Probing a Coral Genome for Components of the Photoprotective Scytonemin Biosynthetic Pathway and the 2-Aminoethylphosphonate Pathway
Abstract
:1. Introduction
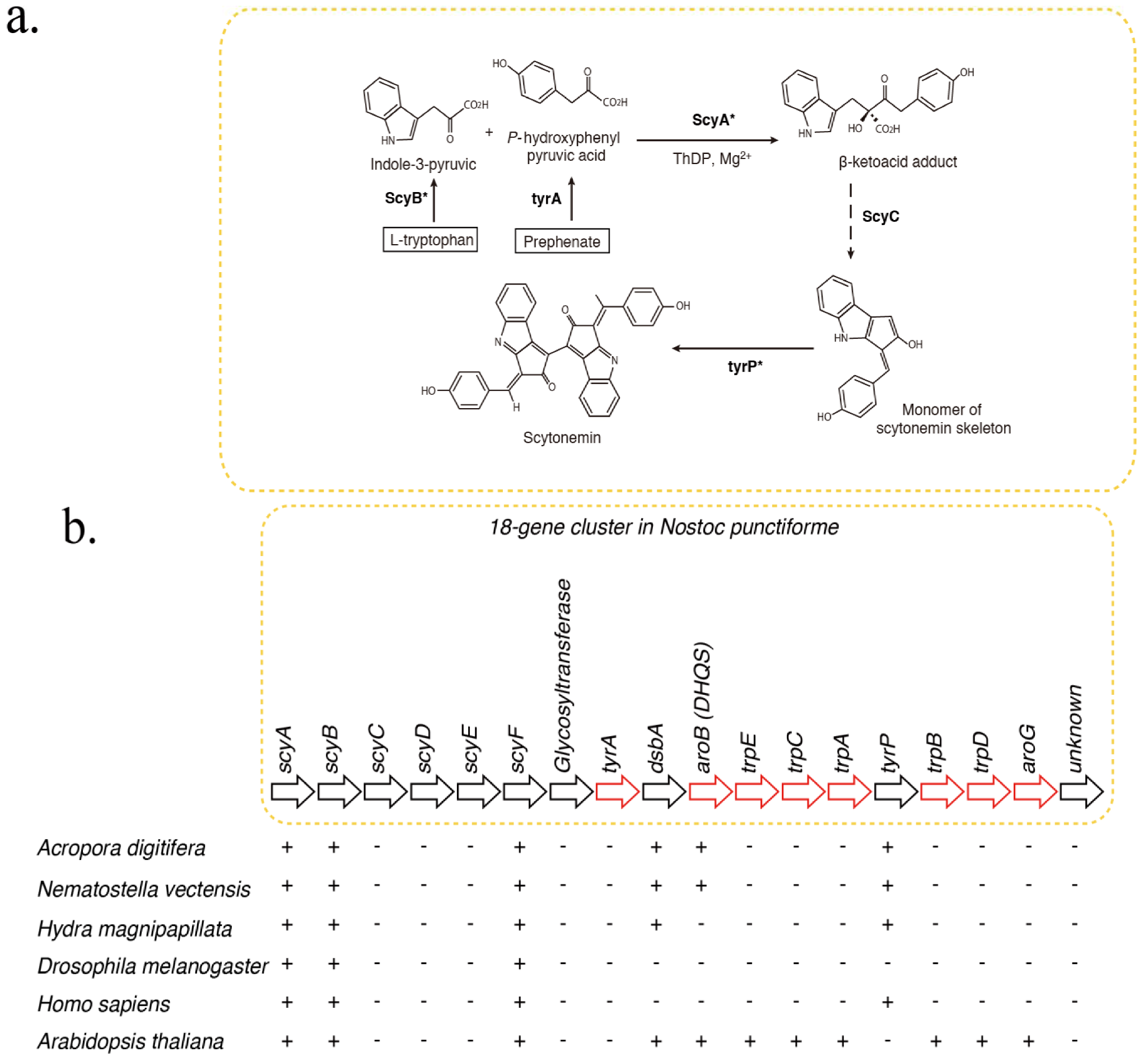
2. Results and Discussion
2.1. scyA (TPP-Dependent Enzyme)
| Gene name | Gene model ID | Intron number | All PFAM domains (in order) * | corresponding to ESTs | scaffold | References |
|---|---|---|---|---|---|---|
| phosphonopyruvate decarboxylase | aug_v2a.20271 | 6 | TPP_enzyme_N, TPP_enzyme_C | + | 12471 | Figure S1 |
| 2-hydroxyacyl-CoA lyase 1 | aug_v2a.06817 | 13 | TPP_enzyme_C | − | 2544 | Figure S1 |
| glutamate dehydrogenase1-1 (gdh1-1) | aug_v2a.22675 | 0 | ELFV_dehydrog_N, ELFV_dehydrog | + | 15779 | Figure S2 |
| glutamate dehydrogenase1-2 (gdh1-2) | aug_v2a.23483 | 1 | ELFV_dehydrog_N, ELFV_dehydrog | + | 16875 | Figure S2 |
| glutamate dehydrogenase2-1 (gdh2-1) | aug_v2a.13667 | 6 | ELFV_dehydrog_N, ELFV_dehydrog | + | 5605 | Figure S2 |
| glutamate dehydrogenase2-2(gdh2-2) | aug_v2a.16277 | 7 | ELFV_dehydrog_N, ELFV_dehydrog | − | 7525 | Figure S2 |
| DSBA domain containing gene-1 | aug_v2a.12085 | 21 | Dynein_Heavy, DSBA, DSBA | + | 4763 | Figure S3 |
| DHQS-like (aroB-like) | aug_v2a.14548 | 2 | DHQ_synthase | + | 6105 | [15] |
| TyrP1 | aug_v2a.08070 | 2 | TSP_1, TSP_1, TSP_1, TSP_1, Tyrosinase | + | 3066 | Figure S4 |
| TyrP2 | aug_v2a.10437 | 12 | Tyrosinase | + | 4001 | Figure S4 |
2.2. scyB (GDH Subfamly)
2.3. scyF (NHL Repeat Containing)
2.4. dsbA
2.5. TyrP
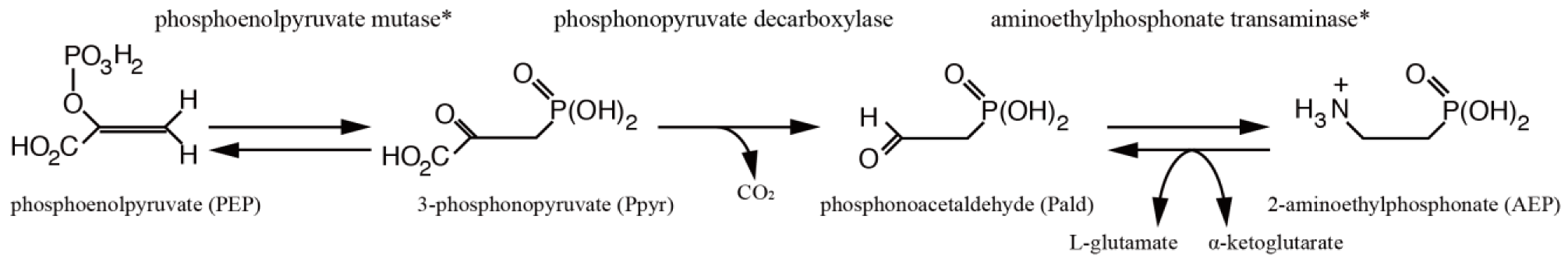
2.6. Genes for AEP Pathway
| Gene name | Gene model ID | Intron number | All PFAM domains (in order) * | corresponding to ESTs | scaffold | References |
|---|---|---|---|---|---|---|
| phosphoenolpyruvate mutase | aug_v2a.19072 | 7 | PEP_mutase | + | 11028 | Figure S5 |
| 2-aminoethylphosphonate transaminase | aug_v2a.21804 | 4 | − | + | 14440 | Figure S6 |
3. Experimental Section
3.1. Gene Search
3.2. Molecular Phylogenetic Analysis
4. Conclusion
Acknowledgments
References
- Hoegh-Guldberg, O.; Mumby, P.J.; Hooten, A.J.; Steneck, R.S.; Greenfield, P.; Gomez, E.; Harvell, C.D.; Sale, P.F.; Edwards, A.J.; Caldeira, K.; et al. Coral reefs under rapid climate change and ocean acidification. Science 2007, 318, 1737–1742. [Google Scholar]
