Evolution and Distribution of Saxitoxin Biosynthesis in Dinoflagellates
Abstract
:1. Introduction
1.1. Harmful Algal Blooms and Saxitoxin-Synthesis in Two Kingdoms of Life
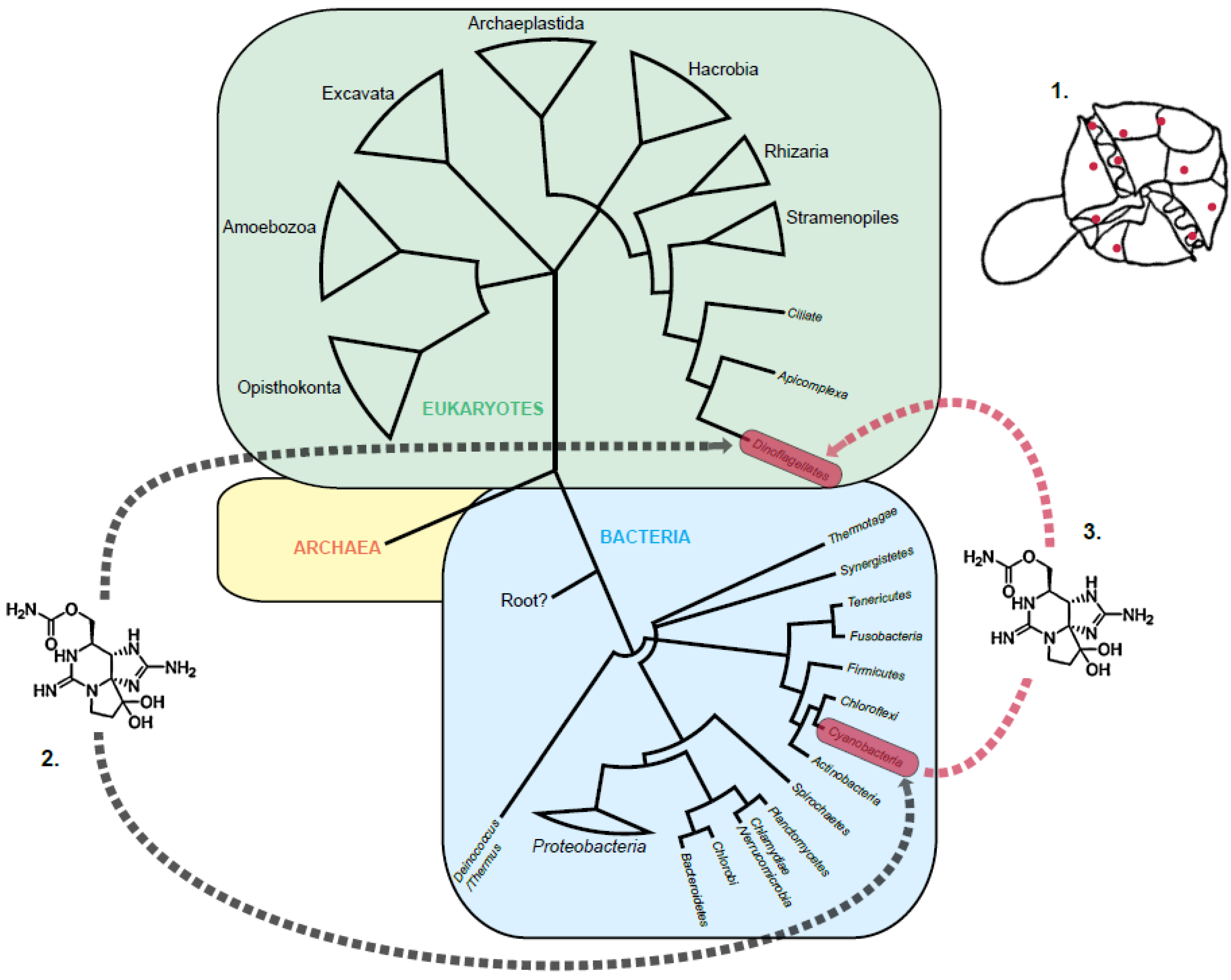
1.2. The STX Genes in Cyanobacteria and Dinoflagellates
2. Re-Evaluation of the Three Theories of STX Evolution in Dinoflagellates
2.1. Co-Cultured Bacteria
2.2. Convergent Evolution
2.3. Horizontal Gene Transfer
3. The Source of STX in Dinoflagellates
3.1. A Proteobacterial Source of STX
3.2. A Cyanobacterial Source of STX
3.3. Multiple Sources of STX
4. The Distribution of STX in Dinoflagellates
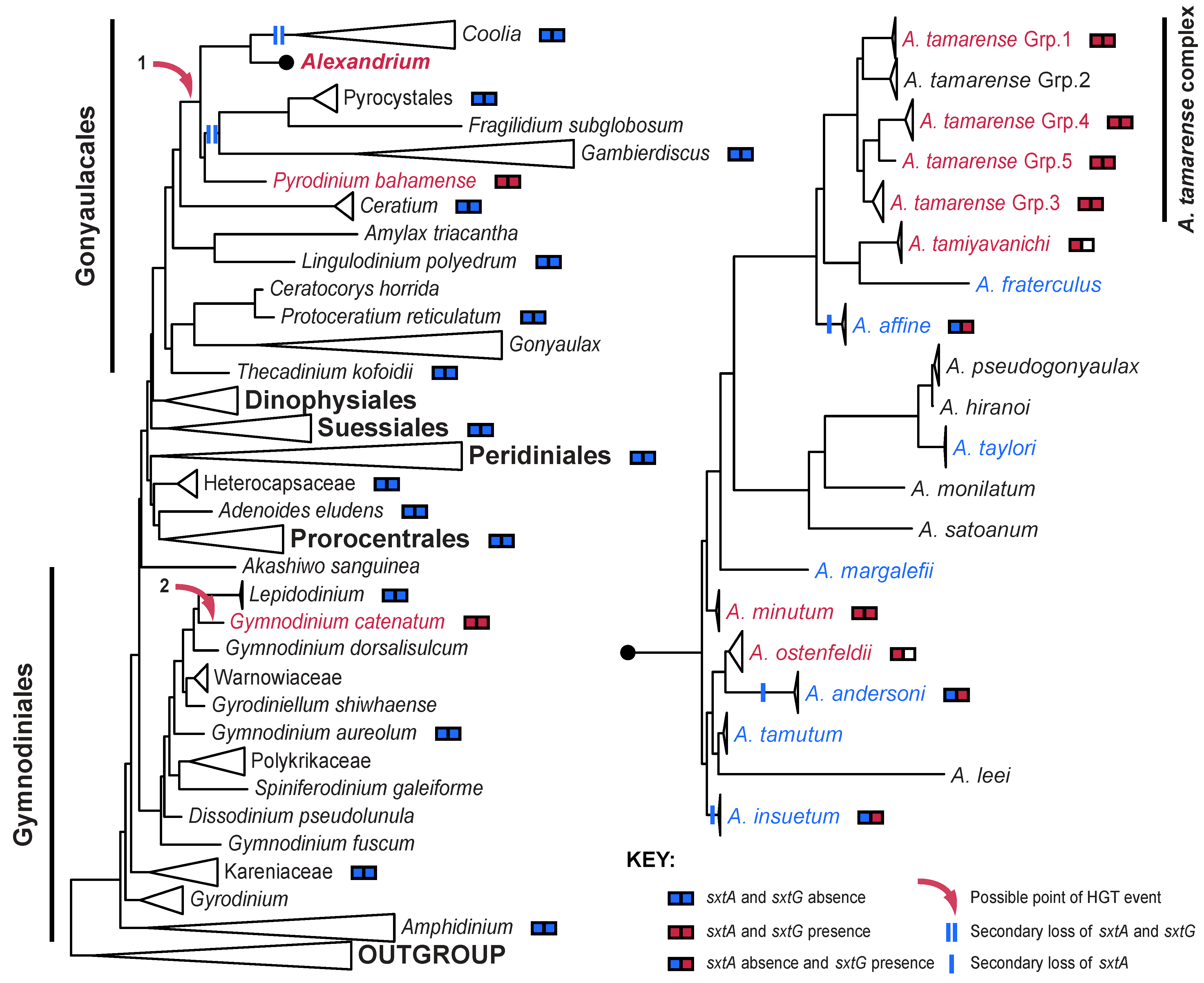
5. Conclusions
5.1. A Revised Theory of STX Evolution in Dinoflagellates
5.2. Outstanding Questions for Future Research
Appendix: Glossary and Definitions
| Alkaloid | An organic heterocyclic secondary metabolite containing nitrogen as the key atom. Many alkaloids have potent pharmacological effects, e.g., caffeine and nicotine. |
| Horizontal gene transfer (HGT) | Also referred to as lateral gene transfer (LGT). Refers to the movement of genetic information across mating barriers, between distantly related organisms. In contrast, vertical inheritance or gene transfer occurs between a parent and its offspring. |
| High performance liquid chromatography (HPLC) | A chromatographic technique used to separate a mixture of compounds to its individual components. Resulting components can be identified and quantified. |
| Monocistronic | An mRNA that only has a single open reading frame (ORF) and is thus translated into a single protein. This is associated with translation in eukaryotes. |
| Monocistronic | A taxonomic grouping that contains the common ancestor and all of its decedents. |
| Non-Ribosomal peptide synthetase (NRPS) | Synthesizes non-ribosomal peptides (NRP) independent of ribosomes and mRNA. Each NRPS synthesizes a single peptide, though they can also work in conjunction with polyketide synthases (PKSs) to give hybrid products. Modular type I PKSs and NPRSs form megasynthetases that generally follow a collinearity rule [58], where one module extends a growing acyl or peptidyl chain by one particular unit. This enables the prediction of the chemical structure of their metabolite products [58]. All hybrid NRPS/PKS investigated so far are modular enzymes [59]. |
| Paraphyletic | A taxonomic group that includes some, but not all, of the descendants of a common ancestor. |
| Paraphyletic | Taxonomic grouping that has no shared common ancestry. |
| Polyketide | A class of secondary metabolites that contain alternating carbonyl and methylene groups via a polyketide synthase (PKS) process. |
