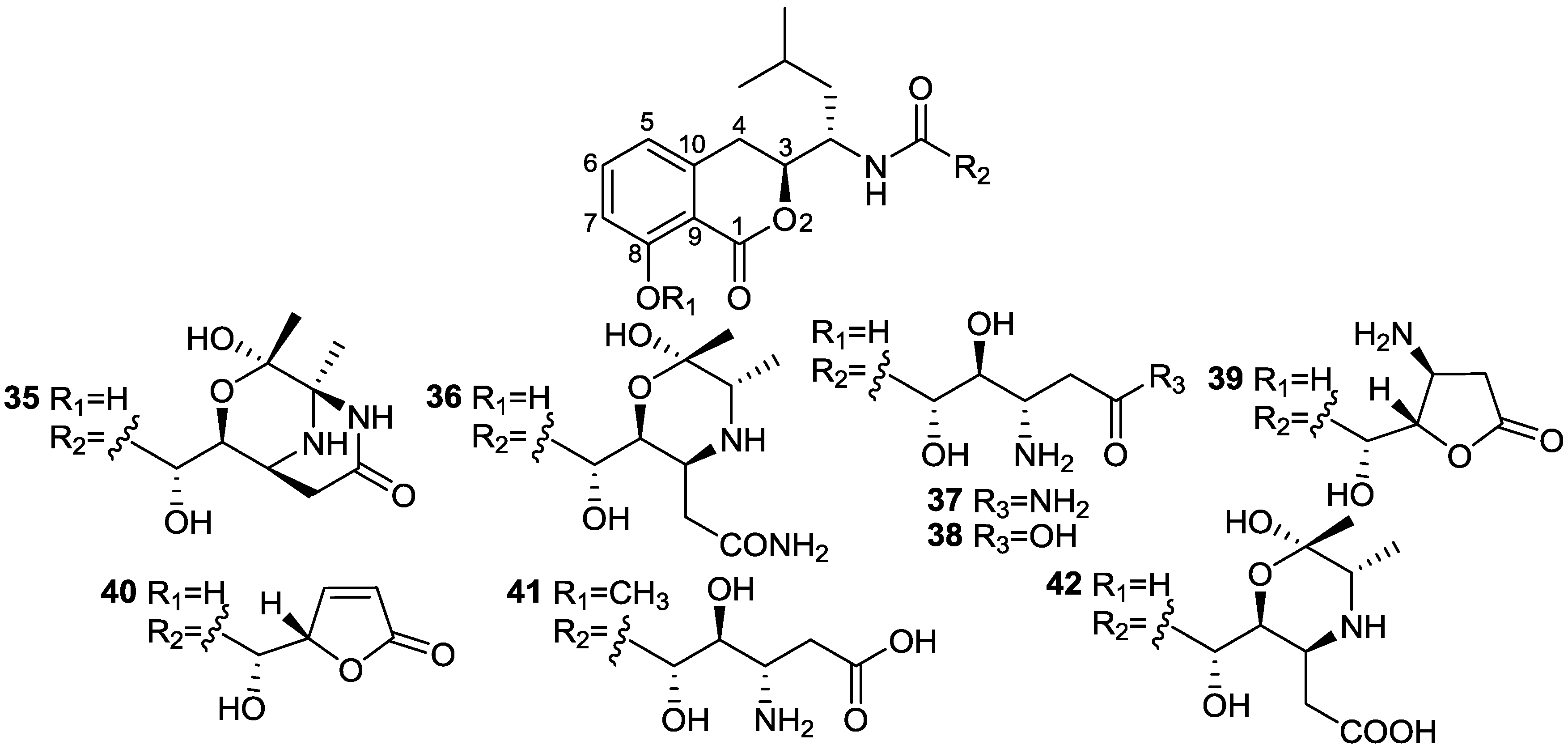
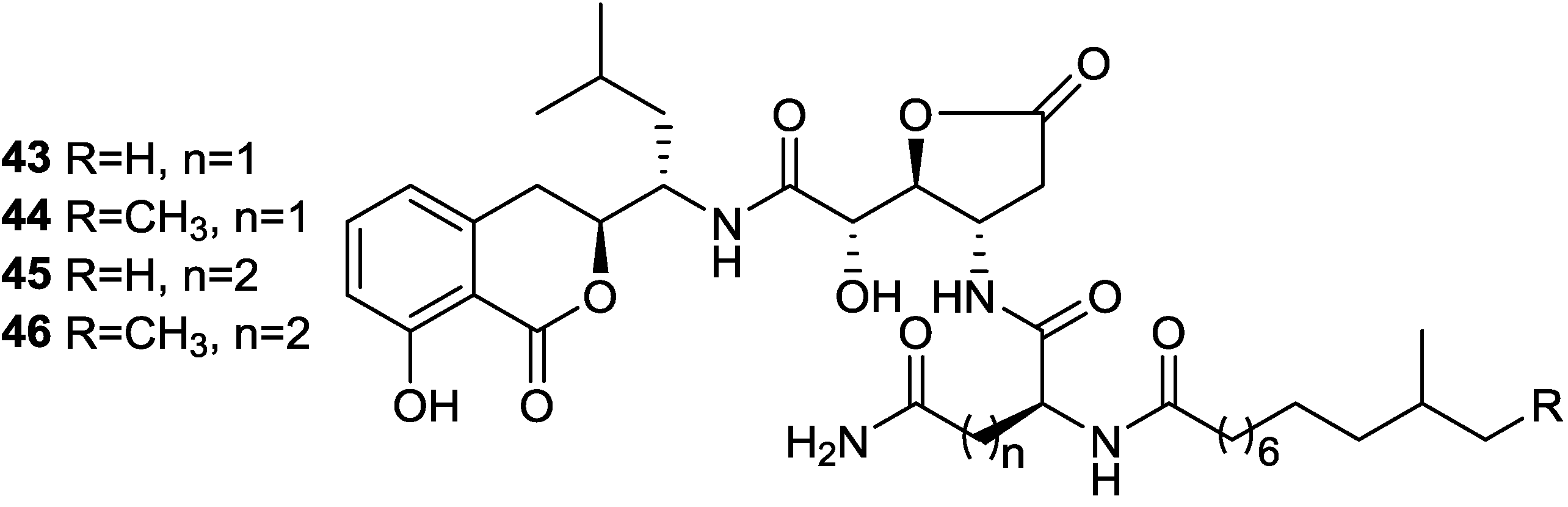
2.5. Fatty Acids
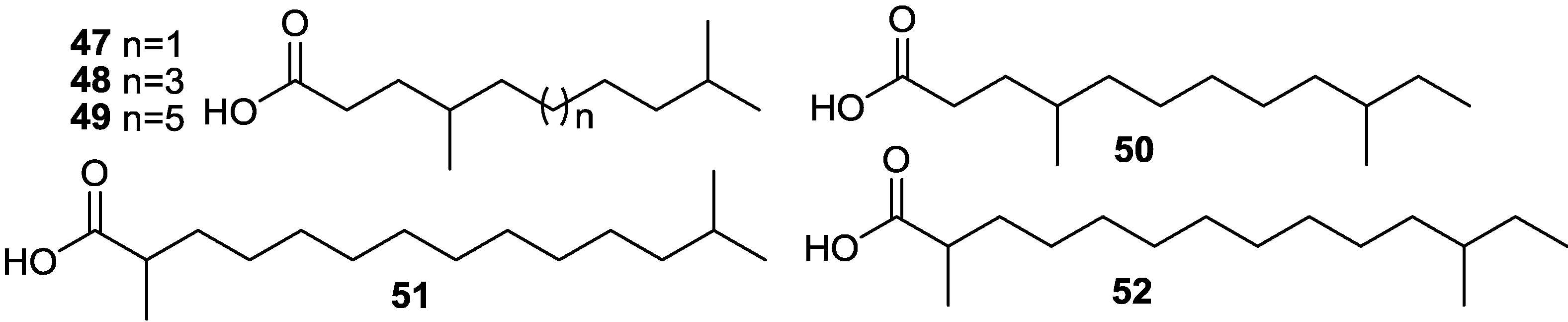
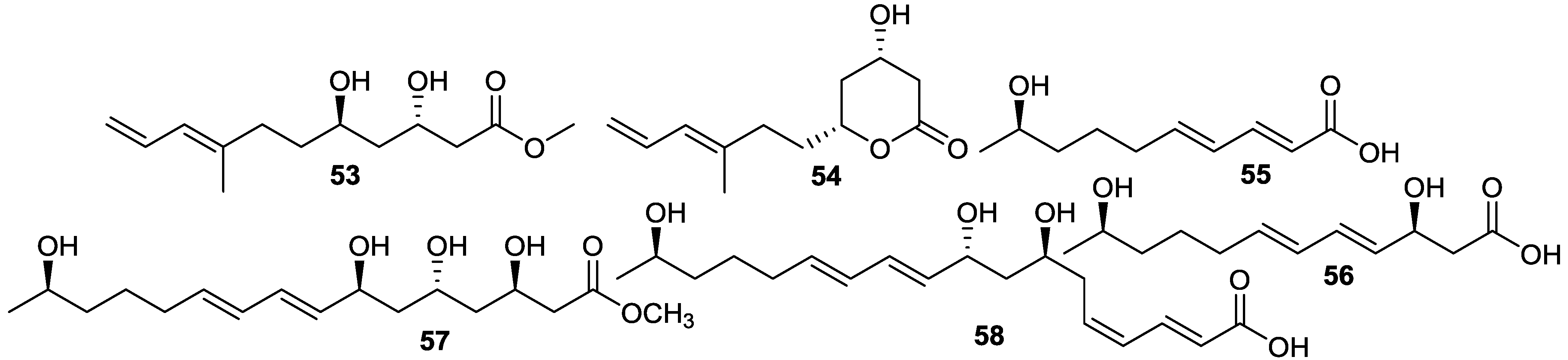
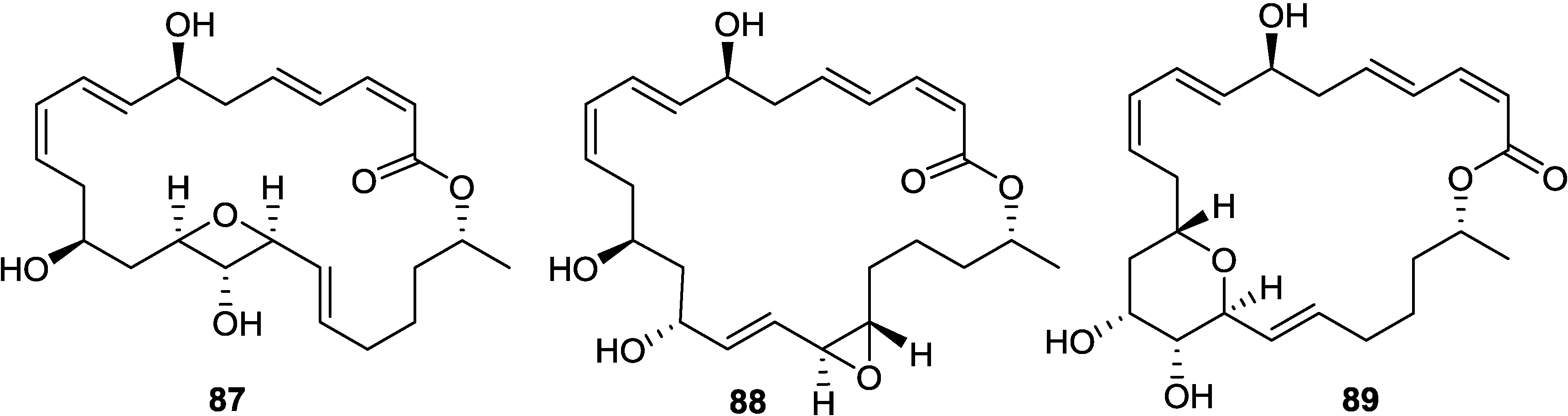
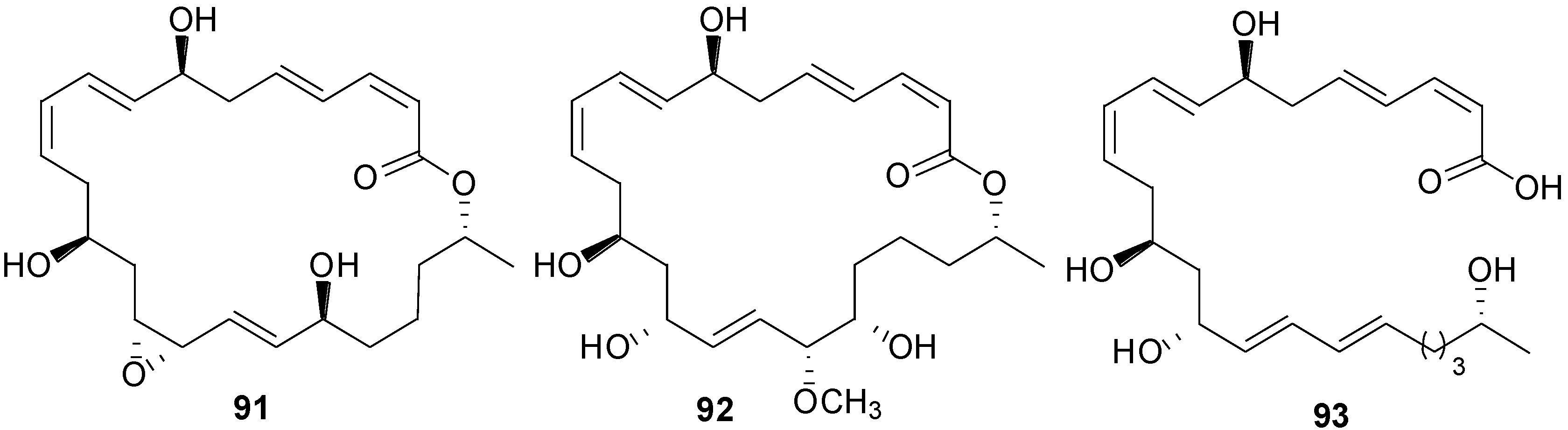
2.6. Macrolactins
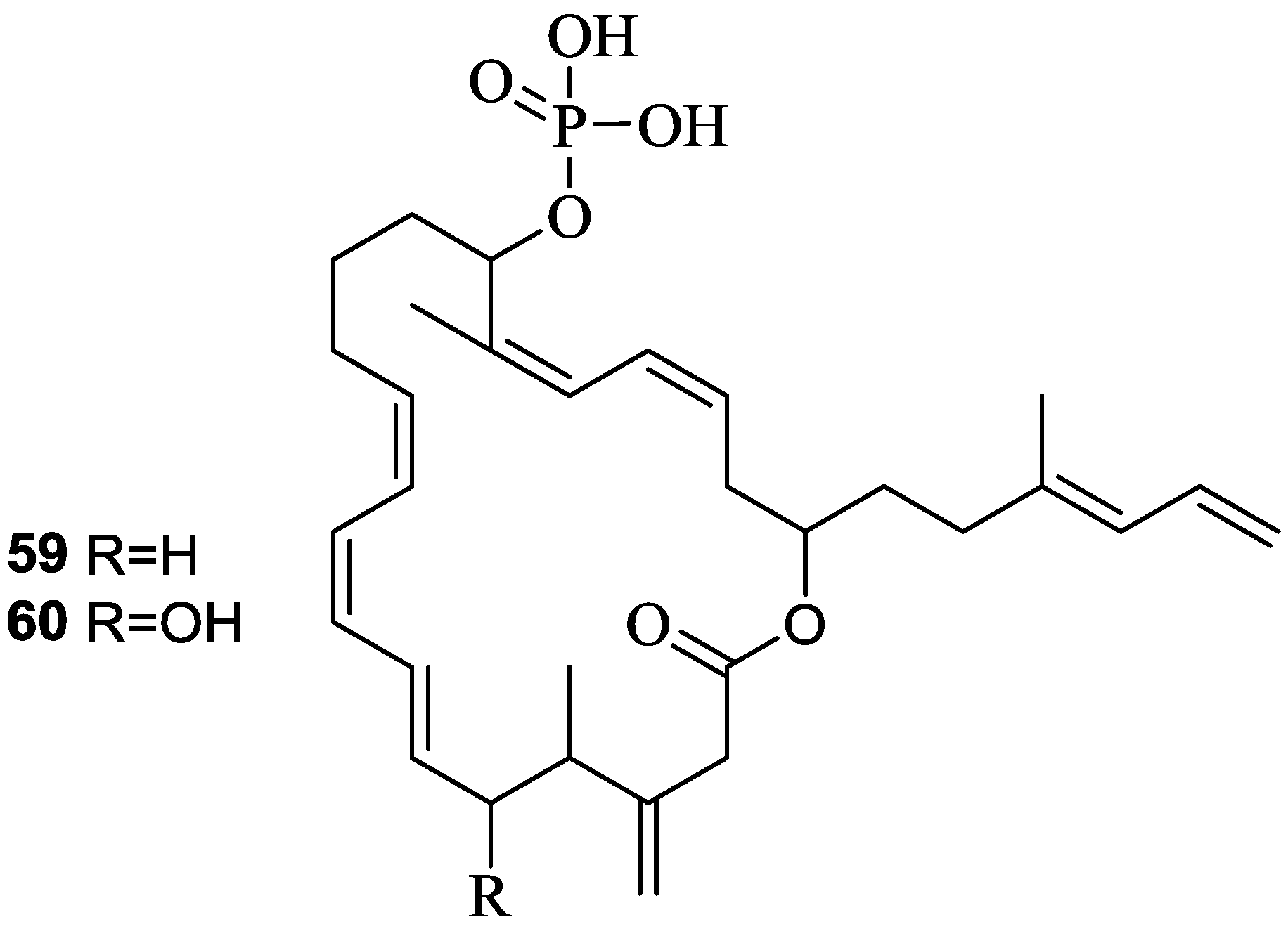
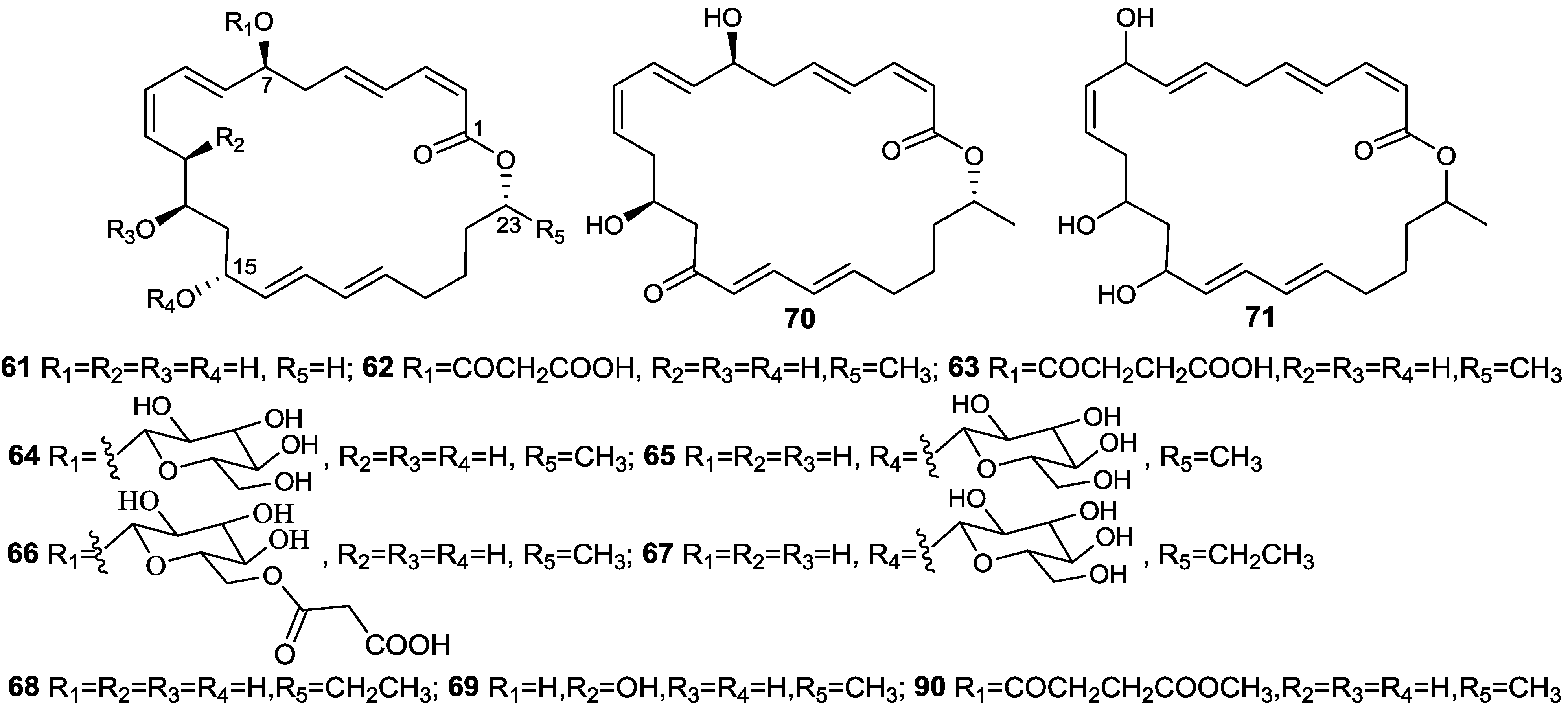
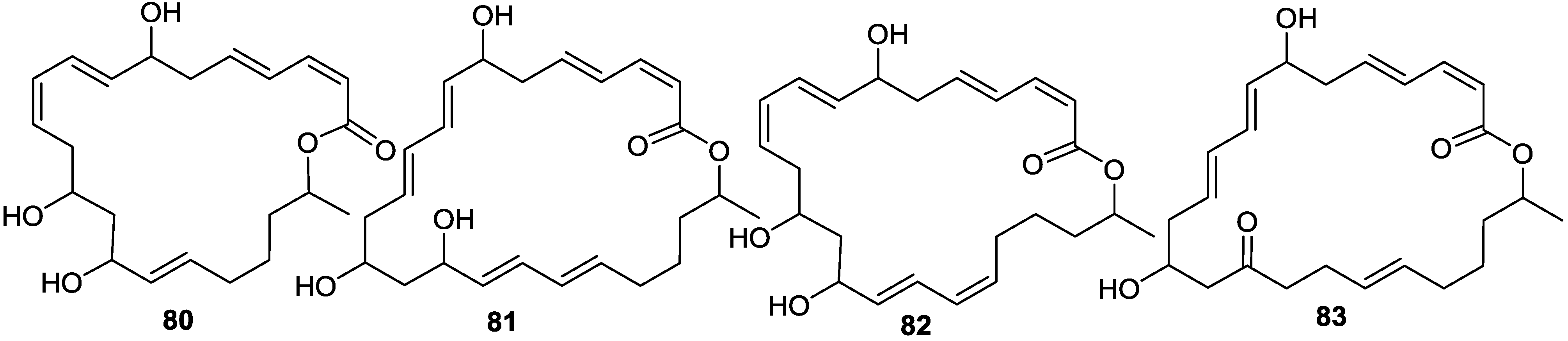
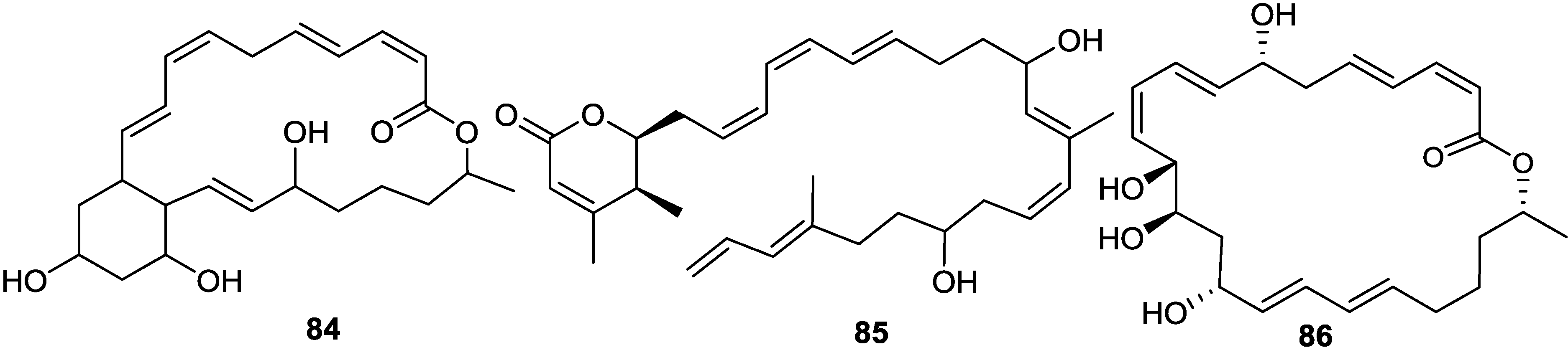
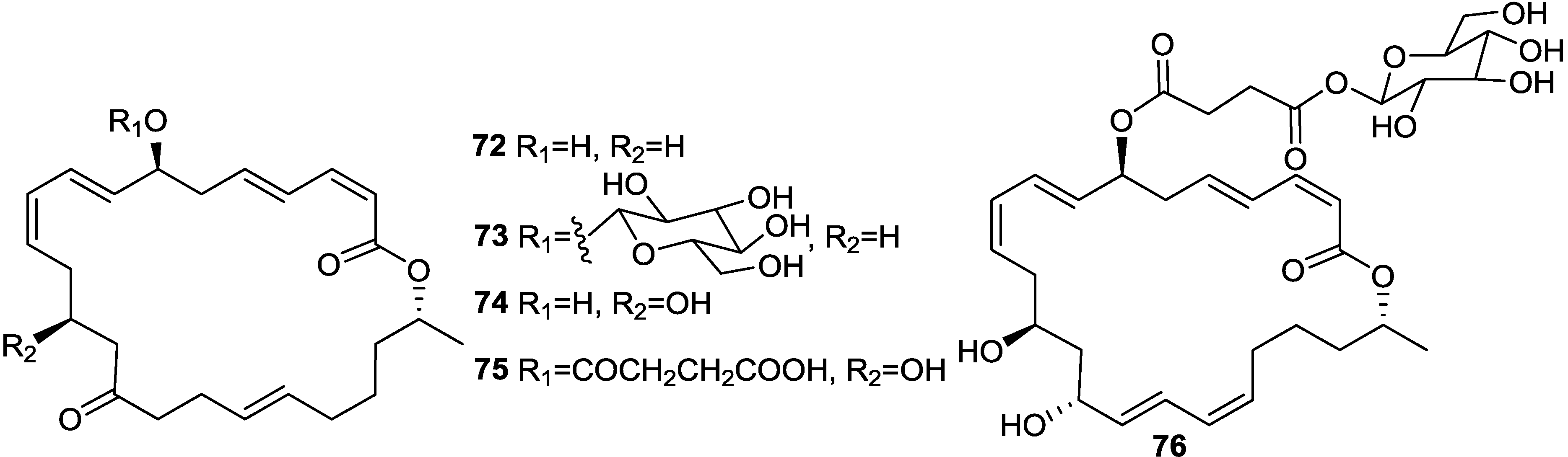
3. Detoxification of Heavy Metals
4. Marine Bacillus Strains as Potential Biocontrol Agents
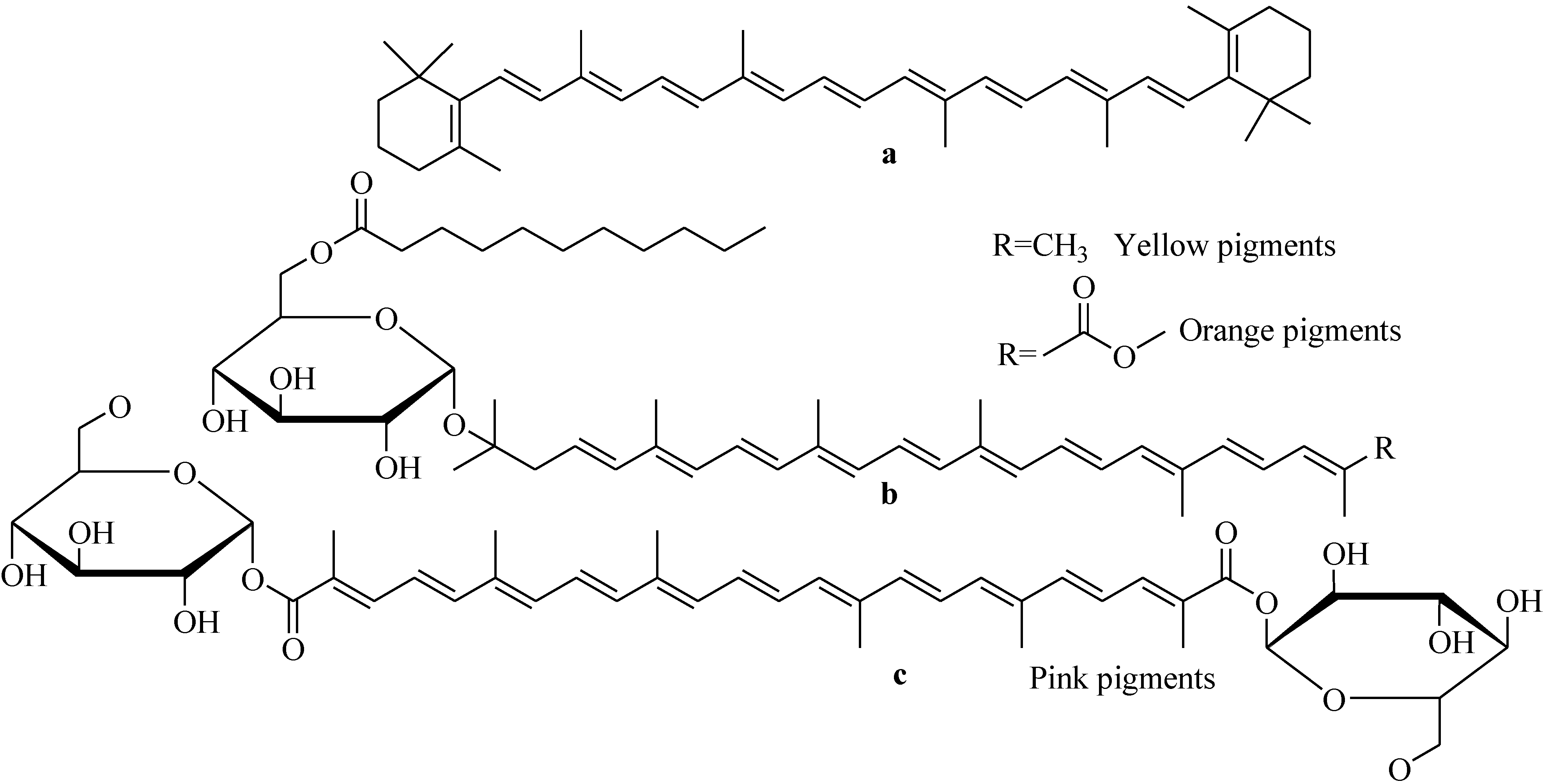
5. Marine Bacillus Isolates as a Potential Source of Natural Carotenoids
6. Conclusions and Future Perspectives
Acknowledgments
Conflict of Interest
References
- Lebar, M.D.; Heimbegner, J.L.; Baker, B.J. Cold-water marine natural products. Nat. Prod. Rep. 2007, 24, 774–797. [Google Scholar] [CrossRef]
- Fenical, W. Chemical studies of marine bacteria: Developing a new resource. Chem. Rev. 1993, 93, 1673–1683. [Google Scholar] [CrossRef]
- Laatsch, H. Marine Bacterial Metabolites. In Frontiers in Marine Biotechnology; Proksch, P., Muller, W.E.G., Eds.; Horizon Bioscience: Norfolk, UK, 2006; pp. 225–288. [Google Scholar]
- Schwartsmann, G.; da Rocha, A.B.; Berlinck, R.G.S.; Jimeno, J. Marine organisms as a source of new anticancer agents. Lancet Oncol. 2001, 2, 221–225. [Google Scholar] [CrossRef]
- Jha, R.K.; Zi-rong, X. Biomedical compounds from marine organisms. Mar. Drugs 2004, 2, 123–146. [Google Scholar] [CrossRef]
- Nathan, C. Antibiotics at the crossroads. Nature 2004, 431, 899–902. [Google Scholar] [CrossRef]
- Von Nussbaum, F.; Brands, M.; Hinzen, B.; Weigand, S.; Häbich, D. Antibacterial natural products in medicinal chemistry—Exodus or revival. Angew. Chem. Int. Ed. 2006, 45, 5072–5129. [Google Scholar]
- Li, J.W.-H.; Vederas, J.C. Drug discovery and natural products: End of an era or an endless frontier? Science 2009, 325, 161–165. [Google Scholar] [CrossRef]
- Islam, M.T.; von Tiedemann, A.; Laatsch, H. Protein kinase C is likely to be involved in zoosporogenesis and maintenance of flagellar motility in the peronosporomycete zoospores. Mol. Plant Microbe Interact. 2011, 24, 938–947. [Google Scholar] [CrossRef]
- Islam, M.T.; Hashidoko, Y.; Deora, A.; Ito, T.; Tahara, S. Suppression of damping-off disease in host plants by the rhizoplane bacterium Lysobacter sp. Strain SB-K88 is linked to plant colonization and antibiosis against soilborne peronosporomycetes. Appl. Environ. Microbiol. 2005, 71, 3786–3796. [Google Scholar] [CrossRef]
- Islam, M.T. Potentials for Biological Control of Plant Disease by Lysobacter spp., with Special Reference to Strain SB-K88. In Bacteria in Agrobiology: Plant Growth Responses; Maheshwari, D.K., Ed.; Springer-Verlag: Berlin/Heidelberg, Germany, 2011; pp. 335–364. [Google Scholar]
- Islam, M.T.; Hossain, M.M. Biological Control of Peronosporomycete Phytopathogens by Antagonistic Bacteria. In Bacteria in Agrobiology: Plant Disease Management; Maheshwari, D.K., Ed.; Springer-Verlag: Berlin/Heidelberg, Germany, 2013; pp. 167–218. [Google Scholar]