- Weis, V.M. Cellular mechanisms of Cnidarian bleaching: Stress causes the collapse of symbiosis. J.Exp. Biol. 2008, 211, 3059–3066. [Google Scholar] [CrossRef]
- Shick, J.M.; Romaine-Lioud, S.; Ferrier-Pages, C.; Gattuso, J.P. Ultraviolet-B radiation stimulates shikimate pathway-dependent accumulation of mycosporine-like amino acids in the coral Stylophora pistillata despite decreases in its population of symbiotic dinoflagellates. Limnol. Oceanogr. 1999, 44, 1667–1682. [Google Scholar] [CrossRef]
- Reef, R.; Dunn, S.; Levy, O.; Dove, S.; Shemesh, E.; Brickner, I.; Leggat, W.; Hoegh-Guldberg, O. Photoreactivation is the main repair pathway for UV-induced DNA damage in coral planulae. J. Exp. Biol. 2009, 212, 2760–2766. [Google Scholar]
- Gordon, B.R.; Leggat, W. Symbiodinium-invertebrate symbioses and the role of metabolomics. Mar. Drugs 2010, 8, 2546–2568. [Google Scholar] [CrossRef]
- Rastogi, R.P.; Richa; Sinha, R.P.; Singh, S.P.; Hader, D.-P. Photoprotective compounds from marine organisms. J. Ind. Microbiol. Biotechnol. 2010, 37, 537–558. [Google Scholar]
- Dubinsky, Z. Coral Reef; Elsevier: New York, NY, USA, 1990. [Google Scholar]
- Stat, M.; Morris, E.; Gates, R.D. Functional diversity in coral-dinoflagellate symbiosis. Proc. Natl. Acad. Sci. USA 2008, 105, 9256–9261. [Google Scholar] [CrossRef]
- Banaszak, A.T.; LaJeunesse, T.C.; Trench, R.K. The synthesis of mycosporine-like amino acids (MAAs) by cultured, symbiotic dinoflagellates. J. Exp. Mar. Biol. Ecol. 2000, 249, 219–233. [Google Scholar]
- Villarreal-Chiu, J.F.; Quinn, J.P.; McGrath, J.W. The genes and enzymes of phosphonate metabolism by bacteria, and their distribution in the marine environment. Front. Microbiol. 2012, 3, 19:1–19:13. [Google Scholar]
- Putnam, N.H.; Srivastava, M.; Hellsten, U.; Dirks, B.; Chapman, J.; Salamov, A.; Terry, A.; Shapiro, H.; Lindquist, E.; Kapitonov, V.V.; et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 2007, 317, 86–94. [Google Scholar]
- Starcevic, A.; Akthar, S.; Dunlap, W.C.; Shick, J.M.; Hranueli, D.; Cullum, J.; Long, P.F. Enzymes of the shikimic acid pathway encoded in the genome of a basal metazoan, Nematostella vectensis, have microbial origins. Proc. Natl. Acad. Sci. USA 2008, 105, 2533–2537. [Google Scholar]
- Chapman, J.A.; Kirkness, E.F.; Simakov, O.; Hampson, S.E.; Mitros, T.; Weinmaier, T.; Rattei, T.; Balasubramanian, P.G.; Borman, J.; Busam, D.; et al. The dynamic genome of Hydra. Nature 2010, 464, 592–596. [Google Scholar]
