Sponge-Derived Kocuria and Micrococcus spp. as Sources of the New Thiazolyl Peptide Antibiotic Kocurin
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification and Diversity of Microccocaceae Sponge Isolates
 , PKS-II
, PKS-II  or NRPS
or NRPS  . Strains with antimicrobial activity against methicillin-resistant S. aureus MB5393 are highlighted in red.
. Strains with antimicrobial activity against methicillin-resistant S. aureus MB5393 are highlighted in red.
 , PKS-II
, PKS-II  or NRPS
or NRPS  . Strains with antimicrobial activity against methicillin-resistant S. aureus MB5393 are highlighted in red.
. Strains with antimicrobial activity against methicillin-resistant S. aureus MB5393 are highlighted in red. 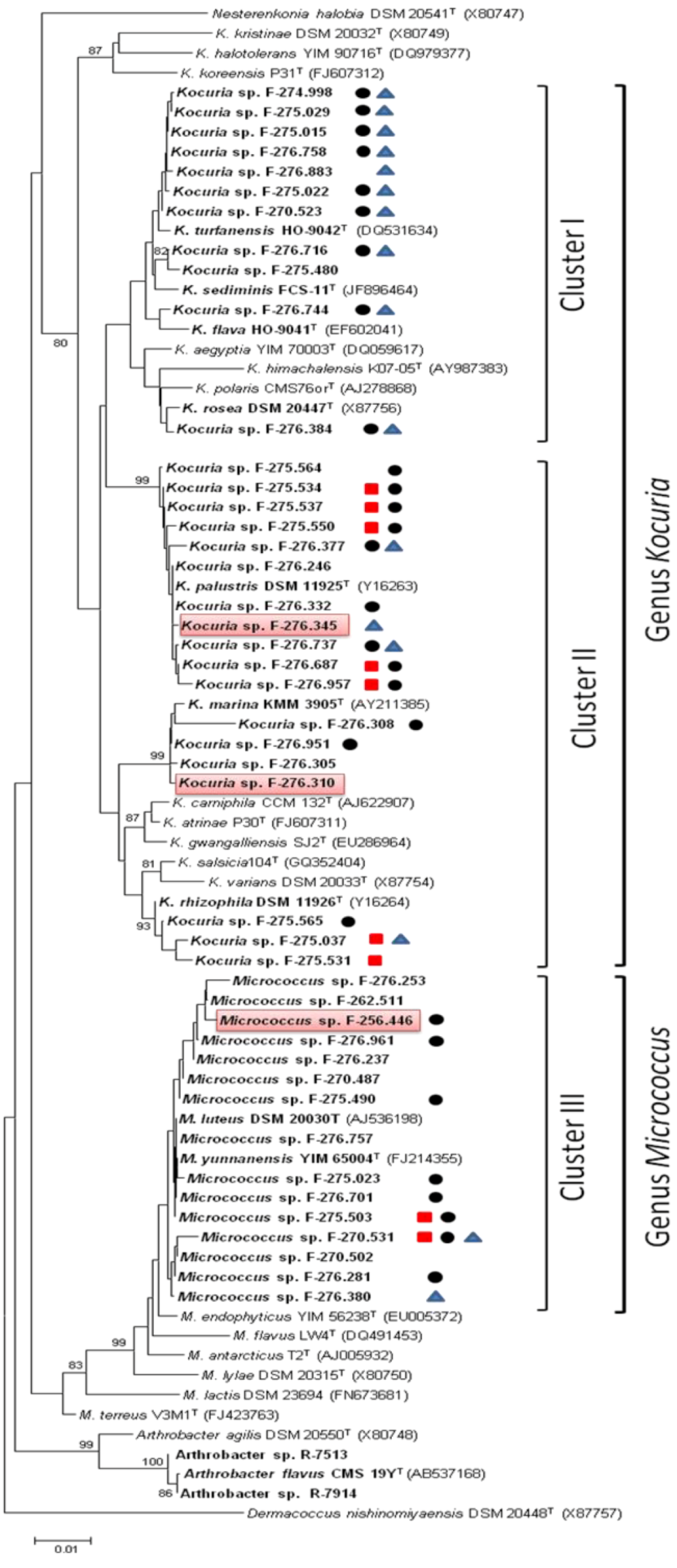
2.2. Detection of PKS and NRPS Genes
| Genera | Strains | PKS-I | PKS-II | NRPS | |||
|---|---|---|---|---|---|---|---|
| Total | (%) | Total | (%) | Total | % | ||
| Kocuria spp. | 29 | 7 | 24.1 | 21 | 72.4 | 14 | 48.3 |
| Micrococcus spp. | 15 | 2 | 13.3 | 8 | 53.3 | 2 | 13.3 |
| Total | 44 | 9 | 20.5 | 29 | 65.9 | 16 | 36.4 |
2.3. Evaluation of Antimicrobial Activities
| Antibacterial activities in agar diffusion assays (mm) | ||||
|---|---|---|---|---|
| Extract volume | Extract volume | |||
| Strain code | Identification | 10 μL | 10 μL | 20 μL |
| (96 well plate format) | (Nunc plate) | |||
| F-276,310 | Kocuriamarina | 5 | 6 | 8 |
| F-276,345 | Kocuriapalustris | 5 | 6 | 8 |
| F-256,446 | Micrococcus sp. | 4 | 5 | 7 |
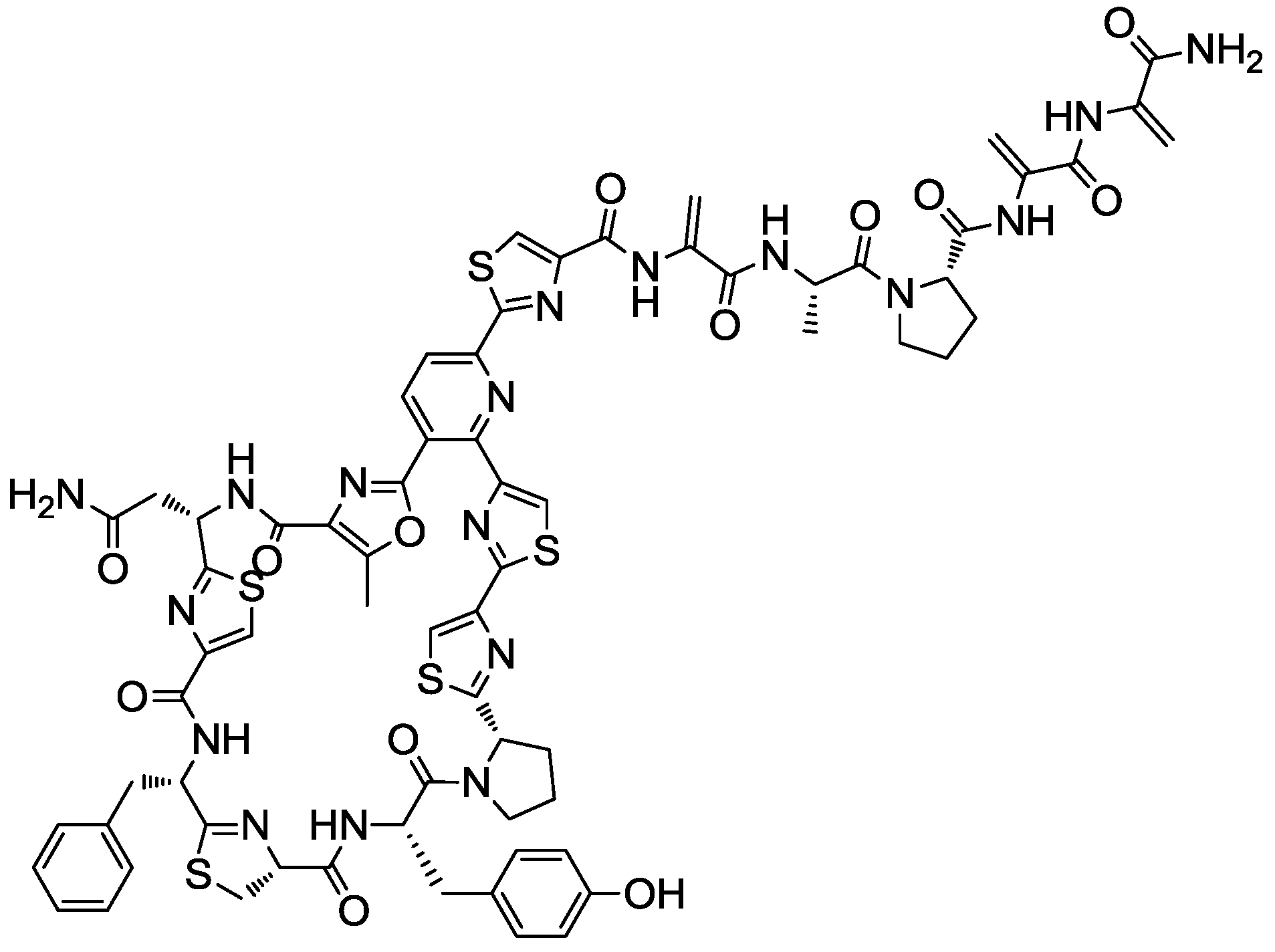
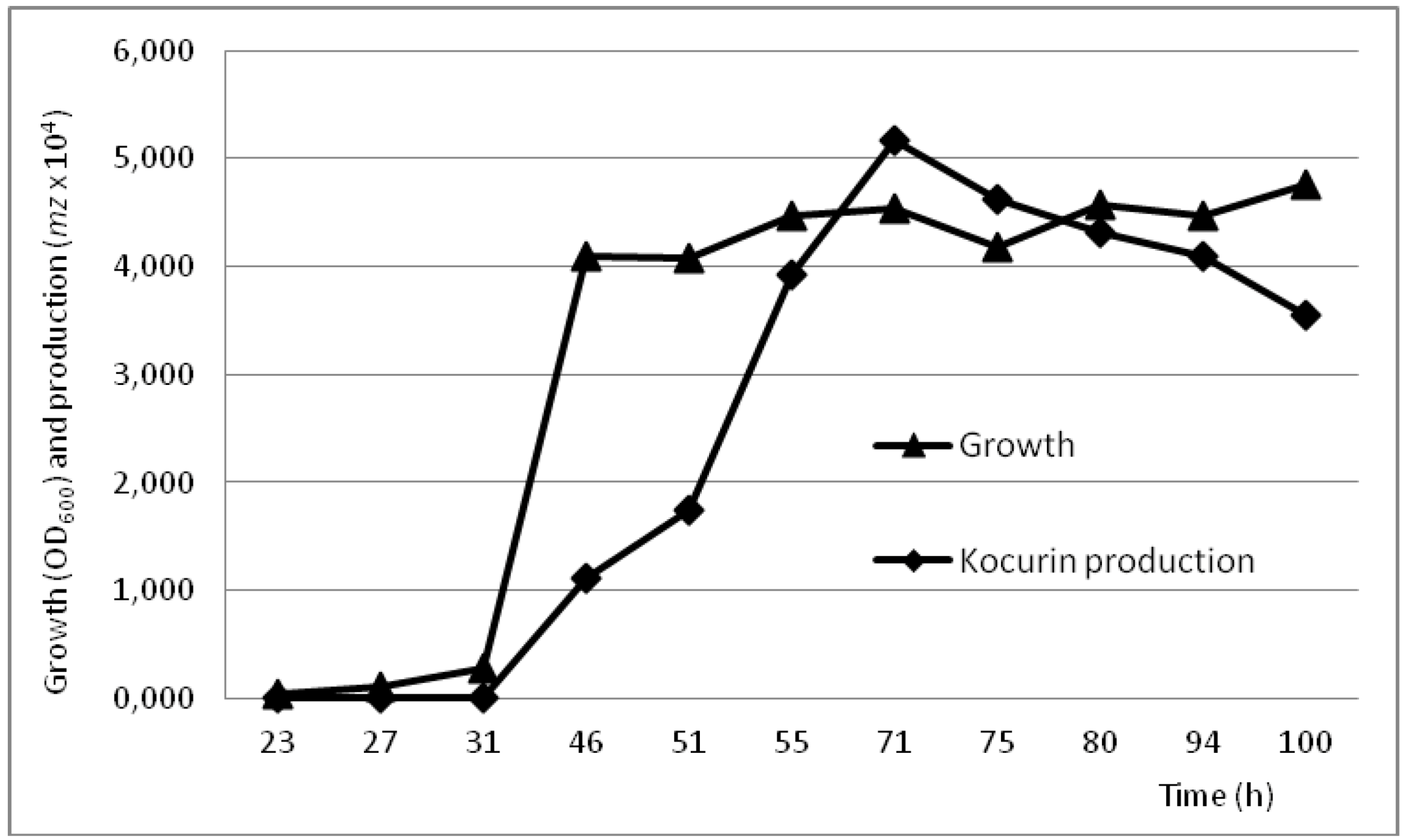
2.4. Production Conditions of the Thiazolyl Peptide Kocurin
2.5. Detection of Biosynthetic Pathways and Production of Bioactivities in Micrococcaceae
3. Experimental Section
3.1. Environmental Sampling
3.2. Strain Culture and DNA Extraction
3.3. Analysis of Fatty Acid by Gas Chromatography
3.4. Phylogenetic Analysis
3.5. PCR Amplification
3.6. Microfermentation and Extraction of Marine Microbial Secondary Metabolites