- Whitman, W.B.; Coleman, D.C.; Wiebe, W.J. Prokaryotes: The unseen majority. Proc. Natl. Acad. Sci.USA 1998, 95, 6578–6583. [Google Scholar] [CrossRef]
- Kennedy, J.; Marchest, J.R.; Dobson, A.D.W. Marine metagenomics: Strategies for the discovery of novel enzymes with biotechnological applications from marine environments. Microb. Cell Fact. 2008, 7, 27–28. [Google Scholar] [CrossRef]
- Rampelotto, P.H. Resistance of microorganisms to extreme environmental conditions and its contribution to astrobiology. Sustainability 2010, 2, 1602–1623. [Google Scholar] [CrossRef]
- Sayem, S.M.A.; Manzo, E.; Ciavatta, L.; Tramice, A.; Cordone, A.; Zanfardino, A.; de Felice, M.; Varcamonti, M. Anti-biofilm activity of an exopolysaccharide from a sponge-associated strain of Bacillus licheniformis. Microb. Cell Fact. 2011, 10, 74. [Google Scholar] [CrossRef]
- Paul, V.J.; Arthur, K.E.; Williams, R.R.; Ross, C.; Sharp, K. Chemical defenses: From compounds to communities. Biol. Bull. 2007, 213, 226–251. [Google Scholar] [CrossRef]
- Jensen, P.R.; Fenical, W. Strategies for the discovery of secondary metabolites from marine bacteria: Ecological perspectives. Annu. Rev. Microbiol. 1994, 48, 559–584. [Google Scholar] [CrossRef]
- Feling, R.H.; Buchanan, G.O.; Mincer, T.J.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Salinosporamide A: A highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus salinospora. Angew. Chem. Int. Ed. Engl. 2003, 20, 355–357. [Google Scholar]
- Chen, X.H.; Koumoutsi, A.; Scholz, R.; Eisenreich, A.; Schneider, K.; Heinemeyer, I.; Morgenstern, B.; Voss, B.; Hess, W.R.; Reva, O.; et al. Comparative analysis of the complete genome sequence of the plant growth—Promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 2007, 25, 1007–1014. [Google Scholar] [CrossRef]
- Kunst, F.; Ogasawara, N.; Moszer, I.; Albertini, A.M.; Alloni, G.; Azevedo, V.; Bertero, M.G.; Bessières, P.; Bolotin, A.; Borchert, S.; et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 1997, 390, 249–256. [Google Scholar] [CrossRef]
- Fan, L.; Bo, S.; Chen, H.; Ye, W.; Kleinschmidt, K.; Baumann, H.I.; Imhoff, J.F.; Kleine, M.; Cai, D. Genome Sequence of Bacillus subtilis subsp. spizizenii gtP20b, isolated from the Indian Ocean. J. Bacteriol. 2011, 193, 1276–1277. [Google Scholar] [CrossRef]
- Hamdache, A.; Lamarti, A.; Aleu, J.; Collado, I.G. Non-peptide metabolites from the genus Bacillus. J. Nat. Prod. 2011, 74, 893–899. [Google Scholar] [CrossRef]
- Baruzzi, F.; Quintieri, L.; Morea, M.; Caputo, L. Antimicrobial Compounds Produced by Bacillus spp. and Applications in Food. In Science against Microbial Pathogens: Communicating Current Research and Technological Advances; Vilas, A.M., Ed.; Formatex: Badajoz, Spain, 2011; pp. 1102–1111. [Google Scholar]
- Shoda, M. Bacterial control of plant diseases. Biosci. Bioeng. 2000, 89, 515–521. [Google Scholar] [CrossRef]
- Stein, T. Bacillus subtilis antibiotics: Structures, syntheses and specific functions. Mol. Microbiol. 2005, 56, 845–857. [Google Scholar]
