Efficient Heterologous Transformation of Chlamydomonas reinhardtii npq2 Mutant with the Zeaxanthin Epoxidase Gene Isolated and Characterized from Chlorella zofingiensis
Abstract
:Abbreviations
| gDNA | Genomic DNA |
| UPGMA | Unweighted Pair Group Method with Arithmetic Mean |
| PSII | Photosystem II |
| qPCR | Quantitative Polymerase Chain Reaction |
1. Introduction
2. Results
2.1. Isolation and Characterization of the ZEP Gene and Deduced Protein Sequence of C. zofingiensis
| Primer | Sequence (5′→3′) |
|---|---|
| Partial ZEP fragment | |
| ZEP-1F | tggtgggcgccgayggnathhg |
| ZEP-1R | cgcccacgtcggtgswnacraarta |
| 5′ and 3′ RACE | |
| NGSP-ZEP-5′R | gatcttggaccagataccatcagcacccaca |
| GSP-ZEP-5′R | agcatgtatagccgctgtagttggg |
| NGSP-ZEP-3′F | cagatttcacgccagcagacattgacat |
| GSP-ZEP-3′F | gcggctatacatgctacacagggatct |
| Genomic DNA amplification | |
| G-ZEP-1F | atggggacttcagtacctgca |
| G-ZEP-1R | aacagccagcacaagacctcc |
| G-ZEP-2F | caagaccagaaagcgtcacggc |
| G-ZEP-2R | tgctgcacacctgtgacagggt |
| G-ZEP-3F | gatcctgattgatgccgtggcg |
| G-ZEP-3R | tgctccatagcatctgccagat |
| G-ZEP-4F | atttgggtcagggtggctgcat |
| G-ZEP-4R | ggtgcgaaaagggtatgaagaacg |
| G-ZEP-5R | cgctctacacttggctgtagtg |
| G-ZEP-6F | atcaatggcatggagtacagga |
| G-ZEP-6R | tagcacgatgctgcaagatcacgt |
| G-ZEP-7F | ctggtgtgcagattggttttgccg |
| G-ZEP-7R | Agtattcactcactatgaactgatgcc |
| G-ZEP-8F | ccattgaagatgcttatcagttggcagc |
| G-ZEP-8R | ggcatacatgctctgagcaggcataag |
| PCR for Chamydomonas reinhardtii transformation a | |
| pStpZEPxhoF | cgcctcgagcatggggacttcagtacctgcaaat |
| pStpZEPecoR | ccgaattcgtggctgttatatttgtgctgttg |
| CzZEP expression | |
| RT-Czzep-F | cgaacgtgatcttacagccat |
| RT-Czzep-R | gatacgatctcctgtgatgcagcca |
| CBLP expression | |
| RT-cblp-F | cgccacccagtcctccatcaaga |
| RT-cblp-R | Ctaggcgcggctgggcatttac |
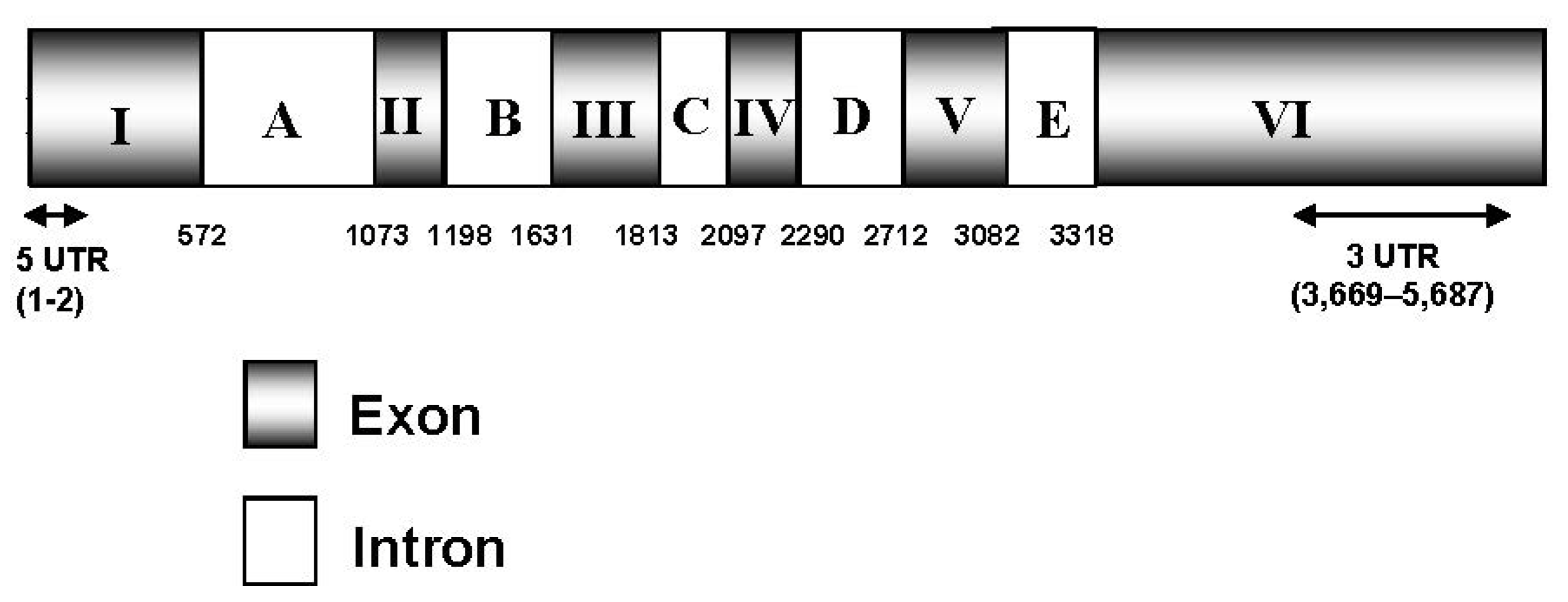

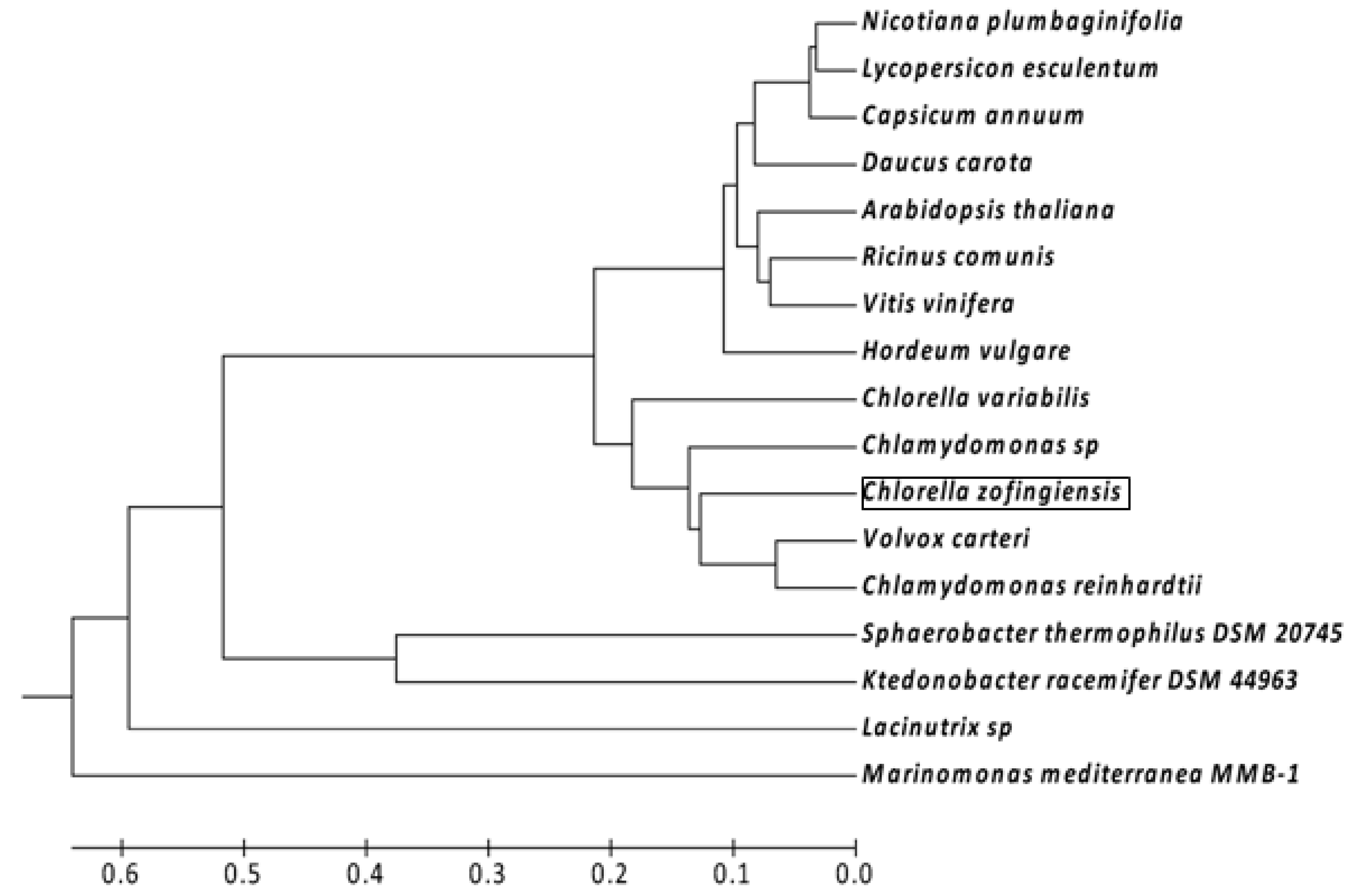
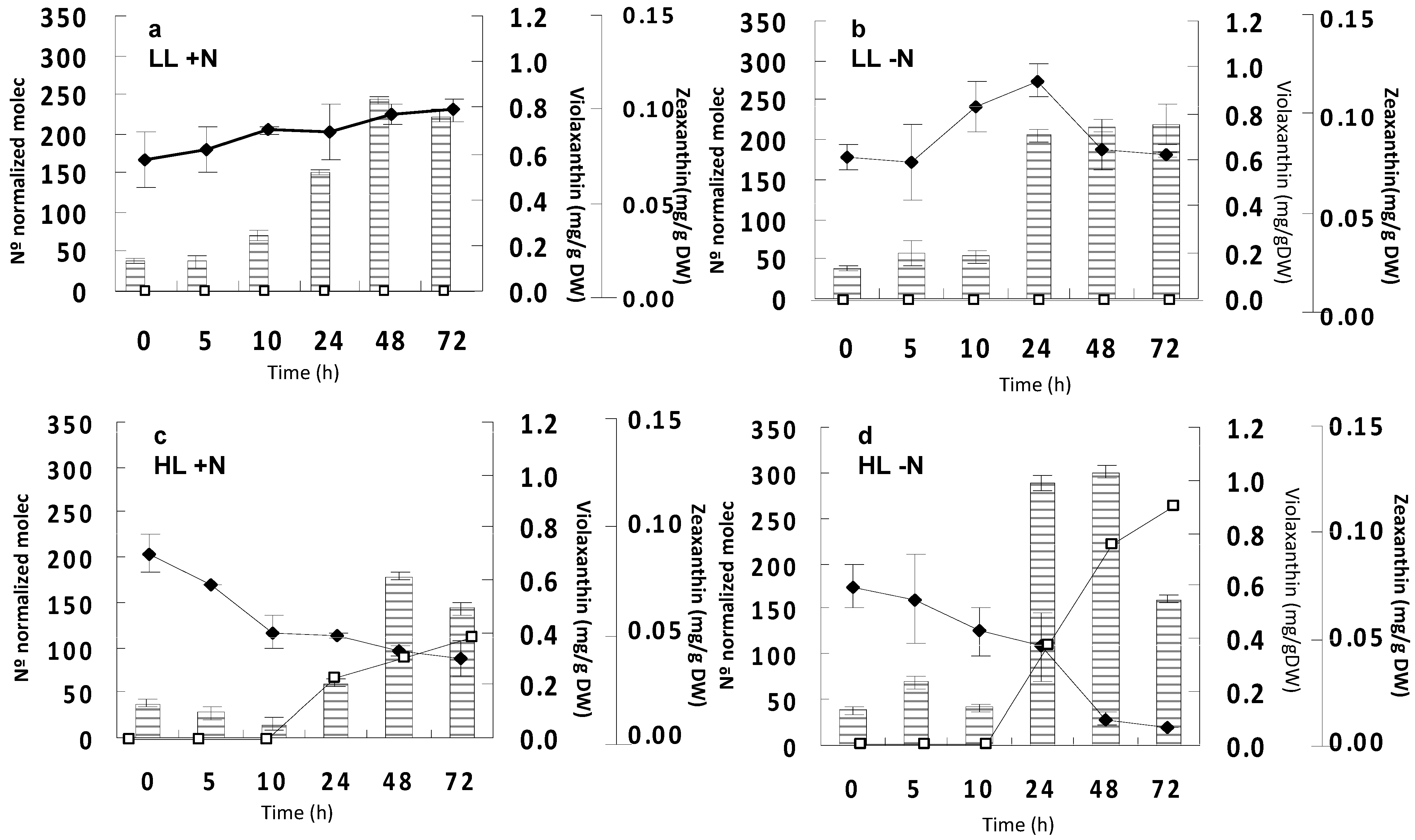
2.2. Effect of Irradiance and Nitrogen on the Expression of Czzep Gene
2.3. Functional Analysis of the ZEP from C. zofingiensis by Heterologous Genetic Complementation in C. reinhardtii npq2 Mutant