3.7. Production of Thiazolyl Peptides
3.8. Evaluation of Antimicrobial Activity
4. Conclusions
Acknowledgments
References
- Hentschel, U.; Usher, K.M.; Taylor, M.W. Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 2006, 55, 167–177. [Google Scholar] [CrossRef]
- Taylor, M.W.; Hill, R.T.; Piel, J.; Thacker, R.W.; Hentschel, U. Soaking it up: The complex lives of marine sponges and their microbial associates. ISME J. 2007, 1, 187–190. [Google Scholar] [CrossRef]
- Webster, N.S.; Taylor, M.W. Marine sponges and their microbial symbionts: Love and other relationships. Environ. Microbiol. 2012, 14, 335–346. [Google Scholar] [CrossRef]
- Berdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef]
- Mincer, T.J.; Jensen, P.R.; Kauffman, C.A.; Fenical, W. Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl. Environ. Microbiol. 2002, 68, 5005–5011. [Google Scholar] [CrossRef]
- Fenical, W.; Jensen, P.R. Developing a new resource for drug discovery: Marine actinomycete bacteria. Nat. Chem. Biol. 2006, 2, 666–673. [Google Scholar] [CrossRef]
- Lam, K.S. Discovery of novel metabolites from marine actinomycetes. Curr. Opin. Microbiol. 2006, 9, 245–251. [Google Scholar] [CrossRef]
- Gontang, E.A.; Fenical, W.; Jensen, P.R. Phylogenetic diversity of Gram-positive bacteria cultured from marine sediments. Appl. Environ. Microbiol. 2007, 73, 3272–3282. [Google Scholar] [CrossRef]
- Maldonado, L.A.; Stach, J.E.; Ward, A.C.; Bull, A.T.; Goodfellow, M. Characterization of Micromonosporae from aquatic environments using molecular taxonomic methods. Antonie Van Leeuwenhoek 2008, 94, 289–298. [Google Scholar] [CrossRef]
- Maldonado, L.A.; Fenical, W.; Jensen, P.R.; Kauffman, C.A.; Mincer, T.J.; Ward, A.C.; Bull, A.T.; Goodfellow, M. Goodfellow, M. Salinispora arenicola gen. nov., sp. nov. and Salinispora tropica sp. nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int. J. Syst. Evol. Microbiol. 2005, 55, 1759–1766. [Google Scholar]