- Dash, H.R.; Neelam, M.; Chakraborty, J.; Kumari, S.; Das, S. Marine bacteria: Potential candidates for enhanced bioremediation. Appl. Microbiol. Biotechnol. 2013, 97, 561–571. [Google Scholar] [CrossRef]
- Nies, D.H. Microbial heavy metal resistance. Appl. Microbiol. Biotechnol. 1999, 51, 730–750. [Google Scholar] [CrossRef]
- Trischman, J.A.; Jensen, P.R.; Fenical, W. Halobacillin: A cytotoxic cyclic acylpeptide of the iturin class produced by a marine Bacillus. Tetrahedron Lett. 1994, 35, 5571–5574. [Google Scholar] [CrossRef]
- Pettit, G.R.; Knight, J.C.; Herald, D.L.; Pettit, R.K.; Hogan, F.; Mukku, V.J.R.V.; Hamblin, J.S.; Dodson, M.J., II; Chapuis, J.C. Antineoplastic agents. 570. Isolation and structure elucidation of bacillistatins 1 and 2 from a marine Bacillus silvestris. J. Nat. Prod. 2009, 72, 366–371. [Google Scholar] [CrossRef]
- Azumi, M.; Ogawa, K.; Fujita, T.; Takeshita, M.; Furumai, T.; Igarashi, Y.; Yoshida, R. Bacilosarcins A and B, novel bioactive isocoumarins with unusual heterocyclic cores from the marine-derived bacterium Bacillus subtilis. Tetrahedron 2008, 64, 6420–6425. [Google Scholar] [CrossRef]
- Barsby, T.; Kelly, M.T.; Andersen, R.J. Tupuseleiamides and basiliskamides, new acyldipeptides and antifungal polyketides produced in culture by a Bacillus laterosporus isolate obtained from a tropical marine habitat. J. Nat. Prod. 2002, 65, 1447–1451. [Google Scholar] [CrossRef]
- Gustafson, K.; Roman, M.; Fenical, W. The macrolactins, a novel class of antiviral and cytotoxic macrolides from a deep-sea marine bacterium. J. Am. Chem. Soc. 1989, 111, 7519–7524. [Google Scholar] [CrossRef]
- Mondol, M.A.M.; Kim, J.H.; Lee, M.A.; Tareq, F.S.; Lee, H.S.; Lee, Y.J.; Shin, H.J. Ieodomycins A–D, antimicrobial fatty acids from a marine Bacillus sp. J. Nat. Prod. 2011, 74, 1606–1612. [Google Scholar] [CrossRef]
- Damalas, C.A.; Eleftherohorinos, I.G. Pesticide exposure, safety issues, and risk assessment indicators. Int. J. Environ. Res. Public Health. 2011, 8, 1402–1419. [Google Scholar] [CrossRef]
- Ongena, M.; Jacques, P. Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 2008, 16, 115–125. [Google Scholar]
- Raaijmakers, J.M.; de Bruijin, I.; de Kock, M.J. Cyclic lipopeptide production by plant-associated Pseudomonas spp.: Diversity, activity, biosynthesis, and regulation. Mol. Plant Microbe Interact. 2006, 19, 699–710. [Google Scholar] [CrossRef]
- Arguelles-Arias, A.; Ongena, M.; Halimi, B.; Lara, Y.; Brans, A.; Joris, B.; Fickers, P. Bacillus amyloliquefaciens GA1 as a source of potent antibiotics and other secondary metabolites for biocontrol of plant pathogens. Microb. Cell Fact. 2009, 8. [Google Scholar] [CrossRef]
- Malfanova, N.; Franzil, L.; Lugtenberg, B.; Chebotar, V.; Ongena, M. Cyclic lipopeptide profile of the plant-beneficial endophytic bacterium Bacillus subtilis HC8. Arch. Microbiol. 2012, 194, 893–899. [Google Scholar] [CrossRef]
- Tsuge, K.; Akiyama, T.; Shoda, M. Cloning, sequencing, and characterization of the iturin A operon. J. Bacterial. 2001, 183, 6265–6273. [Google Scholar] [CrossRef]