2.3.1. Transformation of C. reinhardtii npq2 Mutant with zep Gene from C. zofingiensis and Screening of the Obtained Transformants
2.3.2. Analysis of Carotenoid Content, Maximum Quantum Efficiency and mRNA Level of Czzep Gene in Both npq2 Mutant of C. reinhardtii and Czzep-Transformed Strains of npq2 Mutant


| Strains | Fv/Fm | % Fv/Fm recovered |
|---|---|---|
| WT4A+ | 0.65 ± 0.014 | --- |
| npq2 | 0.485 ± 0.003 | --- |
| npq2-1 | 0.585 ± 0.017 | 89.2% |
| npq2-4 | 0.65 ± 0.014 | 100% |
| npq2-5 | 0.555 ± 0.003 | 84.6% |
| npq2-7 | 0.585 ± 0.007 | 89.2% |
| npq2-8 | 0.6 ± 0.007 | 92.3% |
| npq2-9 | 0.585 ± 0 | 89.2% |
| npq2-10 | 0.605 ± 0.003 | 93.1% |
| npq2-13 | 0.58 ± 0 | 89.2% |
| npq2-14 | 0.61 ± 0.007 | 93.8% |
3. Discussion
4. Experimental Section
4.1. Strains and Culture Conditions
4.2. Genomic DNA and RNA Isolation and cDNA Preparation
4.3. Cloning of C. zofingiensis zep cDNA and Genomic Gene
4.4. Nucleotide Sequence Accession Numbers
4.5. Sequencing and Phylogenetic Analysis
4.6. Southern Blot Analysis
4.7. Chlamydomonas Nuclear Transformation
4.8. Quantitative RT-PCR
4.9. Analytical Methods
4.9.1. Cell Concentration and Dry Weight Determinations
4.9.2. Carotenoid Extraction and HPLC Analysis
4.9.3. Photochemical Efficiency Analysis (Fv/Fm Measurements)
5. Conclusions
Acknowledgements
References
- Niyogi, K.K. Photoprotection revisited: Genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 333–359. [Google Scholar] [CrossRef]
- Li, Z.; Wakao, S.; Fischer, B.B.; Niyogi, K. Sensing and responding to excess light. Annu. Rev. Plant Biol. 2009, 60, 239–260. [Google Scholar] [CrossRef]
- Freer, A.A.; Prince, S.M.; Sauer, K.; Papiz, M.Z.; Hawthornthwaite-Lawless, A.M.; McDermott, G.; Cogdell, R.J.; Isaacs, N.W. Pigment-protein interactions and energy transfer in the antennacomplex of the photosynthetic bacterium Rhodopseudomonas acidophila. Structure 1996, 4, 449–462. [Google Scholar] [CrossRef]
- Niyogi, K.K.; Shih, C.; Chow, W.S.; Pogson, B.J.; DellaPenna, D.; Björkman, O. Photoprotection in a zeaxanthin- and lutein-deficient double mutant of Arabidopsis. Photosynth. Res. 2001, 67, 139–145. [Google Scholar] [CrossRef]