- Montalvo, N.; Mohamed, N.; Enticknap, J.; Hill, R. Novel actinobacteria from marine sponges. Antonie Van Leeuwenhoek 2005, 87, 29–36. [Google Scholar] [CrossRef]
- Schneemann, I.; Nagel, K.; Kajahn, I.; Labes, A.; Wiese, J.; Imhoff, J.F. Comprehensive investigation of marine actinobacteria associated with the sponge Halichondria panicea. Appl. Environ. Microbiol. 2010, 76, 3702–3714. [Google Scholar] [CrossRef]
- Ayuso-Sacido, A.; Genilloud, O. New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: Detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb. Ecol. 2005, 49, 10–24. [Google Scholar] [CrossRef]
- Gontang, E.A.; Gaudencio, S.P.; Fenical, W.; Jensen, P.R. Sequence-based analysis of secondary-metabolite biosynthesis in marine actinobacteria. Appl. Environ. Microbiol. 2010, 76, 2487–2499. [Google Scholar] [CrossRef]
- Metsa-Ketela, M.; Halo, L.; Munukka, E.; Hakala, J.; Mantsala, P.; Ylihonko, K. Molecular evolution of aromatic polyketides and comparative sequence analysis of polyketide ketosynthase and 16S ribosomal DNA genes from various Streptomyces species. Appl. Environ. Microbiol. 2002, 68, 4472–4479. [Google Scholar] [CrossRef]
- Penesyan, A.; Kjelleberg, S.; Egan, S. Development of novel drugs from marine surface associated microorganisms. Mar. Drugs 2010, 8, 438–459. [Google Scholar] [CrossRef]
- Freel, K.C.; Nam, S.-J.; Fenical, W.; Jensen, P.R. Evolution of secondary metabolite genes in three closely related marine actinomycete species. Appl. Environ. Microbiol. 2011, 77, 7261–7270. [Google Scholar] [CrossRef]
- Fenical, W.; Jensen, P.R.; Palladino, M.A.; Lam, K.S.; Lloyd, G.K.; Potts, B.C. Discovery and development of the anticancer agent salinosporamide A (NPI-0052). Bioorg. Med. Chem. 2009, 17, 2175–2180. [Google Scholar] [CrossRef]
- Lane, A.L.; Moore, B.S. A sea of biosynthesis: Marine natural products meet the molecular age. Nat. Prod. Rep. 2011, 28, 411–428. [Google Scholar] [CrossRef]
- Zhao, X.Q. Genome-based studies of marine microorganisms to maximize the diversity of natural products discovery for medical treatments. Evid. Based Complement. Altern. Med. 2011, 2011, 384572. [Google Scholar] [CrossRef]