- Berrue, F.; Ibrahim, A.; Boland, P.; Kerr, R.G. Newly isolated marine Bacillus pumilus (SP21): A source of novel lipoamides and other antimicrobial agents. Pure Appl. Chem. 2009, 81, 1027–1031. [Google Scholar] [CrossRef]
- Cooper, D.G.; MacDonald, C.R.; Duff, S.J.B.; Kosaric, N. Enhanced production of surfactin from Bacillus subtilis by continuous product removal and metal cation additions. Appl. Environ. Microbiol. 1981, 42, 408–412. [Google Scholar]
- Ma, Z.; Wang, N.; Hu, J.; Wang, S. Isolation and characterization of a new iturinic lipopeptide, mojavensin A produced by a marine-derived bacterium Bacillus mojavensis. J. Antibiot. 2012, 65, 317–322. [Google Scholar] [CrossRef]
- Zhang, H.L.; Hua, H.M.; Pei, Y.H.; Yao, S. Three new cytotoxic cyclic acylpeptides from marine Bacillus sp. Chem. Pharm. Bull. 2004, 52, 1029–1030. [Google Scholar] [CrossRef]
- Barsby, T.; Kelly, M.T.; Gagne, S.M.; Andersen, R.J. Bogorol A produced in culture by a marine Bacillus sp. reveals a novel template for cationic peptide antibiotics. Org. Lett. 2001, 3, 437–440. [Google Scholar] [CrossRef]
- Gerard, J.; Haden, P.; Kelly, M.T.; Andersen, R.J. Loloatin B, cyclic decapeptide antibiotic, produced in culture by a tropical marine bacterium. Tetrahedron Lett. 1996, 37, 7201–7294. [Google Scholar] [CrossRef]
- Jeong, S.Y.; Ishida, K.; Ito, Y.; Okada, S.; Murakami, M. Bacillamide, a novel algicide from the marine bacterium, Bacillus sp. SY-1, against the harmful dinoflagellate, Cochlodinium polykrikoides. Tetrahedron Lett. 2003, 44, 8005–8007. [Google Scholar] [CrossRef]
- Gao, C.-H.; Tian, X.-P.; Qi, S.-H.; Luo, X.-M.; Wang, P.; Zhang, S. Antibacterial and antilarval compounds from marine gorgonian-associated bacterium Bacillus amyloliquefaciens SCSIO 00856. J. Antibiot. 2010, 63, 191–193. [Google Scholar] [CrossRef]
- Liu, R.F.; Zhang, D.J.; Li, Y.G.; Tao, L.M.; Tian, L. A new antifungal cyclic lipopeptide from Bacillus marinus B-9987. Helv. Chim. Acta 2010, 93, 2419–2425. [Google Scholar] [CrossRef]
- Fickers, P. Antibiotic compounds from Bacillus: Why are they so amazing? Am. J. Biochem. Biotechnol. 2012, 8, 40–46. [Google Scholar] [CrossRef]
- Li, D.; Carr, G.; Zhang, Y.; Williams, D.E.; Amlani, A.; Bottriell, H.; Mui, A.L.F.; Andersen, R. Turnagainolides A and B, cyclic depsipeptides produced in culture by a Bacillus sp.: Isolation, structure elucidation, and synthesis. J. Nat. Prod. 2011, 74, 1093–1099. [Google Scholar] [CrossRef]
- Cane, D.E.; Walsh, C.T. The parallel and convergent universes of polyketide synthases and nonribosomal peptide synthetases. Chem. Biol. 1999, 6, 319–325. [Google Scholar] [CrossRef]
- Tareq, F.S.; Kim, J.-H.; Lee, M.A.; Lee, H.-S.; Lee, Y.-J.; Lee, J.-S.; Shin, H.J. Ieodoglucomides A and B from a marine-derived bacterium Bacillus licheniformis. Org. Lett. 2012, 14, 1464–1467. [Google Scholar] [CrossRef]
- Yu, L.-L.; Li, Z.-Y.; Peng, C.-S.; Li, Z.-Y.; Guo, Y.-W. Neobacillamide A, a novel thiazole-containing alkaloid from the marine bacterium Bacillus vallismortis C89, associated with South China Sea Sponge Dysidea avar. Helv. Chim. Acta 2009, 92, 607–612. [Google Scholar] [CrossRef]
- Okazaki, H.; Kishi, T.; Beppu, T.; Arima, K. A new antibiotic baciphelacin. J. Antibiot. 1975, 28, 717–719. [Google Scholar] [CrossRef]