| Type I polyketide synthases (PKSs) | PKSs are multi-domain enzymes or enzyme complexes that synthesize polyketides, a class of secondary metabolites. PKSs are classified into three groups, with Type I PKSs being multifunctional enzymes organized into large modules. At the genomic level, modular PKSs can be encoded as single domain enzymes in dinoflagellates [24]. |
| Secondary metabolites | Metabolic compounds that are non-essential for the primary metabolic functions of an organism. They can largely be divided into three classes: alkaloids, terpenoids and phenolics. |
| Spliced leader (SL) trans-splicing | A short RNA fragment from a small noncoding RNA (SL RNA) is transplanted to the 5′ end of independently transcribed pre-mRNAs yielding mature mRNAs. The process converts a polycistronic transcript into translatable monocistronic mRNAs [60]. |
| SxtA | First gene of STX-synthesis. It has four catalytic domains: A SAM-dependent methyltransferase (A1), GCN5-related N-acetyltransferase (A2), acyl carrier protein (A3) and a class II aminotransferase (A4) [21]. |
| SxtG | An amidinotransferase enzyme that encodes the second step is saxitoxin biosynthesis [21]. |
| Toxicity | The degree to which a substance can damage/harm an organism or its parts; this is therefore a species-specific definition. |
Acknowledgements
Conflict of Interest
References
- Taylor, F.J.R.; Hoppenrath, M.; Saldarriaga, J.F. Dinoflagellate diversity and distribution. Biodivers. Conserv. 2008, 17, 407–418. [Google Scholar] [CrossRef]
- Hallegraeff, G.M. A review of harmful algal blooms and their apparent global increase. Phycologia 1993, 32, 79–99. [Google Scholar] [CrossRef]
- Wiese, M.; D’Agostino, P.M.; Mihali, T.K.; Moffitt, M.C.; Neilan, B.A. Neurotoxic alkaloids: Saxitoxin and its analogs. Mar. Drugs 2010, 8, 2185–2211. [Google Scholar] [CrossRef]
- Catterall, W.A.; Morrow, C.S.; Hartshorne, R.P. Neurotoxin binding to receptor sites associated with voltage sensitive sodium channels in intact, lysed, and detergent solubilized brain membranes. J. Biol. Chem. 1979, 254, 1379–1387. [Google Scholar]
- Llewellyn, L.E. Saxitoxin, a toxic marine natural product that targets a multitude of receptors. Nat. Prod. Rep. 2006, 23, 200–222. [Google Scholar] [CrossRef]
- Hallegraeff, G.M. Harmful Algal Blooms: A Global Overview. In Manual of Harmful Marine Microalgae; Hallegraeff, G.M., Anderson, D.M., Cembella, A.D., Eds.; International Oceanographic Commission (IOC) Manual and Guides UNESCO: Paris, France, 1995; pp. 1–22. [Google Scholar]
