Biosynthesis of Polyunsaturated Fatty Acids in the Oleaginous Marine Diatom Fistulifera sp. Strain JPCC DA0580
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culturing Fistulifera sp. Strain JPCC DA0580
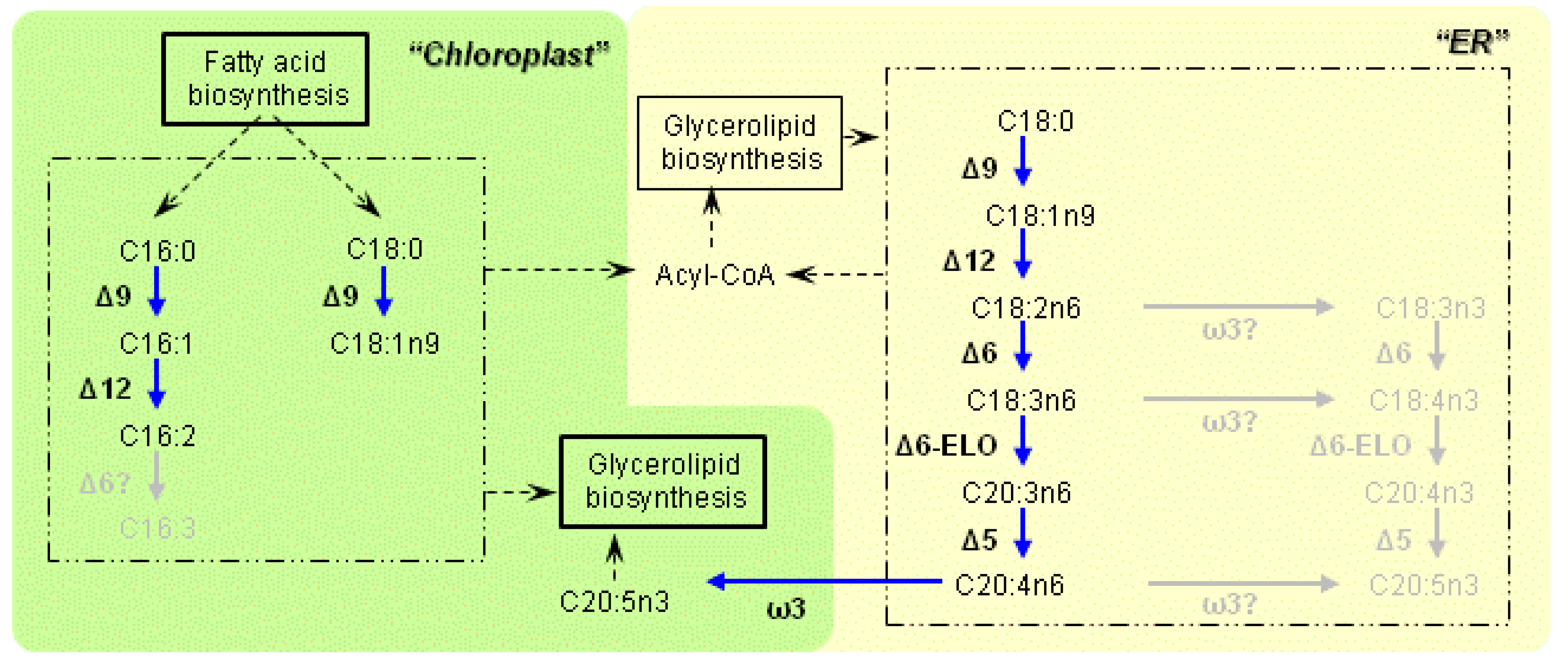
2.2. Pathway Prediction
2.3. Transcriptome Analysis
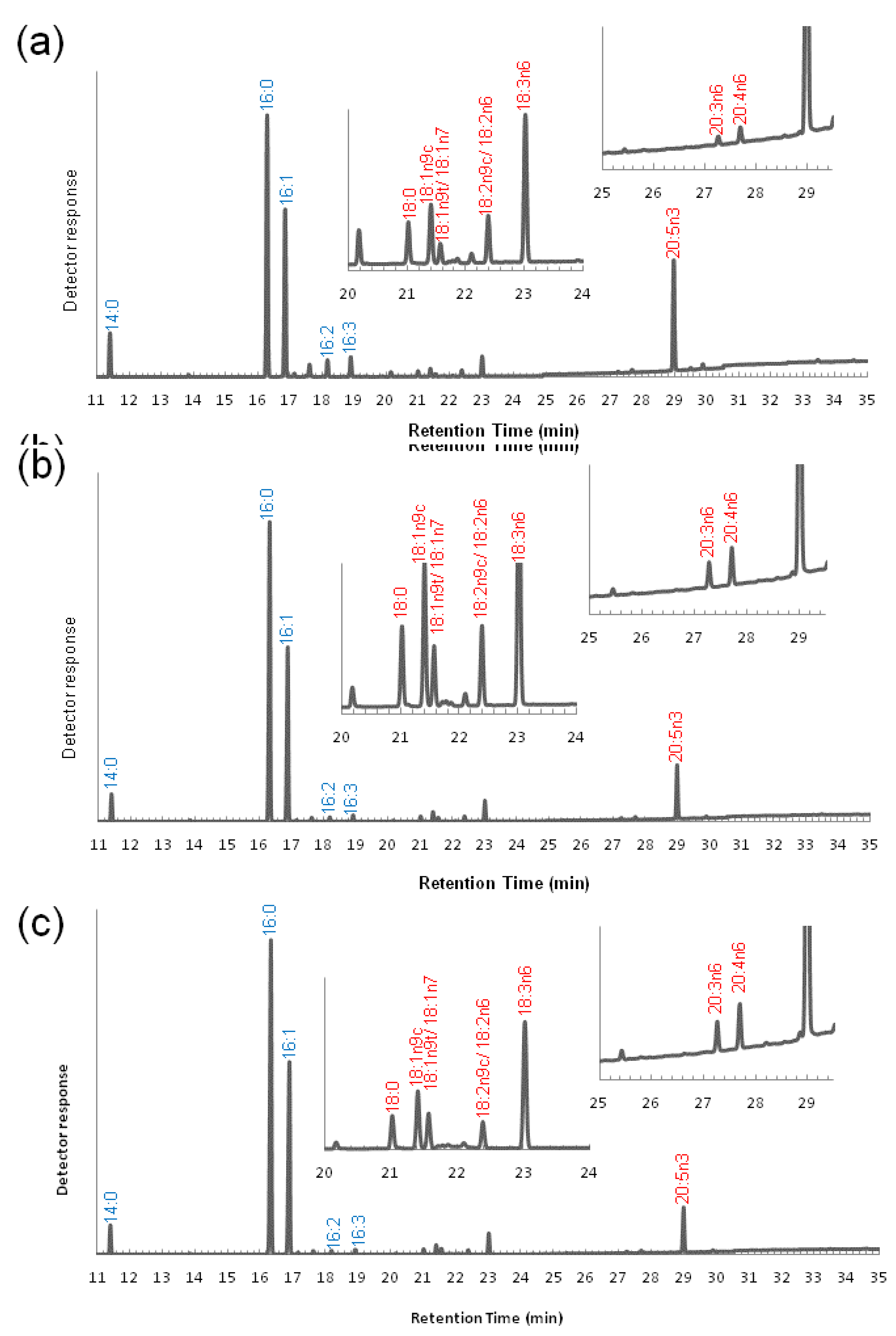
2.4. Methyl Esterification and GC-MS Analysis
3. Results
3.1. Characteristics of the Enzymes Necessary for PUFA Synthesis
| Desaturase | Organisms | Accession No. | Predicted localization | Cytochrome b5 domain | Conserved histidine boxes | No. of predicted TMHs | |||
|---|---|---|---|---|---|---|---|---|---|
| First | Second | Third | TMHMM | HMMTOP | |||||
| Δ12 (*1) desaturase | Fso | AB858393 (g13618) | ER (*4) | − | HECGH | HAKHH | HVVHH | 5 | 6 |
| AB858398 (g3281) | ER (*4) | − | HECGH | HAKHH | HVVHH | 5 | 6 | ||
| AB858392 (g13836) | Chloro | − | HECGH | HAVHH | HVAHH | 5 | 5 | ||
| AB858391 (g14210) | Chloro | − | HECGH | HAVHH | HVAHH | 5 | 5 | ||
| Ptr | XM_002186103 | ER (*4) | − | HECGH | HAKHH | HVVHH | 4 | 5 | |
| XM_002182796 | Chloro | − | HECGH | HAVHH | HVAHH | 5 | 5 | ||
| Tps | XM_002292035 | ER (*4) | − | HECGH | HAKHH | HVAHH | 4 | 2 | |
| XM_002288140 | Chloro | − | HECGH | HAVHH | HVAHH | 4 | 3 | ||
| ω3 desaturase | Fso | AB858396 (g8143) | Chloro | − | HDAGH | HKKHH | HVIHH | 6 | 4 |
| AB858395 (g9395) | Chloro | − | HDAGH | HKKHH | HVIHH | 6 | 4 | ||
| Ptr | XM_002185462 | ER (*4) | − | HDAGH | HLKHH | HLVHH | 6 | 6 | |
| Tps | XM_002291021 | ER (*4) | − | HDAGH | HRKHH | HVVHH | 2 | 2 | |
| Δ6 desaturase (*2) | Fso | AB858389 (g18158) | ER (*4) | + | HDFLHH | WKNKHNGHH | QVDDHHLFP | 4 | 4 |
| AB858388 (g18269) | ER (*4) | + | HDFLHH | WKNKHNGHH | QVDDHHLFP | 4 | 4 | ||