- Itoh, J.; Omoto, S.; Shomura, T.; Nishizawa, N.; Miyado, S.; Yuda, Y.; Shibata, U.; Inouye, S. Amicoumacin-A, a new antibiotic with strong antiinflammatory and antiulcer activity. J. Antibiot. 1981, 34, 611–613. [Google Scholar] [CrossRef]
- Itoh, J.; Omoto, S.; Shomura, T.; Nishizawa, N.; Miyado, S.; Yuda, Y.; Shibata, U.; Inouye, S. Chemical structures of amicoumacins produced by Bacillus pumilus. Agric. Biol. Chem. 1982, 46, 1255–1259. [Google Scholar] [CrossRef]
- Shimojima, Y.; Hayashi, H.; Ooka, T.; Shibukawa, M. (Studies on AI-77s, microbial products with pharmacological activity) structures and the chemical nature of AI-77s. Tetrahedron Lett. 1982, 23, 5435–5438. [Google Scholar]
- Shimojima, Y.; Hayashi, H.; Ooka, T.; Shibukawa, M.; Iitaka, Y. Studies on AI-77s, microbial products with gastroprotective activity. Structures and the chemical nature of AI-77s. Tetrahedron 1984, 40, 2519–2527. [Google Scholar] [CrossRef]
- McInerney, B.V.; Taylor, W.C.; Lacey, M.J.; Akhurst, R.J.; Gregson, R.P. Biologically active metabolites from Xenorhabdus spp., Part 2. Benzopyran-1-one derivatives with gastroprotective activity. J. Nat. Prod. 1991, 54, 785–795. [Google Scholar] [CrossRef]
- Omura, S.; Iwai, Y.; Takahashi, Y.; Sadakane, N.; Nakagawa, A.; Oiwa, H.; Hasegawa, Y.; Ikai, T. Herbimycin, a new antibiotic produced by a strain of Streptomyces. J. Antibiot. 1979, 32, 255–261. [Google Scholar] [CrossRef]
- Li, Y.; Xu, Y.; Liu, L.; Han, Z.; Lai, P.Y.; Guo, X.; Zhang, X.; Lin, W.; Qian, P.Y. Five new amicoumacins isolated from a marine-derived bacterium Bacillus subtilis. Mar. Drugs 2012, 10, 319–328. [Google Scholar] [CrossRef]
- Fulco, A.J. Fatty acid metabolism in bacteria. Prog. Lipid Res. 1983, 22, 133–160. [Google Scholar] [CrossRef]
- Fang, J.; Kato, C. FAS or PKS, lipid biosynthesis and stable carbon isotope fractionation in deep-sea piezophilic bacteria. Commun. Curr. Res. Educ. Top. Trends Appl. Microbiol. 2007, 1, 190–200. [Google Scholar]
- Kolattukudy, P.E.; Bohnet, S.; Sasaki, G.; Rogers, L. Developmental changes in the expression of S-acyl fatty acid synthase thioesterase gene and lipid composition in the uropygial gland of mallard ducks (Anas platyrhynchous). Arch. Biochem. Biophys. 1991, 284, 201–206. [Google Scholar] [CrossRef]
- Carballeira, N.M.; Ilieva, M.; Miranda, C.; Tzvetkova, I.; Lozano, C.M.; Nechev, J.T.; Ivanova, A.; Stefanov, K. Characterization of novel methyl-branched chain fatty acids from a halophilic Bacillus species. J. Nat. Prod. 2001, 64, 256–259. [Google Scholar] [CrossRef]
- Mondol, M.A.M.; Tareq, F.S.; Kim, J.-H.; Lee, M.A.; Lee, H.-S.; Lee, Y.-J.; Lee, J.-S.; Shin, H.J. New antimicrobial compounds from a marine-derived Bacillus sp. J. Antibiot. 2013, 66, 89–95. [Google Scholar] [CrossRef]
- Idriss, E.E.S.; Bochow, H.; Ross, H.; Borriss, R.Z. Use of Bacillus subtilis as biocontrol agent. VI. Phytohormone-like action of culture filtrates prepared from plant growth-promoting Bacillus amyloliquefaciens FZB24, FZB42, FZB45 and Bacillus subtilis FZB37. J. Plant Dis. Prot. 2004, 111, 583–597. [Google Scholar]