- Pogson, B.J.; Niyogi, K.K.; Björkman, O.; DellaPenna, D. Altered xanthophyll compositions adversely affect chlorophyll accumulation and nonphotochemical quenching in Arabidopsis mutants. Proc. Natl. Acad. Sci. USA 1998, 95, 13324–13329. [Google Scholar]
- Young, A.J.; Phillip, D.; Savill, J. Carotenoids in higher plant photosynthesis. In Handbook of Photosynthesis; Pessaraki, M., Ed.; Marcel Dekker Inc.: New York, NY, USA, 1997; pp. 575–596. [Google Scholar]
- Demmig-Adams, B.; Adams, W.W.; Winter, K.; Meyer, A.; Schreiber, U.; Pereira, J.S.; Kriiger, A.; Czygan, F.C.; Lange, O.L. Photochemical efficiency of photosystem II, photon yield of O2 evolution, photosynthetic capacity, and carotenoid composition during the midday depression of net CO2 uptake in Arbutus unedo growing in Portugal. Planta 1989, 177, 377–387. [Google Scholar] [CrossRef]
- Goss, R.; Richter, M.; Wild, A. Role of DpH in the mechanism of zeaxanthin-dependent amplification of qE. J. Photochem. Photobiol. B Biol. 1995, 27, 147–152. [Google Scholar]
- Goss, R.; Jakob, T. Regulation and function of xanthophyll cycle-dependent photoprotection in algae. Photosynth. Res. 2010, 106, 103–122. [Google Scholar] [CrossRef]
- Stransky, H.; Hager, A. Das Carotinoidmuster und die Verbreitung des lichtinduzierten Xanthophyllcyclus in verschiedenen Algenklassen. Arch. Microbiol. 1970, 71, 164–190. [Google Scholar]
- Olaizola, M.; Roche, J.; Kolber, Z.; Falkowski, P.G. Nonphotochemical fluorescence quenching and the diadinoxanthin cycle in a marine diatom. Photosynth. Res. 1994, 41, 357–370. [Google Scholar] [CrossRef]
- Lohr, M.; Wilhelm, C. Algae displaying the diadinoxanthin cycle also possess the violaxanthin cycle. Proc. Natl. Acad. Sci. USA 1999, 96, 8784–8789. [Google Scholar] [CrossRef]
- Lohr, M.; Wilhelm, C. Xanthophyll synthesis in diatoms: quantification of putative intermediates and comparison of pigment conversion kinetics with rate constants derived from a model. Planta 2001, 212, 382–391. [Google Scholar] [CrossRef]