| Ptr | XM_002182865 | ER | + | HDFLHH | WKNKHNGHH | QVDDHHLFP | 2 | 3 | |
| Tps | XM_002291493 | ER (*4) | + | HDFLHH | WKNKHNGHH | QVDDHHLFP | 4 | 4 | |
| Δ5 desaturase (*2) | Fso | AB858387 (g19653) | ER (*4) | + | HDANH | WQEQHWTHH | QVEHHLFP | 1 | 6 |
| AB858397 (g7119) | ER (*4) | + | HDANH | WQEQHWTHH | QVEHHLFP | 3 | 6 | ||
| Ptr | XM_002185696 | ER | + | HDANH | WQEQHWTHH | QVEHHLFP | 4 | 5 | |
| XM_002182822 | ER (*4) | + | HDANH | WIQKHWTHH | QVEHHLFP | 4 | 6 | ||
| Tps | XM_002296831 | ER (*4) | + | HDANH | WLAQHWTHH | QVEHHLFP | 5 | 8 | |
| XM_002288806 | ER (*4) | + | HDANH | WMAQHWTHH | QVEHHLFP | 4 | 6 | ||
| Elongase | Organisms | Accession No. | Predicted localization | Conserved elongase motif | No. of predicted TMHs | ||||
| TMHMM | HMMTOP | ||||||||
| Δ6 poly unsaturated elongase (*3) | Fso | AB858394 (g11153) | ER (*4) | QLSFLHVYHH | 5 | 7 | |||
| AB858390 (g17615) | ER (*4) | QLSFLHVYHH | 5 | 7 | |||||
| Ptr | XM_002180392 | ER (*4) | QLSFLHVYHH | 7 | 7 | ||||
| XM_002182520 | ER (*4) | QLSFLHVYHH | 5 | 6 | |||||
| Tps | XM_002288445 | ER (*4) | QLSFLHVYHH | 7 | 7 | ||||
3.2. Variation of the Fatty Acid Composition in Fistulifera sp. during Cultivation
3.3. Transcriptome Analysis of the Enzymes for Fatty Acid Desaturation
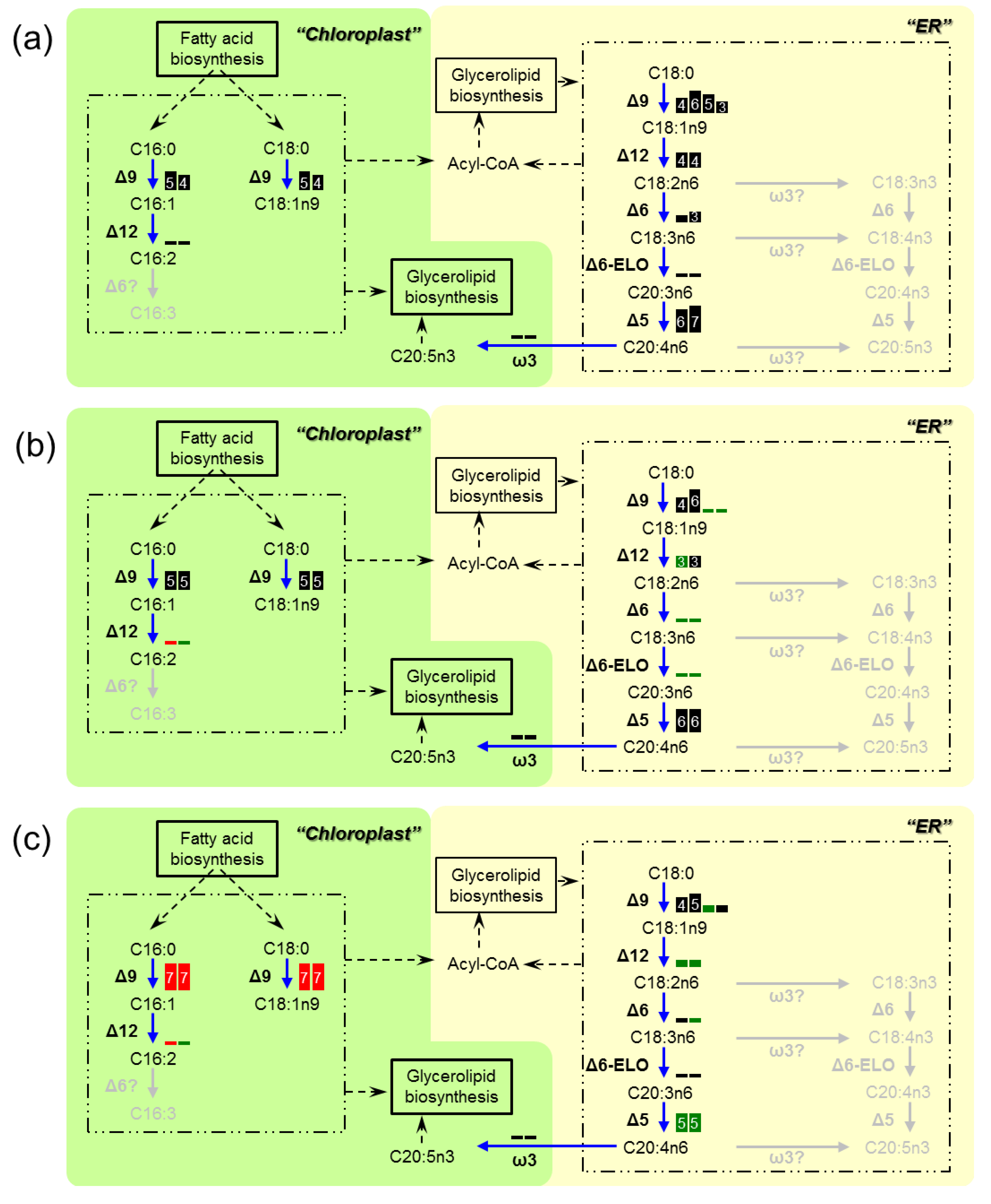
4. Discussion
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Hu, Q.; Sommerfeld, M.; Jarvis, E.; Ghirardi, M.; Posewitz, M.; Seibert, M.; Darzins, A. Microalgal triacylglycerols as feedstocks for biofuel production: Perspectives and advances. Plant J. 2008, 54, 621–639. [Google Scholar] [CrossRef]
- Chisti, Y. Biodiesel from microalgae. Biotechnol. Adv. 2007, 25, 294–306. [Google Scholar] [CrossRef]