- Hoagland, P.; Scatasta, S. The Economic Effects of Harmful Algal Blooms. In Ecology of Harmful Algae; Graneli, E., Turner, T., Eds.; Springer-Verlag: Dordrecht, The Netherlands, 2006; pp. 391–402. [Google Scholar]
- Adl, S.M.; Simpson, A.G.B.; Lane, C.E.; Lukes, J.; Bass, D.; Bowser, S.S.; Brown, M.W.; Burki, F.; Dunthorn, M.; Hampl, V.; et al. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 2012, 59, 429–493. [Google Scholar] [CrossRef]
- Wu, D.Y.; Hugenholtz, P.; Mavromatis, K.; Pukall, R.; Dalin, E.; Ivanova, N.N.; Kunin, V.; Goodwin, L.; Wu, M.; Tindall, B.J.; et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 2009, 462, 1056–1060. [Google Scholar] [CrossRef]
- Usup, G.; Ahmad, A.; Matsuoka, K.; Lim, P.T.; Leaw, C.P. Biology, ecology and bloom dynamics of the toxic marine dinoflagellate Pyrodinium bahamense. Harmful Algae 2012, 14, 301–312. [Google Scholar] [CrossRef]
- Gárate-Lizárraga, I.; Bustillos-Guzmán, J.J.; Morquecho, L.; Band-Schmidt, C.J.; Alonso-Rodriguez, R.; Erler, K.; Luckas, B.; Reyes-Salinas, A.; Góngora-González, D.T. Comparative paralytic shellfish toxin profiles in the strains of Gymnodinium catenatum Graham from the Gulf of California, Mexico. Mar. Pollut. Bull. 2005, 50, 211–217. [Google Scholar] [CrossRef]
- Hallegraeff, G.M. Harmful Algal Blooms: A Global Overview. In Manual on Harmful Marine Microalgae; Hallegraeff, G.M., Anderson, D.M., Cembella, A.D., Eds.; Imprimerie Landais: Paris, France, 2005; pp. 25–49. [Google Scholar]
- Cembella, A.D. Ecophysiology and Metabolism of Paralytic Shellfish Toxins in Marine Microalgae. In Physiological Ecology of Harmful Algal Blooms; Anderson, D.M., Cembella, A.D., Hallegraeff, G.M., Eds.; Springer: Berlin, Germany, 1998; pp. 381–403. [Google Scholar]
- Yang, I.; John, U.; Beszteri, S.; Glockner, G.; Krock, B.; Goesmann, A.; Cembella, A.D. Comparative gene expression in toxic versus non-toxic strains of the marine dinoflagellate Alexandrium minutum. BMC Genomics 2010, 11. [Google Scholar] [CrossRef]
- Orr, R.J.S.; Stüken, A.; Rundberget, T.; Eikrem, W.; Jakobsen, K.S. Improved phylogenetic resolution of toxic and non-toxic Alexandrium strains using a concatenated rDNA approach. Harmful Algae 2011, 10, 676–688. [Google Scholar] [CrossRef]
- Murray, S.A.; Mihali, T.K.; Neilan, B.A. Extraordinary conservation, gene loss, and positive selection in the evolution of an ancient neurotoxin. Mol. Biol. Evol. 2011, 28, 1173–1182. [Google Scholar] [CrossRef]
- Kodama, M.; Ogata, T.; Sato, S. Bacterial production of saxitoxin. Agric. Biol. Chem. 1988, 52, 1075–1077. [Google Scholar] [CrossRef]