- Rojas, J.L.; Martín, J.; Tormo, J.R.; Vicente, F.; Brunati, M.; Ciciliato, I.; Losi, D.; van Trappen, S.; Mergaert, J.; Swings, J.; et al. Bacterial diversity from benthic mats of Antarctic lakes as a source of new bioactive metabolites. Mar. Genomics 2009, 2, 33–41. [Google Scholar] [CrossRef]
- Bala, M.; Kaur, C.; Kaur, I.; Khan, F.; Mayilraj, S. Kocuria sediminis sp. nov., isolated from a marine sediment sample. Antonie Van Leeuwenhoek 2012, 101, 469–478. [Google Scholar]
- Hentschel, U.; Schmid, M.; Wagner, M.; Fieseler, L.; Gernert, C.; Hacker, J. Isolation and phylogenetic analysis of bacteria with antimicrobial activities from the Mediterranean sponges Aplysina aerophoba and Aplysina cavernicola. FEMS Microbiol. Ecol. 2001, 35, 305–312. [Google Scholar]
- Jukes, T.H.; Cantor, C. Evolution of Protein Molecules. In Mammalian Protein Metabolism; Munro, H.N., Allison, J.B., Eds.; Academic Press: New York, NY, USA, 1969; pp. 121–132. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Stackebrandt, E.; Koch, C.; Gvozdiak, O.; Schumann, P. Taxonomic dissection of the genus Micrococcus: Kocuria gen. nov., Nesterenkonia gen. nov., Kytococcus gen. nov., Dermacoccus gen. nov., and Micrococcus Cohn 1872 gen. emend. Int. J. Syst. Bacteriol. 1995, 45, 682–692. [Google Scholar]
- Kovacs, G.; Burghardt, J.; Pradella, S.; Schumann, P.; Stackebrandt, E.; Marialigeti, K. Kocuria palustris sp. nov. and Kocuria rhizophila sp. nov., isolated from the rhizoplane of the narrow-leaved cattail (Typha angustifolia). Int. J. Syst. Bacteriol. 1999, 49, 167–173. [Google Scholar]
- Reddy, G.S.N.; Prakash, J.S.S.; Prabahar, V.; Matsumoto, G.I.; Stackebrandt, E.; Shivaji, S. Kocuria polaris sp. nov., an orange-pigmented psychrophilic bacterium isolated from an Antarctic cyanobacterial mat sample. Int. J. Syst. Evol. Microbiol. 2003, 53, 183–187. [Google Scholar]