- Siefermann, D.; Yamamoto, H. NADPH and oxygen-dependent epoxidation of zeaxanthin in isolated chloroplasts. Biochem. Biophys. Res. Commun. 1975, 62, 456–461. [Google Scholar] [CrossRef]
- Bouvier, F.; d’Harlingue, A.; Hugueney, P.; Marin, E.; Marion-Poll, A.; Camara, B. Xanthophyll biosynthesis: Cloning, expression, functional reconstitution, and regulation of b-cyclohexenyl carotenoid epoxidase from pepper (Capsicum annuum). J. Biol. Chem. 1996, 271, 28861–28867. [Google Scholar]
- Holt, N.E.; Fleming, G.R.; Niyogi, K.K. Toward an understanding of the mechanism of nonphotochemical quenching in green plants. Biochemistry 2004, 43, 8281–8289. [Google Scholar]
- Büch, K.; Stransky, H.; Hager, A. FAD is a further essential cofactor of the NAD(P)H and O2-dependent zeaxanthin-epoxidase. FEBS Lett. 1995, 376, 45–48. [Google Scholar] [CrossRef]
- Marin, E.; Nussaume, L.; Quesada, A.; Gonneau, M.; Sotta, B.; Hugueney, P.; Frey, A.; Marion-Poll, A. Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 1996, 15, 2331–2342. [Google Scholar]
- Audran, C.; Borel, C.; Frey, A.; Sotta, B.; Meyer, C.; Simonneau, T.; Marion-Poll, A. Expression Studies of the Zeaxanthin Epoxidase Gene in Nicotiana plumbaginifolia. Plant Physiol. 1998, 118, 1021–1028. [Google Scholar] [CrossRef]
- Rock, C.D.; Zeevaart, J.A.D. The aba mutant of Arabidopsis thaliana is impaired in epoxy-carotenoid biosynthesis. Proc. Natl. Acad. Sci. USA 1991, 88, 7496–7499. [Google Scholar]
- Niyogi, K.K.; Bjirkman, O.; Grossman, A.R. Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell 1997, 9, 1369–1380. [Google Scholar]
- Eonseon, J.; Polle, J.W.; Lee, H.K.; Hyun, S.M.; Chang, M.J. Xanthophylls in microalgae: from biosynthesis to biotechnological mass production and application. Microbiol. Biotechnol. 2003, 13, 165–174. [Google Scholar]
- Cordero, B.F.; Couso, I.; León, R.; Rodríguez, H.; Vargas, M.A. Enhancement of carotenoids biosynthesis in Chlamydomonas reinhardtii by nuclear transformation using a phytoene synthase gene isolated from Chlorella zofingiensis. Appl. Microbiol. Biotechnol. 2011, 91, 341–351. [Google Scholar]