- Knutzon, D.S.; Thompson, G.A.; Radke, S.E.; Johnson, W.B.; Knauf, V.C.; Kridl, J.C. Modification of Brassica seed oil by antisense expression of a stearoyl-acyl carrier protein desaturase gene. Proc. Natl. Acad. Sci. USA 1992, 89, 2624–2628. [Google Scholar] [CrossRef]
- Singh, A.K.; Fu, D.Q.; El-Habbak, M.; Navarre, D.; Ghabrial, S.; Kachroo, A. Silencing genes encoding omega-3 fatty acid desaturase alters seed size and accumulation of bean pod mottle virus in soybean. Mol. Plant. Microbe Interact. 2011, 24, 506–515. [Google Scholar] [CrossRef]
- Liu, Q.; Singh, S.P.; Green, A.G. High-stearic and high-oleic cottonseed oils produced by hairpin RNA-mediated post-transcriptional gene silencing. Plant Physiol. 2002, 129, 1732–1743. [Google Scholar] [CrossRef]
- Arao, T.; Yamada, M. Biosynthesis of polyunsaturated fatty acids in the marine diatom, Phaeodactylum tricornutum. Phytochemistry 1994, 35, 1177–1181. [Google Scholar] [CrossRef]
- Shiran, D.; Khozin, I.; Heimer, Y.M.; Cohen, Z. Biosynthesis of eicosapentaenoic acid in the microalga Porphyridium cruentum. I. The use of externally supplied fatty acids. Lipids 1996, 31, 1277–1282. [Google Scholar] [CrossRef]
- Khozin, I.; Adlerstein, D.; Bigongo, C.; Heimer, Y.M.; Cohen, Z. Elucidation of the biosynthesis of eicosapentaenoic acid in the microalga Porphyridium cruentum (II. Studies with radiolabeled precursors). Plant Physiol. 1997, 114, 223–230. [Google Scholar]
- Deckelbaum, R.J.; Torrejon, C. The omega-3 fatty acid nutritional landscape: Health benefits and sources. J. Nutr. 2012, 142, 587S–591S. [Google Scholar] [CrossRef]
- Arao, T.; Kawaguchi, A.; Yamada, M. Positional distribution of fatty acids in lipids of the marine diatom Phaeodactylum tricornutum. Phytochemistry 1987, 26, 2573–2576. [Google Scholar] [CrossRef]