- Kim, S.B.; Nedashkovskaya, O.I.; Mikhailov, V.V.; Han, S.K.; Kim, K.O.; Rhee, M.S.; Bae, K.S. Kocuria marina sp. nov., a novel actinobacterium isolated from marine sediment. Int. J. Syst. Evol. Microbiol. 2004, 54, 1617–1620. [Google Scholar]
- Liu, H.; Xu, Y.; Ma, Y.; Zhou, P. Characterization of Micrococcus antarcticus sp. nov., a psychrophilic bacterium from Antarctica. Int. J. Syst. Evol. Microbiol. 2000, 50, 715–719. [Google Scholar]
- Hodges, T.; Slattery, M.; Olson, J. Unique actinomycetes from marine caves and coral reef sediments provide novel pks and nrps biosynthetic gene clusters. Mar. Biotechnol. 2012, 14, 270–280. [Google Scholar] [CrossRef]
- Pathom-aree, W.; Stach, J.; Ward, A.; Horikoshi, K.; Bull, A.; Goodfellow, M. Diversity of actinomycetes isolated from Challenger Deep sediment (10,898 m) from the Mariana Trench. Extremophiles 2006, 10, 181–189. [Google Scholar] [CrossRef]
- Engelhardt, K. Assessment of the Antibiotic Production Potential of Marine Derived Actinomycetes via Bioactivity Screening and Targeted Genetic Analysis. Ph.D. Thesis, Norwegian University of Science and Technology, Trondheim, Norway, October 2010. [Google Scholar]
- Jiang, S.; Li, X.; Zhang, L.; Sun, W.; Dai, S.; Xie, L.; Liu, Y.; Lee, K. Culturable actinobacteria isolated from marine sponge Iotrochota sp. Mar. Biol. 2008, 153, 945–952. [Google Scholar]
- Xin, Y.; Kanagasabhapathy, M.; Janussen, D.; Xue, S.; Zhang, W. Phylogenetic diversity of Gram-positive bacteria cultured from Antarctic deep-sea sponges. Polar Biol. 2011, 34, 1501–1512. [Google Scholar]
- Li, J.; Zhao, G.-Z.; Huang, H.-Y.; Qin, S.; Zhu, W.-Y.; Zhao, L.-X.; Xu, L.-H.; Zhang, S.; Li, W.-J.; Strobel, G. Isolation and characterization of culturable endophytic actinobacteria associated with Artemisia annua L. Antonie Van Leeuwenhoek 2012, 101, 515–527. [Google Scholar]
- Takarada, H.; Sekine, M.; Kosugi, H.; Matsuo, Y.; Fujisawa, T.; Omata, S.; Kishi, E.; Shimizu, A.; Tsukatani, N.; Tanikawa, S.; et al. Complete genome sequence of the soil actinomycete Kocuria rhizophila. J. Bacteriol. 2008, 190, 4139–4146. [Google Scholar]