- Huang, J.C.; Chen, F.; Sandmann, G. Stress-related differential expression of multiple β-carotene ketolase genes in the unicellular green alga Haematococcus pluvialis. J. Biotechnol. 2006, 122, 176–185. [Google Scholar]
- Huang, J.C.; Liu, J.; Li, Y.; Chen, F. Isolation and characterization of the phytoene desaturase gene as a potential selective marker for genetic engineering of the astaxanthin-producing green alga Chlorella zofingiensis (Chlorophyta). J. Phycol. 2008, 44, 684–690. [Google Scholar] [CrossRef]
- Li, Y.; Huang, J.; Sandmann, G.; Chen, F. Glucose sensing and the mitochondrial alternative pathway are involved in the regulation of astaxanthin biosynthesis in the dark-grown Chlorella zofingiensis (Chlorophyceae). Planta 2008, 228, 735–743. [Google Scholar] [CrossRef]
- Cordero, B.F.; Obraztsova, I.; Martín, L.; Couso, I.; León, R.; Vargas, M.A.; Rodríguez, H. Isolation and characterization of a lycopene β-cyclase gene from the astaxanthin-producing green alga Chlorella zofingiensis (Chlorophyta). J. Phycol. 2010, 46, 1229–1238. [Google Scholar] [CrossRef]
- Cordero, B.F.; Obraztsova, I.; Couso, I.; León, R.; Vargas, M.A.; Rodriguez, H. Enhancement of lutein production in Chlorella sorokiniana (Chorophyta) by improvement of culture conditions and random mutagenesis. Mar. Drugs 2011, 9, 1607–1624. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Hurry, V.; Anderson, J.M.; Chow, W.S.; Osmond, C.B. Accumulation of zeaxanthin in abscisic acid-deficient mutants of Arabidopsis does not affect chlorophyll fluorescence quenching or sensitivity to photoinhibition in vivo. Plant Physiol. 1997, 113, 639–648. [Google Scholar]
- Tardy, F.; Havaux, M. Photosynthesis, chlorophyll fluorescence, light-harvesting system and photoinhibition resistance of a zeaxanthin-accumulating mutant of Arabidopsis thaliana. J. Photochem. Photobiol. 1996, 34, 87–94. [Google Scholar] [CrossRef]
- Baroli, I.; Do, A.D.; Yamane, T.; Niyogi, K.K. Zeaxanthin accumulation in the absence of a functional xanthophyll cycle protects Chlamydomonas reinhardtii from photooxidative stress. Plant Cell 2003, 15, 992–1008. [Google Scholar] [CrossRef]