- Forêt, S.; Knack, B.; Houliston, E.; Momose, T.; Manuel, M.; Quéinnec, E.; Hayward, D.C.; Ball, E.E.; Miller, D.J. New tricks with old genes: The genetic bases of novel cnidarian traits. Trends Genet. 2010, 26, 154–158. [Google Scholar]
- Shinzato, C.; Shoguchi, E.; Kawashima, T.; Hamada, M.; Hisata, K. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 2011, 476, 320–323. [Google Scholar]
- Balskus, E.P.; Walsh, C.T. The genetic and molecular basis for sunscreen biosynthesis in cyanobacteria. Science 2010, 329, 1653–1656. [Google Scholar]
- Gao, Q.; Garcia-Pichel, F. Microbial ultraviolet sunscreens. Nat. Rev. Microbiol. 2011, 9, 791–802. [Google Scholar] [CrossRef]
- Soule, T.; Stout, V.; Swingley, W.D.; Meeks, J.C.; Garcia-Pichel, F. Molecular genetics and genomic analysis of scytonemin biosynthesis in Nostoc punctiforme ATCC 29133. J. Bacteriol. 2007, 189, 4465–4472. [Google Scholar] [CrossRef]
- Soule, T.; Palmer, K.; Gao, Q.J.; Potrafka, R.M.; Stout, V.; Garcia-Pichel, F. A comparative genomics approach to understanding the biosynthesis of the sunscreen scytonemin in cyanobacteria. BMC Genomics 2009, 10, 336:1–336:10. [Google Scholar]
- Lesser, M.P.; Mazel, C.H.; Gorbunov, M.Y.; Falkowski, P.G. Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science 2004, 305, 997–1000. [Google Scholar]
- Gantar, M.; Kaczmarsky, L.T.; Stanic, D.; Miller, A.W.; Richardson, L.L. Antibacterial activity of marine and black band disease cyanobacteria against coral-associated bacteria. Mar. Drugs 2011, 9, 2089–2105. [Google Scholar] [CrossRef]
- Sunagawa, S.; DeSantis, T.Z.; Piceno, Y.M.; Brodie, E.L.; DeSalvo, M.K.; Voolstra, C.R.; Weil, E.; Andersen, G.L.; Medina, M. Bacterial diversity and White Plague Disease-associated community changes in the Caribbean coral Montastraea faveolata. ISME J. 2009, 3, 512–521. [Google Scholar] [CrossRef]
- Horiguchi, M.; Kandatsu, M. Isolation of 2-aminoethane phosphonic acid from rumen protozoa. Nature 1959, 184, 901–902. [Google Scholar] [CrossRef]
- Balskus, E.P.; Walsh, C.T. Investigating the initial steps in the biosynthesis of cyanobacterial sunscreen scytonemin. J. Am. Chem. Soc. 2008, 130, 15260. [Google Scholar]
- Costelloe, S.J.; Ward, J.M.; Dalby, P.A. Evolutionary analysis of the TPP-dependent enzyme family. J. Mol. Evol. 2008, 66, 36–49. [Google Scholar] [CrossRef]