- Ayuso, A.; Clark, D.; Gonzalez, I.; Salazar, O.; Anderson, A.; Genilloud, O. A novel actinomycete strain de-replication approach based on the diversity of polyketide synthase and nonribosomal peptide synthetase biosynthetic pathways. Appl. Microbiol. Biotechnol. 2005, 67, 795–806. [Google Scholar]
- Fenical, W. Chemical studies of marine bacteria: Developing a new resource. Chem. Rev. 1993, 93, 1673–1683. [Google Scholar]
- Selvin, J. Exploring the antagonistic producer streptomyces MSI051: Implications of polyketide synthase gene type II and a ubiquitous defense enzyme phospholipase A2 in the host sponge Dendrilla nigra. Curr. Microbiol. 2009, 58, 459–463. [Google Scholar]
- Suay, I.; Arenal, F.; Asensio, F.J.; Basilio, A.; Angeles Cabello, M.; Teresa Díez, M.; García, J.B.; González del Val, A.; Gorrochategui, J.; Hernández, P.; Peláez, F.; Vicente, M.F. Screening of basidiomycetes for antimicrobial activities. Antonie Van Leeuwenhoek 2000, 78, 129–140. [Google Scholar]
- Martín, J.; Sousa, T.; Crespo, G.; Palomo, S.; González, I.; Tormo, J.R.; de la Cruz, M.; Anderson, M.; Hill, R.T.; Vicente, F.; et al. Kocurin, the true structure of PM 181104, an anti-MRSA thiazolyl peptide from the marine-derived bacterium Kocuria palustris. Mar. Drugs 2013, 11, 387–398. [Google Scholar]
- Jensen, P.R.; Williams, P.G.; Oh, D.-C.; Zeigler, L.; Fenical, W. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl. Environ. Microbiol. 2007, 73, 1146–1152. [Google Scholar]
- Marwick, J.D.; Wright, P.C.; Burgess, J. Grant, Bioprocess intensification for production of novel marine bacterial antibiotics through bioreactor operation and design. Mar. Biotechnol. 1999, 1, 495–507. [Google Scholar]
- Bagley, M.C.; Dale, J.W.; Merritt, E.A.; Xiong, X. Thiopeptide antibiotics. Chem. Rev. 2005, 105, 685–714. [Google Scholar]
- Morris, R.P.; Leeds, J.A.; Naegeli, H.U.; Oberer, L.; Memmert, K.; Weber, E.; LaMarche, M.J.; Parker, C.N.; Burrer, N.; Esterow, S.; et al. Ribosomally synthesized thiopeptide antibiotics targeting elongation factor Tu. J. Am. Chem. Soc. 2009, 131, 5946–5955. [Google Scholar]