- Jin, E.; Yokthongwattana, K.; Polle, J.E.W.; Melis, A. Role of the reversible xanthophyll cycle in the photosystem II damage and repair cycle in Dunaliella salina. Plant Physiol. 2003, 132, 352–364. [Google Scholar] [CrossRef]
- Burbidge, A.; Grieve, T.M.; Jackson, A.; Thompson, A.; Taylor, I.B. Structure and expression of a cDNA encoding a zeaxanthin epoxidase, isolated from a wilt-related tomato (Lycopersicon esculentum Mill.) library. J. Exp. Bot. 1997, 48, 1749–1750. [Google Scholar]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Maréchal-Drouard, L.; et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 2007, 318, 245–250. [Google Scholar]
- Blanc, G.; Duncan, G.; Agarkova, I.; Borodovsky, M.; Gurnon, J.; Kuo, A.; Lindquist, E.; Lucas, S.; Pangilinan, J.; Polle, J.; et al. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 2010, 22, 2943–2955. [Google Scholar] [CrossRef]
- Coesel, S.; Oborník, M.; Varela, J.; Falciatore, A.; Bowler, C. Evolutionary origins and functions of the carotenoid biosynthetic pathway in marine diatoms. PLoS ONE 2008, 3, e2896. [Google Scholar] [CrossRef]
- Frommolt, R.; Werner, S.; Paulsen, H.; Goss, R.; Wilhelm, C.; Zauner, S.; Maier, U.G.; Grossman, A.R.; Bhattacharya, D.; Lohr, M. Ancient recruitment by chromists of green algal genes encoding enzymes for carotenoid biosynthesis. Mol. Biol. Evol. 2008, 25, 2653–2667. [Google Scholar] [CrossRef]
- Cunningham, F.X.; Gantt, E. Genes and enzymes of carotenoid biosynthesis in plants. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 557–583. [Google Scholar] [CrossRef]
- Bugos, R.C.; Hieber, A.D.; Yamamoto, H.Y. Xanthophyll cycle enzymes are members of the lipocalin family, the first identified from plants. J. Biol. Chem. 1998, 273, 15321–15324. [Google Scholar] [CrossRef]
- Bohne, F.; Linden, H. Regulation of carotenoids biosynthesis in response to light in Chlamydomonas reinhardtii. Biochim. Biophys. Acta 2002, 1579, 26–34. [Google Scholar] [CrossRef]