- Bowler, C.; Allen, A.E.; Badger, J.H.; Grimwood, J.; Jabbari, K.; Kuo, A.; Maheswari, U.; Martens, C.; Maumus, F.; Otillar, R.P.; et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 2008, 456, 239–244. [Google Scholar] [CrossRef]
- Miyahara, M.; Aoi, M.; Inoue-Kashino, N.; Kashino, Y.; Ifuku, K. Highly efficient transformation of the diatom Phaeodactylum tricornutum by multi-pulse electroporation. Biosci. Biotechnol. Biochem. 2013, 77, 874–876. [Google Scholar]
- Apt, K.E.; Kroth-Pancic, P.G.; Grossman, A.R. Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Mol. Gen. Genet. 1996, 252, 572–579. [Google Scholar]
- Matsumoto, M.; Sugiyama, H.; Maeda, Y.; Sato, R.; Tanaka, T.; Matsunaga, T. Marine diatom, Navicula sp. strain JPCC DA0580 and marine green alga, Chlorella sp. strain NKG400014 as potential sources for biodiesel production. Appl. Biochem. Biotechnol. 2010, 161, 483–490. [Google Scholar] [CrossRef]
- Tanaka, T.; Fukuda, Y.; Yoshino, T.; Maeda, Y.; Muto, M.; Matsumoto, M.; Mayama, S.; Matsunaga, T. High-throughput pyrosequencing of the chloroplast genome of a highly neutral-lipid-producing marine pennate diatom, Fistulifera sp. strain JPCC DA0580. Photosynth. Res. 2011, 109, 223–229. [Google Scholar] [CrossRef]
- Nojima, D.; Yoshino, T.; Maeda, Y.; Tanaka, M.; Nemoto, M.; Tanaka, T. Proteomics analysis of oil body-associated proteins in the oleaginous diatom. J. Proteome Res. 2013, 12, 5293–5301. [Google Scholar] [CrossRef]
- Muto, M.; Fukuda, Y.; Nemoto, M.; Yoshino, T.; Matsunaga, T.; Tanaka, T. Establishment of a genetic transformation system for the marine pennate diatom Fistulifera sp. strain JPCC DA0580—A high triglyceride producer. Mar. Biotechnol. 2013, 15, 48–55. [Google Scholar] [CrossRef]