- Plumley, F.G. Purification of an enzyme involved in saxitoxin synthesis. J. Phycol. 2001, 37, 926–928. [Google Scholar] [CrossRef]
- Schopf, J.W. The Fosil Record: Tracing the Roots of the Cyanobacterial Lineage. In The Ecology of Cyanobacteria: Their Diversity in Time and Space; Whitton, B.A., Potts, M., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2000; pp. 13–35. [Google Scholar]
- Moustafa, A.; Loram, J.E.; Hackett, J.D.; Anderson, D.M.; Plumley, F.G.; Bhattacharya, D. Origin of saxitoxin biosynthetic genes in cyanobacteria. PLoS One 2009, 4, e5758. [Google Scholar]
- Kellmann, R.; Mihali, T.K.; Jeon, Y.J.; Pickford, R.; Pomati, F.; Neilan, B.A. Biosynthetic intermediate analysis and functional homology reveal a saxitoxin gene cluster in cyanobacteria. Appl. Environ. Microbiol. 2008, 74, 4044–4053. [Google Scholar] [CrossRef]
- Stüken, A.; Orr, R.J.S.; Kellmann, R.; Murray, S.A.; Neilan, B.A.; Jakobsen, K.S. Discovery of nuclear-encoded genes for the neurotoxin saxitoxin in dinoflagellates. PLoS One 2011, 6, e20096. [Google Scholar]
- Hackett, J.D.; Wisecaver, J.H.; Brosnahan, M.L.; Kulis, D.M.; Anderson, D.M.; Bhattacharya, D.; Plumley, F.G.; Erdner, D.L. Evolution of saxitoxin synthesis in cyanobacteria and dinoflagellates. Mol. Biol. Evol. 2013, 30, 70–78. [Google Scholar] [CrossRef]
- Kellmann, R.; Stüken, A.; Orr, R.J.S.; Svendsen, H.M.; Jakobsen, K.S. Biosynthesis and molecular genetics of polyketides in marine dinoflagellates. Mar. Drugs 2010, 8, 1011–1048. [Google Scholar] [CrossRef]
- Shimizu, Y. Microalgal metabolites: A new perspective. Annu. Rev. Microbiol. 1996, 50, 431–465. [Google Scholar] [CrossRef]
- Harlow, L.D.; Koutoulis, A.; Hallegraeff, G.M. S-Adenosylmethionine synthetase genes from eleven marine dinoflagellates. Phycologia 2007, 46, 46–53. [Google Scholar] [CrossRef]
- Hackett, J.D.; Scheetz, T.E.; Yoon, H.S.; Soares, M.B.; Bonaldo, M.F.; Casavant, T.L.; Bhattacharya, D. Insights into a dinoflagellate genome through expressed sequence tag analysis. BMC Genomics 2005, 6. [Google Scholar] [CrossRef]
- Orr, R.J.S.; Stüken, A.; Murray, S.A.; Jakobsen, K.S. Evolutionary acquisition and loss of saxitoxin biosynthesis in dinoflagellates: The second “core” gene—sxtG. Appl. Environ. Microbiol. 2013, 79, 2128–2136. [Google Scholar] [CrossRef]
- Silva, E.S. Intracellular bacteria: The origin of dinoflagellate toxicity. J. Environ. Pathol. Toxicol. 1990, 10, 124–128. [Google Scholar]
- Green, D.H.; Llewellyn, L.E.; Negri, A.P.; Blackburn, S.I.; Bolch, C.J.S. Phylogenetic and functional diversity of the cultivable bacterial community associated with the paralytic shellfish poisoning dinoflagellate Gymnodinium catenatum. FEMS Microbiol. Ecol. 2004, 47, 345–357. [Google Scholar] [CrossRef]