- Krebs, B.; Höding, B.; Kübart, S.M.; Workie, A.; Junge, H.; Schmiedeknecht, G.; Grosch, P.; Bochow, H.; Heves, M.Z. Use of Bacillus subtilis as biocontrol agent: Activities and characterization of Bacillus subtilis strains. J. Plant Dis. Prot. 1998, 105, 181–197. [Google Scholar]
- Koumoutsi, A.; Chen, X.-H.; Henne, A.; Liesegang, H.; Hitzeroth, G.; Franke, P.; Vater, J.; Borriss, R. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J. Bacteriol. 2004, 184, 1084–1096. [Google Scholar]
- Chen, X.H.; Vater, J.; Piel, J.; Franke, P.; Scholz, R.; Schneider, K.; Koumoutsi, A.; Hitzeroth, G.; Grammel, N.; Strittmatter, A.W.; et al. Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB42. J. Bacteriol. 2006, 188, 4024–4036. [Google Scholar] [CrossRef]
- Schneider, K.; Chen, X.-H.; Vater, J.; Franke, P.; Nicholson, G.; Borriss, R.; Süssmuth, R.D. Macrolactin is the polyketide biosynthesis product of the pks2 cluster of Bacillus amyloliquefaciens FZB42. J. Nat. Prod. 2007, 70, 1417–1423. [Google Scholar] [CrossRef]
- Zimmerman, S.B.; Schwartz, C.D.; Monaghan, R.L.; Pelak, B.A.; Weissberger, B.; Gilfillan, E.C.; Mochales, S.; Hernandez, S.; Currie, S.A.; Tejera, E.; et al. Difficidin and oxydifficidin: Novel broad spectrum antibacterial antibiotics produced by Bacillus subtilis. I. Production, taxonomy and antibacterial activity. J. Antibiot. 1987, 40, 1677–1681. [Google Scholar] [CrossRef]
- Chen, X.H.; Scholz, R.; Borriss, M.; Junge, H.; Moegel, G.; Kunz, S.; Borriss, R. Difficidin and bacilysin produced by plant-associated Bacillus amyloliquefaciens are efficient in controlling fire blight disease. J. Biotechnol. 2009, 140, 38–44. [Google Scholar] [CrossRef]
- Zweerink, M.M.; Edison, A. Difficidin and oxydifficidin: Novel broad spectrum antibacterial antibiotics produced by Bacillus subtilis. III. Mode of action of difficidin. J. Antibiot. 1987, 40, 1692–1697. [Google Scholar] [CrossRef]
- Lu, X.L.; Xu, Q.Z.; Liu, X.Y.; Cao, X.; Ni, K.Y.; Jiao, B.H. Marine Drugs—Macrolactins. Chem. Biodivers. 2008, 5, 1669–1674. [Google Scholar]
- Zheng, C.-J.; Lee, S.; Lee, C.-H.; Kim, W.-G.J. Macrolactins O–R, glycosylated 24-membered lactones from Bacillus sp. AH159-1. J. Nat. Prod. 2007, 70, 1632–1635. [Google Scholar] [CrossRef]
- Nagao, T.; Adachi, K.; Sakai, M.; Nishijima, M.; Sano, H. Novel macrolactins as antibiotic lactones from a marine bacterium. J. Antibiot. 2001, 54, 333–339. [Google Scholar] [CrossRef]
- Yoo, J.; Zheng, C.; Lee, S.; Kwak, J.; Kim, W. Macrolactin N, a new peptide deformylase inhibitor produced by Bacillus subtilis. Bioorg. Med. Chem. Lett. 2006, 16, 4889–4892. [Google Scholar] [CrossRef]
- Xue, C.M.; Tian, L.; Xu, M.J.; Deng, Z.W.; Lin, W.H. A new 24-membered lactones and a new polyene δ-lactone from the marine bacterium Bacillus marinus. J. Antibiot. 2008, 61, 668–674. [Google Scholar] [CrossRef]
- Jaruchoktaweechai, C.; Suwanborirux, K.; Tanasupawatt, S.; Kittakoop, P.; Menasveta, P. New macrolactins from a marine Bacillus sp. Sc026. J. Nat. Prod. 2000, 63, 984–986. [Google Scholar] [CrossRef]