- Guillard, R.R.; Ryther, J.H. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can. J. Microbiol. 1962, 8, 229–239. [Google Scholar] [CrossRef]
- Tanaka, T.; Maeda, Y.; Veluchamy, A.; Tanaka, M.; Bowler, C.; Muto, M.; Sunaga, Y.; Tanaka, M.; Yoshino, T.; Taniguchi, T.; Fukuda, Y.; Nemoto, M.; Matsumoto, M.; Wong, P.S.; Aburatani, S.; Fujibuchi, W. Whole Genome and transcriptome analyses of the oleaginous diatom Fistulifera sp. JPCC DA0580 reveal insights into simultaneous growth and oil accumulation. 2013; to be submitted for publication. [Google Scholar]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef]
- Gschloessl, B.; Guermeur, Y.; Cock, J.M. HECTAR: A method to predict subcellular targeting in heterokonts. BMC Bioinf. 2008, 9. [Google Scholar] [CrossRef]
- Bendtsen, J.D.; Nielsen, H.; von Heijne, G.; Brunak, S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004, 340, 783–795. [Google Scholar] [CrossRef]
- Kroth, P.G.; Chiovitti, A.; Gruber, A.; Martin-Jezequel, V.; Mock, T.; Parker, M.S.; Stanley, M.S.; Kaplan, A.; Caron, L.; Weber, T.; et al. A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS One 2008, 3, e1426. [Google Scholar] [CrossRef]
- Gruber, A.; Vugrinec, S.; Hempel, F.; Gould, S.B.; Maier, U.-G.; Kroth, P.G. Protein targeting into complex diatom plastids: Functional characterisation of a specific targeting motif. Plant Mol. Biol. 2007, 64, 519–530. [Google Scholar] [CrossRef]
- Claros, M.G.; Vincens, P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996, 241, 779–786. [Google Scholar]