- McCourt, J.A.; Duggleby, R.G. Acetohydroxyacid synthase and its role in the biosynthetic pathway for branched-chain amino acids. Amino Acids 2006, 31, 173–210. [Google Scholar]
- Zhang, G.; Dai, J.; Lu, Z.; Dunaway-Mariano, D. The phosphonopyruvate decarboxylase from Bacteroides fragilis. J. Biol. Chem. 2003, 278, 41302–41308. [Google Scholar]
- Miñambres, B.; Olivera, E.R.; Jensen, R.A.; Luengo, J.M. A new class of glutamate dehydrogenases (GDH). Biochemical and genetic characterization of the first member, the AMP-requiring NAD-specific GDH of Streptomyces clavuligerus. J. Biol. Chem. 2000, 275, 39529–39542. [Google Scholar]
- Andersson, J.O.; Roger, A.J. Evolution of glutamate dehydrogenase genes: Evidence for lateral gene transfer within and between prokaryotes and eukaryotes. BMC Evol. Biol. 2003, 3, 14:1–14:10. [Google Scholar]
- Slack, F.J.; Ruvkun, G. A novel repeat domain that is often associated with RING finger and B-box motifs. Trends Biochem. Sci. 1998, 23, 474–475. [Google Scholar] [CrossRef]
- Bardwell, J.C.A.; Mcgovern, K.; Beckwith, J. Identification of a protein required for disulfide bond formation in vivo. Cell 1991, 67, 581–589. [Google Scholar] [CrossRef]
- Hu, S.H.; Peek, J.A.; Rattigan, E.; Taylor, R.K.; Martin, J.L. Structure of TcpG, the DsbA protein folding catalyst from Vibrio cholerae. J. Mol. Biol. 1997, 268, 137–146. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, Q.; Ni, J. In silico identification of the sea squirt selenoproteome. BMC Genomics 2010, 11, 289:1–289:14. [Google Scholar]
- Halaouli, S.; Asther, M.; Sigoillot, J.C.; Hamdi, M.; Lomascolo, A. Fungal tyrosinases: New prospects in molecular characteristics, bioengineering and biotechnological applications. J. Appl. Microbiol. 2006, 100, 219–232. [Google Scholar]
- Metcalf, W.W.; van der Donk, W.A. Biosynthesis of phosphonic and phosphinic acid natural products. Annu. Rev. Biochem. 2009, 78, 65–94. [Google Scholar]
- Koyanagi, R.; Takeuchi, T.; Hisata, K.; Gyoja, F.; Shoguchi, E.; Satoh, N.; Kawashima, T. An integrated genome viewer for community-based annotation of genomes. Zool. Sci. 2013, in press. [Google Scholar]
- Oist Marine Genomics Unit Genome Project. Available online: http://marinegenomics.oist.jp/acropora_digitifera (accessed on 18 August 2011).