- Young, T.S.; Walsh, C.T. Identification of the thiazolyl peptide GE37468 gene cluster from Streptomyces ATCC 55365 and heterologous expression in Streptomyces lividans. Proc. Natl. Acad. Sci. USA 2011, 108, 13053–13058. [Google Scholar]
- Ferrari, P.; Colombo, L.; Stella, S.; Selva, E.; Zerilli, L.F. Antibiotic GE37468 A: A novel inhibitor of bacterial protein synthesis II. Structure elucidation. J. Antibiot. 1995, 48, 1304–1311. [Google Scholar]
- Marinelli, F.; Gastaldo, L.; Toppo, G.; Quarta, C. Antibiotic GE37468 A: A new inhibitor of bacterial protein synthesis. III. Strain and fermentation study. J. Antibiot. 1996, 49, 880–885. [Google Scholar]
- Selva, E.; Beretta, G.; Montanini, N.; Saddler, G.S.; Gastaldo, L.; Ferrari, P.; Ripamonti, F.; Goldstein, B.P.; Berti, M.; Montanaro, L.; et al. Antibiotic GE2270 a: A novel inhibitor of bacterial protein synthesis. I. Isolation and characterization. J. Antibiot. (Tokyo) 1991, 44, 693–701. [Google Scholar]
- Xi, L.; Ruan, J.; Huang, Y. Diversity and biosynthetic potential of culturable actinomycetes associated with marine sponges in the China seas. Int. J. Mol. Sci. 2012, 13, 5917–5932. [Google Scholar]
- Abdelmohsen, U.R.; Pimentel-Elardo, S.M.; Hanora, A.; Radwan, M.; Abou-El-Ela, S.H.; Ahmed, S.; Hentschel, U. Isolation, phylogenetic analysis and anti-infective activity screening of marine sponge-associated actinomycetes. Mar. Drugs 2010, 8, 399–412. [Google Scholar]
- Innis, M.A.; Gelfand, D.H.; Sninsky, J.J.; White, T.J. PCR Protocols: A Guide to Methods and Amplifications; Academic Press: San Diego, CA, USA, 1990; pp. 3–12. [Google Scholar]
- Miller, L.T. Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxy acids. J. Clin. Microbiol. 1982, 16, 584–586. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar]