- Wichels, A.; Hummert, C.; Elbrachter, M.; Luckas, B.; Schutt, C.; Gerdts, G. Bacterial diversity in toxic Alexandrium tamarense blooms off the Orkney Isles and the Firth of Forth. Helgol. Mar. Res. 2004, 58, 93–103. [Google Scholar] [CrossRef]
- Martins, C.A.; Alvito, P.; Tavares, M.J.; Pereira, P.; Doucette, G.; Franca, S. Reevaluation of production of paralytic shellfish toxin by bacteria associated with dinoflagellates of the Portuguese coast. Appl. Environ. Microbiol. 2003, 69, 5693–5698. [Google Scholar] [CrossRef]
- Lu, Y.H.; Chai, T.J.; Hwang, D.F. Isolation of bacteria from toxic dinoflagellate Alexandrium minutum and their effects on algae toxicity. J. Nat. Toxins 2000, 9, 409–417. [Google Scholar]
- Baker, T.R.; Doucette, G.J.; Powell, C.L.; Boyer, G.L.; Plumley, F.G. GTX(4) imposters: Characterization of fluorescent compounds synthesized by Pseudomonas stutzeri SF/PS and Pseudomonas/Alteromonas PTB-1, symbionts of saxitoxin-producing Alexandrium spp. Toxicon 2003, 41, 339–347. [Google Scholar] [CrossRef]
- Hold, G.L.; Smith, E.A.; Birkbeck, T.H.; Gallacher, S. Comparison of paralytic shellfish toxin (PST) production by the dinoflagellates Alexandrium lusitanicum NEPCC 253 and Alexandrium tamarense NEPCC 407 in the presence and absence of bacteria. FEMS Microbiol. Ecol. 2001, 36, 223–234. [Google Scholar] [CrossRef]
- Palacios, L.; Reguera, B.; Franco, J.; Marin, I. Phylogenetic diversity of bacteria associated with toxic and non-toxic strains of Alexandrium minutum. Afr. J. Mar. Sci. 2006, 28, 409–414. [Google Scholar] [CrossRef]
- Pichersky, E.; Gang, D.R. Genetics and biochemistry of secondary metabolites in plants: An evolutionary perspective. Trends Plant Sci. 2000, 5, 439–445. [Google Scholar] [CrossRef]
- Lukes, J.; Leander, B.S.; Keeling, P.J. Cascades of convergent evolution: The corresponding evolutionary histories of euglenozoans and dinoflagellates. Proc. Natl. Acad. Sci. USA 2009, 106, 9963–9970. [Google Scholar] [CrossRef]
- Gough, J. Convergent evolution of domain architectures (is rare). Bioinformatics 2005, 21, 1464–1471. [Google Scholar] [CrossRef]
- Jost, M.C.; Hillis, D.M.; Lu, Y.; Kyle, J.W.; Fozzard, H.A.; Zakon, H.H. Toxin-resistant sodium channels: Parallel adaptive evolution across a complete gene family. Mol. Biol. Evol. 2008, 25, 1016–1024. [Google Scholar] [CrossRef]
- De la Cruz, F.; Davies, J. Horizontal gene transfer and the origin of species: Lessons from bacteria. Trends Microbiol. 2000, 8, 128–133. [Google Scholar] [CrossRef]