- Adams, M.D.; Celniker, S.E.; Holt, R.A.; Evans, C.A.; Gocayne, J.D.; Amanatides, P.G.; Scherer, S.E.; Li, P.W.; Hoskins, R.A.; Galle, R.F.; et al. The genome sequence of Drosophila melanogaster. Science 2000, 287, 2185–2195. [Google Scholar]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815.
- National Center for Biotechnology Information. Available online: http://www.ncbi.nlm.nih.gov/guide/ (accessed on 30 December 2011).
- Finn, R.D.; Mistry, J.; Schuster-Bockler, B.; Griffiths-Jones, S.; Hollich, V.; Lassmann, T.; Moxon, S.; Marshall, M.; Khanna, A.; Durbin, R.; et al. Pfam: Clans, web tools and services. Nucl. Acids Res. 2006, 34, D247–D251. [Google Scholar]
- Eddy, S.R. Profile hidden Markov models. Bioinformatics 1998, 14, 755–763. [Google Scholar]
- Kawashima, T.; Kawashima, S.; Tanaka, C.; Murai, M.; Yoneda, M.; Putnam, N.H.; Rokhsar, D.S.; Kanehisa, M.; Satoh, N.; Wada, H. Domain shuffling and the evolution of vertebrates. Genome Res. 2009, 19, 1393–1403. [Google Scholar]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar]
- Lartillot, N.; Philippe, H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 2004, 21, 1095–1109. [Google Scholar]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView Version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar]
- Perriere, G.; Gouy, M. WWW-Query: An on-line retrieval system for biological sequence banks. Biochimie 1996, 78, 364–369. [Google Scholar] [CrossRef]
- Jobb, G.; von Haeseler, A.; Strimmer, K. TREEFINDER: A powerful graphical analysis environment for molecular phylogenetics. BMC Evol. Biol. 2004, 4:1–4:18. [Google Scholar]
- Tanabe, A.S. Kakusan4 and Aminosan: Two programs for comparing nonpartitioned, proportional and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Mol. Ecol. Resour. 2011, 11, 914–921. [Google Scholar] [CrossRef]
- Shinzato, C.; Shoguchi, E.; Tanaka, M.; Satoh, N. Fluorescent protein candidate genes in the coral Acropora digitifera genome. Zool. Sci. 2012, 29, 260–264. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Shoguchi, E.; Tanaka, M.; Takeuchi, T.; Shinzato, C.; Satoh, N. Probing a Coral Genome for Components of the Photoprotective Scytonemin Biosynthetic Pathway and the 2-Aminoethylphosphonate Pathway. Mar. Drugs 2013, 11, 559-570. https://doi.org/10.3390/md11020559
Shoguchi E, Tanaka M, Takeuchi T, Shinzato C, Satoh N. Probing a Coral Genome for Components of the Photoprotective Scytonemin Biosynthetic Pathway and the 2-Aminoethylphosphonate Pathway. Marine Drugs. 2013; 11(2):559-570. https://doi.org/10.3390/md11020559
Chicago/Turabian StyleShoguchi, Eiichi, Makiko Tanaka, Takeshi Takeuchi, Chuya Shinzato, and Nori Satoh. 2013. "Probing a Coral Genome for Components of the Photoprotective Scytonemin Biosynthetic Pathway and the 2-Aminoethylphosphonate Pathway" Marine Drugs 11, no. 2: 559-570. https://doi.org/10.3390/md11020559
APA StyleShoguchi, E., Tanaka, M., Takeuchi, T., Shinzato, C., & Satoh, N. (2013). Probing a Coral Genome for Components of the Photoprotective Scytonemin Biosynthetic Pathway and the 2-Aminoethylphosphonate Pathway. Marine Drugs, 11(2), 559-570. https://doi.org/10.3390/md11020559