- Chun, J.; Lee, J.-H.; Jung, Y.; Kim, M.; Kim, S.; Kim, B.K.; Lim, Y.-W. EzTaxon: A web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2007, 57, 2259–2261. [Google Scholar]
- Higgins, D.G. CLUSTAL W: Multiple alignment of DNA and protein sequences. Methods Mol. Biol. 1994, 25, 307–318. [Google Scholar]
- Duetz, W.A.; Ruedi, L.; Hermann, R.; O’Connor, K.; Buchs, J.; Witholt, B. Methods for intense aeration, growth, storage, and replication of bacterial strains in microtiter plates. Appl. Environ. Microbiol. 2000, 66, 2641–2646. [Google Scholar]
- Obata, H.; Muryoi, N.; Kawahara, H.; Yamade, K.; Nishikawa, J. Identification of a novel ice-nucleating bacterium of Antarctic origin and its ice nucleation properties. Cryobiology 1999, 38, 131–139. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Palomo, S.; González, I.; De la Cruz, M.; Martín, J.; Tormo, J.R.; Anderson, M.; Hill, R.T.; Vicente, F.; Reyes, F.; Genilloud, O. Sponge-Derived Kocuria and Micrococcus spp. as Sources of the New Thiazolyl Peptide Antibiotic Kocurin. Mar. Drugs 2013, 11, 1071-1086. https://doi.org/10.3390/md11041071
Palomo S, González I, De la Cruz M, Martín J, Tormo JR, Anderson M, Hill RT, Vicente F, Reyes F, Genilloud O. Sponge-Derived Kocuria and Micrococcus spp. as Sources of the New Thiazolyl Peptide Antibiotic Kocurin. Marine Drugs. 2013; 11(4):1071-1086. https://doi.org/10.3390/md11041071
Chicago/Turabian StylePalomo, Sara, Ignacio González, Mercedes De la Cruz, Jesús Martín, José Rubén Tormo, Matthew Anderson, Russell T. Hill, Francisca Vicente, Fernando Reyes, and Olga Genilloud. 2013. "Sponge-Derived Kocuria and Micrococcus spp. as Sources of the New Thiazolyl Peptide Antibiotic Kocurin" Marine Drugs 11, no. 4: 1071-1086. https://doi.org/10.3390/md11041071
APA StylePalomo, S., González, I., De la Cruz, M., Martín, J., Tormo, J. R., Anderson, M., Hill, R. T., Vicente, F., Reyes, F., & Genilloud, O. (2013). Sponge-Derived Kocuria and Micrococcus spp. as Sources of the New Thiazolyl Peptide Antibiotic Kocurin. Marine Drugs, 11(4), 1071-1086. https://doi.org/10.3390/md11041071