- Gonzalez, N.H.; Felsner, G.; Schramm, F.D.; Klingl, A.; Maier, U.G.; Bolte, K. A single peroxisomal targeting signal mediates matrix protein import in diatoms. PLoS One 2011, 6, e25316. [Google Scholar]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Zhi-Liang, H.; Bao, J.; Reecy, J. CateGOrizer: A web-based program to batch analyze gene ontology classification categories. Online J. Bioinf. 2008, 9, 108–112. [Google Scholar]
- Armbrust, E.V.; Berges, J.A.; Bowler, C.; Green, B.R.; Martinez, D.; Putnam, N.H.; Zhou, S.; Allen, A.E.; Apt, K.E.; Bechner, M.; et al. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science 2004, 306, 79–86. [Google Scholar] [CrossRef]
- Los, D.A.; Murata, N. Structure and expression of fatty acid desaturases. Biochim. Biophys. Acta 1998, 1394, 3–15. [Google Scholar] [CrossRef]
- Hashimoto, K.; Yoshizawa, A.C.; Okuda, S.; Kuma, K.; Goto, S.; Kanehisa, M. The repertoire of desaturases and elongases reveals fatty acid variations in 56 eukaryotic genomes. J. Lipid Res. 2008, 49, 183–191. [Google Scholar] [CrossRef]
- Muto, M.; Kubota, C.; Tanaka, M.; Satoh, A.; Matsumoto, M.; Yoshino, T.; Tanaka, T. Identification and functional analysis of delta-9 desaturase, a key enzyme in PUFA synthesis, isolated from the oleaginous diatom Fistulifera. PLoS One 2013, 8, e73507. [Google Scholar]
- Domergue, F.; Spiekermann, P.; Lerchl, J.; Beckmann, C.; Kilian, O.; Kroth, P.G.; Boland, W.; Zahringer, U.; Heinz, E. New insight into Phaeodactylum tricornutum fatty acid metabolism. Cloning and functional characterization of plastidial and microsomal delta12-fatty acid desaturases. Plant Physiol. 2003, 131, 1648–1660. [Google Scholar] [CrossRef]
- Domergue, F.; Lerchl, J.; Zähringer, U.; Heinz, E. Cloning and functional characterization of Phaeodactylum tricornutum front-end desaturases involved in eicosapentaenoic acid biosynthesis. Eur. J. Biochem. 2002, 269, 4105–4113. [Google Scholar] [CrossRef]