- Keeling, P.J.; Palmer, J.D. Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 2008, 9, 605–618. [Google Scholar] [CrossRef]
- Yoon, H.S.; Hackett, J.D.; van Dolah, F.M.; Nosenko, T.; Lidie, K.L.; Bhattacharya, D. Tertiary endosymbiosis driven genome evolution in dinoflagellate algae. Mol. Biol. Evol. 2005, 22, 1299–1308. [Google Scholar] [CrossRef]
- Morse, D.; Salois, P.; Markovic, P.; Hastings, J.W. A nuclear-encoded form II RuBisCO in dinoflagellates. Science 1995, 268, 1622–1624. [Google Scholar]
- Wong, J.T.Y.; New, D.C.; Wong, J.C.W.; Hung, V.K.L. Histone-like proteins of the dinoflagellate Crypthecodinium cohnii have homologies to bacterial DNA-binding proteins. Eukaryot. Cell 2003, 2, 646–650. [Google Scholar] [CrossRef]
- Singh, S.P.; Hader, D.P.; Sinha, R.P. Bioinformatics evidence for the transfer of mycosporine-like amino acid core (4-deoxygadusol) synthesizing gene from cyanobacteria to dinoflagellates and an attempt to mutate the same gene (YP_324358) in Anabaena variabilis PCC 7937. Gene 2012, 500, 155–163. [Google Scholar] [CrossRef]
- Silva, E.S. Relationship between dinoflagellates and intracellular bacteria. Mar. Algae Pharmacol. Sci. 1982, 2, 269–288. [Google Scholar]
- Hotopp, J.C.D.; Clark, M.E.; Oliveira, D.C.S.G.; Foster, J.M.; Fischer, P.; Torres, M.C.; Giebel, J.D.; Kumar, N.; Ishmael, N.; Wang, S.L.; et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 2007, 317, 1753–1756. [Google Scholar] [CrossRef]
- Stüken, A.; Jakobsen, K.S. The cylindrospermopsin gene cluster of Aphanizomenon sp. strain 10E6: Organization and recombination. Microbiology 2010, 156, 2438–2451. [Google Scholar] [CrossRef]
- Salcedo, T.; Upadhyay, R.J.; Nagasaki, K.; Bhattacharya, D. Dozens of toxin-related genes are expressed in a nontoxic strain of the dinoflagellate Heterocapsa circularisquama. Mol. Biol. Evol. 2012, 29, 1503–1506. [Google Scholar] [CrossRef]
- Orr, R.J.S.; Murray, S.; Stüken, A.; Rhodes, L.; Jakobsen, K.S. When naked became armored: An eight-gene phylogeny reveals monophyletic origin of theca in dinoflagellates. PLoS One 2012, 7, e50004. [Google Scholar]
- Lilly, E.L.; Halanych, K.M.; Anderson, D.M. Species boundaries and global biogeography of the Alexandrium tamarense complex (Dinophyceae). J. Phycol. 2007, 43, 1329–1338. [Google Scholar] [CrossRef]
- Murray, S.A.; Wiese, M.; Neilan, B.A.; Orr, R.J.S.; de Salas, M.; Brett, S.; Hallegraeff, G. A reinvestigation of saxitoxin production and sxtA in the “non-toxic” Alexandrium tamarense Group V clade. Harmful Algae 2012, 18, 96–104. [Google Scholar] [CrossRef]