- Romero-Tabarez, M.; Jansen, R.; Sylla, M.; Lunsdorf, H.; Haubler, S.; Santosa, D.A.; Timmis, K.N.; Molinari, G. 7-O-malonyl macrolactin A, a new macrolactin antibiotic from Bacillus subtilis active against methicillin-resistant Staphylococcus aureus, vancomycin-resistant enterococci, and a small-colony variant of Burkholderia cepacia. Antimicrob. Agents Chemother. 2006, 50, 1701–1709. [Google Scholar] [CrossRef]
- Mondol, M.A.M.; Kim, J.-H.; Lee, M.A.; Lee, H.-S.; Lee, Y.-J.; Shin, H.J. Macrolactin W, a new antibacterial macrolide from a marine Bacillus sp. Bioorg. Med. Chem. Lett. 2011, 21, 3832–3835. [Google Scholar] [CrossRef]
- Mondol, M.A.M.; Tareq, F.S.; Kim, J.-H.; Lee, M.A.; Lee, H.-S.; Lee, Y.-J.; Lee, J.-S.; Shin, H.J. Cyclic ether-containing macrolactins, antimicrobial 24-membered isomeric macrolactones from a marine Bacillus sp. J. Nat. Prod. 2011, 74, 2582–2587. [Google Scholar] [CrossRef]
- Losi, E.; Amrhein, C.; Frankenberger, W.T.J. Environmental biochemistry of chromium. Rev. Environ. Contam. Toxicol. 1994, 136, 91–121. [Google Scholar] [CrossRef]
- Katz, S.A.; Salem, H. The Biological and Environmental Chemistry of Chromium; VCH Publishers, Inc.: New York, NY, USA, 1994. [Google Scholar]
- Anderson, R.A. Essentiality of Cr in humans. Sci. Total Environ. 1989, 86, 75–81. [Google Scholar] [CrossRef]
- Anderson, R.A. Chromium as an essential nutrient for humans. Regul. Toxicol. Pharmacol. 1997, 26, 35–41. [Google Scholar] [CrossRef]
- Costa, M. Toxicity and carcinogenicity of Cr(VI) in animal models and humans. Crit. Rev. Toxicol. 1997, 27, 431–442. [Google Scholar] [CrossRef]
- Zhitkovich, A.; Voitkun, V.; Costa, M. Formation of the aminoacid-DNA complexes by hexavalent and trivalent chromium in vitro: Importance of trivalent chromium and the phosphate group. Biochemistry 1996, 35, 7275–7282. [Google Scholar] [CrossRef]
- Syracuse Research Corporation, Toxicological Profile for Chromium; Prepared for U.S. Department Health and Human Services, Public Health Service, Agency for Toxic Substances and Disease Registry, under Contract No. 205-88-0608; ATSDR: Atlanta, GA, USA, 1993.
- Krishnamurthy, S.; Wilkens, M.M. Environmental chemistry of Cr. Northeast. Geol. 1994, 16, 14–17. [Google Scholar]
- Pawlisz, A.V. Canadian water quality guidelines for Cr. Environ. Toxicol. Water Qual. 1997, 12, 123–161. [Google Scholar] [CrossRef]
- Kavitha, V.; Radhakrishnan, N.; Gnanamani, A.; Mandal, A.B. Management of chromium induced oxidative stress by marine Bacillus licheniformis. Biol. Med. 2011, 3, 16–26. [Google Scholar]
- De Vrind, J.P.M.; Boogerd, F.C.; de Vrind-de Jong, E.W. Manganese reduction by a marine Bacillus species. J. Bacteriol. 1986, 167, 30–34. [Google Scholar]
- Jorge, R.-I.; Soto, L.A.; Federico, P.-O. Heavy-metal accumulation in the hydrothermal vent clam Vesicomya gigas from Guaymas basin, Gulf of California. Deep Sea Res. 2003, 50, 675–761. [Google Scholar] [CrossRef]
- Shoresh, M.; Harman, G.; Mastouri, F. Induced systemic resistance and plant responses to fungal biocontrol agents. Annu. Rev. Phytopathol. 2010, 48, 21–43. [Google Scholar] [CrossRef]