- Satoh, A.; Ichii, K.; Matsumoto, M.; Kubota, C.; Nemoto, M.; Tanaka, M.; Yoshino, T.; Matsunaga, T.; Tanaka, T. A process design and productivity evaluation for oil production by indoor mass cultivation of a marine diatom, Fistulifera sp. JPCC DA0580. Bioresour. Technol. 2013, 137, 132–138. [Google Scholar] [CrossRef]
- Khozin, I.; Cohen, Z. Differential response of microalgae to the substituted pyridazinone, Sandoz 9785, reveal different pathways in the biosynthesis of eicosapentaenoic acid. Phytochemistry 1996, 42, 1025–1029. [Google Scholar] [CrossRef]
Supplementary Files
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Liang, Y.; Maeda, Y.; Sunaga, Y.; Muto, M.; Matsumoto, M.; Yoshino, T.; Tanaka, T. Biosynthesis of Polyunsaturated Fatty Acids in the Oleaginous Marine Diatom Fistulifera sp. Strain JPCC DA0580. Mar. Drugs 2013, 11, 5008-5023. https://doi.org/10.3390/md11125008
Liang Y, Maeda Y, Sunaga Y, Muto M, Matsumoto M, Yoshino T, Tanaka T. Biosynthesis of Polyunsaturated Fatty Acids in the Oleaginous Marine Diatom Fistulifera sp. Strain JPCC DA0580. Marine Drugs. 2013; 11(12):5008-5023. https://doi.org/10.3390/md11125008
Chicago/Turabian StyleLiang, Yue, Yoshiaki Maeda, Yoshihiko Sunaga, Masaki Muto, Mitsufumi Matsumoto, Tomoko Yoshino, and Tsuyoshi Tanaka. 2013. "Biosynthesis of Polyunsaturated Fatty Acids in the Oleaginous Marine Diatom Fistulifera sp. Strain JPCC DA0580" Marine Drugs 11, no. 12: 5008-5023. https://doi.org/10.3390/md11125008
APA StyleLiang, Y., Maeda, Y., Sunaga, Y., Muto, M., Matsumoto, M., Yoshino, T., & Tanaka, T. (2013). Biosynthesis of Polyunsaturated Fatty Acids in the Oleaginous Marine Diatom Fistulifera sp. Strain JPCC DA0580. Marine Drugs, 11(12), 5008-5023. https://doi.org/10.3390/md11125008