- Murray, S.A.; Wiese, M.; Stüken, A.; Brett, S.; Kellmann, R.; Hallegraeff, G.; Neilan, B.A. SxtA-based quantitative molecular assay to identify saxitoxin-producing harmful algal blooms in marine waters. Appl. Environ. Microbiol. 2011, 77, 7050–7057. [Google Scholar] [CrossRef]
- Suikkanen, S.; Kremp, A.; Hautala, H.; Krock, B. Paralytic shellfish toxins or spirolides? The role of environmental and genetic factors in toxin production of the Alexandrium ostenfeldii complex. Harmful Algae 2013, 26, 52–59. [Google Scholar] [CrossRef]
- Hii, K.S.; Lim, P.T.; Tan, T.H.; Leaw, C.P. Characterization of the Saxitoxin Biosynthetic Starting Gene, sxta in the Toxic Dinoflagellate Alexandrium tamiyavanichii. In Proceedins of The 12th Symposium of the Malaysian Society of Applied Biology: Solutions to Global Challenges and Issues, Kuala Terengganu, Malaysia, 1–3 June 2012; pp. 196–202.
- Wang, D.-Z.; Li, C.; Zhang, Y.; Wang, Y.-Y.; He, Z.-P.; Lin, L.; Hong, H.-S. Quantitative proteomic analysis of differentially expressed proteins in the toxicity-lost mutant of Alexandrium catenella (Dinophyceae) in the exponential phase. J. Proteomics 2012, 75, 5564–5577. [Google Scholar] [CrossRef]
- Callahan, B.; Thattai, M.; Shraiman, B.I. Emergent gene order in a model of modular polyketide synthases. Proc. Natl. Acad. Sci. USA 2009, 106, 19410–19415. [Google Scholar] [CrossRef]
- Doekel, S.; Marahiel, M.A. Biosynthesis of natural products on modular peptide synthetases. Metab. Eng. 2001, 3, 64–77. [Google Scholar] [CrossRef]
- Zhang, H.; Hou, Y.B.; Miranda, L.; Campbell, D.A.; Sturm, N.R.; Gaasterland, T.; Lin, S.J. Spliced leader RNA trans-splicing in dinoflagellates. Proc. Natl. Acad. Sci. USA 2007, 104, 4618–4623. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Orr, R.J.S.; Stüken, A.; Murray, S.A.; Jakobsen, K.S. Evolution and Distribution of Saxitoxin Biosynthesis in Dinoflagellates. Mar. Drugs 2013, 11, 2814-2828. https://doi.org/10.3390/md11082814
Orr RJS, Stüken A, Murray SA, Jakobsen KS. Evolution and Distribution of Saxitoxin Biosynthesis in Dinoflagellates. Marine Drugs. 2013; 11(8):2814-2828. https://doi.org/10.3390/md11082814
Chicago/Turabian StyleOrr, Russell J. S., Anke Stüken, Shauna A. Murray, and Kjetill S. Jakobsen. 2013. "Evolution and Distribution of Saxitoxin Biosynthesis in Dinoflagellates" Marine Drugs 11, no. 8: 2814-2828. https://doi.org/10.3390/md11082814
APA StyleOrr, R. J. S., Stüken, A., Murray, S. A., & Jakobsen, K. S. (2013). Evolution and Distribution of Saxitoxin Biosynthesis in Dinoflagellates. Marine Drugs, 11(8), 2814-2828. https://doi.org/10.3390/md11082814