- Baig, D.N.; Mehnaz, S. Determination and distribution of cry-type genes in halophilc Bacillus thuringiensis isolates of Arabian Sea sedimentary rocks. Microbiol. Res. 2010, 165, 376–383. [Google Scholar] [CrossRef]
- Cawoy, H.; Bettiol, W.; Fickers, P.; Ongena, M. Bacillus-Based Biological Control of Plant Diseases; InTech: New York, NY, USA, 2011 October 21; ISBN 978-953-307-459-7. [Google Scholar]
- Hoch, J.; Sonenshein, A.; Losick, A. Bacillus subtilis and Other Gram-Positive Bacteria: Biochemistry, Physiology and Molecular Genetics; American Society for Microbiology: Washington, DC, USA, 1993. [Google Scholar]
- Devaraja, T.; Banerjee, S.; Yusoff, F. A holistic approach for selection of Bacillus spp. as a bioremediator for shrimp postlarvae culture. Turk. J. Biol. 2013, 37, 92–100. [Google Scholar]
- Nells, H.J.; de Leenheen, A.P. Microbial source of carotenoid pigments used in foods and feeds. J. Appl. Bacteriol. 1991, 70, 181–191. [Google Scholar] [CrossRef]
- Stafsnes, M.H.; Josefsen, K.D.; Kildahl-Andersen, G.; Valla, S.; Ellingsen, T.E.; Bruheim, P. Isolation and characterization of marine pigmented bacteria from Norwegian coastal waters and screening for carotenoids with UVA-blue light absorbing properties. J. Microbiol. 2010, 48, 16–23. [Google Scholar] [CrossRef]
- Sy, C.; Gleize, B.; Chamot, S.; Dangles, O.; Carlin, F.; Veyrat, C.C.; Borel, P. Glycosyl carotenoids from marine spore-forming Bacillus sp. strains are readily bioaccessible and bioavailable. Food Res. Int. 2013, 51, 914–923. [Google Scholar] [CrossRef]
- Duc, L.H.; Fraser, P.D.; Tam, N.K.M.; Cutting, S.M. Carotenoids present in halotolerant Bacillus spore formers. FEMS Microbiol. Lett. 2006, 255, 215–224. [Google Scholar]
- Nugraheni, S.A.; Khoeri, M.M.; Kusmita, L.; Widyastut, Y.; Radjasa, O.K. Characterization of carotenoid pigments from bacterial symbionts of seagrass Thalassia hemprichii. J. Coast. Dev. 2010, 14, 51–60. [Google Scholar]
- Desal, C.; Pathak, H.; Madamwar, D. Advances in molecular and “omics” technologies to gauge microbial communities and bioremediation at xenobiotic/anthropogen contaminated sites. Bioresour. Technol. 2010, 101, 1558–1569. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Mondol, M.A.M.; Shin, H.J.; Islam, M.T. Diversity of Secondary Metabolites from Marine Bacillus Species: Chemistry and Biological Activity. Mar. Drugs 2013, 11, 2846-2872. https://doi.org/10.3390/md11082846
Mondol MAM, Shin HJ, Islam MT. Diversity of Secondary Metabolites from Marine Bacillus Species: Chemistry and Biological Activity. Marine Drugs. 2013; 11(8):2846-2872. https://doi.org/10.3390/md11082846
Chicago/Turabian StyleMondol, Muhammad Abdul Mojid, Hee Jae Shin, and Mohammad Tofazzal Islam. 2013. "Diversity of Secondary Metabolites from Marine Bacillus Species: Chemistry and Biological Activity" Marine Drugs 11, no. 8: 2846-2872. https://doi.org/10.3390/md11082846
APA StyleMondol, M. A. M., Shin, H. J., & Islam, M. T. (2013). Diversity of Secondary Metabolites from Marine Bacillus Species: Chemistry and Biological Activity. Marine Drugs, 11(8), 2846-2872. https://doi.org/10.3390/md11082846