- Botella-Pavía, P.; Besumbes, O.; Phillips, M.A.; Carretero-Paulet, L.; Boronat, A.; Rodríguez-Concepción, M. Regulation of carotenoid biosynthesis in plants: Evidence for a key role of hydroxymethylbutenyl diphosphate reductase in controlling the supply of plastidial isoprenoid precursors. Plant J. 2004, 40, 188–199. [Google Scholar] [CrossRef]
- Rise, M.; Cohen, E.; Vishkautsan, M.; Cojocaru, M.; Gottlieb, H.E.; Arad, S.M. Accumulation of secondary carotenoids in Chlorella zofingiensis. J. Plant Physiol. 1994, 144, 287–292. [Google Scholar] [CrossRef]
- Bar, E.; Rise, M.; Vishkantsan, M.; Arad, S. Pigment andstructural changes in Chlorella zofingiensis upon light and nitrogen stress. J. Plant Physiol. 1995, 146, 527–534. [Google Scholar] [CrossRef]
- Orosa, M.; Valero, J.F.; Herrero, C.; Abalde, J. Comparison of the accumulation of astaxanthin in Haematococcus pluvialis and other green microalgae under N-starvation and high light conditions. Biotechnol. Lett. 2001, 23, 1079–1085. [Google Scholar] [CrossRef]
- Huang, J.C.; Wang, Y.; Sandmann, G.; Chen, F. Isolation and characterization of a carotenoid oxygenase gene from Chlorella zofingiensis (Chlorophyta). Appl. Microbiol. Biotechnol. 2006, 71, 473–479. [Google Scholar] [CrossRef]
- Gilmore, N.; Mohanty, H.; Yamamoto, Y. Epoxidation of zeaxanthin and antheraxanthin reverses non-photochemical quenching of photosystem II chlorophyll a fluorescence in the presence of trans-thylakoid delta pH. FEBS Lett. 1994, 350, 271–274. [Google Scholar] [CrossRef]
- Goss, R.; Böhme, K.; Wilhelm, C. The xanthophyll cycle of Mantoniella squamata converts violaxanthin into antheraxanthin but not to zeaxanthin: consequences for the mechanism of enhanced non-photochemical energy dissipation. Planta 1998, 205, 613–621. [Google Scholar] [CrossRef]
- Jahns, P.; Latowski, D.; Strzalka, K. Mechanism and regulation of the violaxanthin cycle: The role of antenna proteins and membrane lipids. Biochim. Biophys. Acta 2009, 1787, 3–14. [Google Scholar] [CrossRef]
- Misawa, N.; Satomi, Y.; Kondo, K.; Yokoyama, A.; Kajiwara, S. Structure and functional analysis of a marine bacterial carotenoid biosynthesis gene cluster and astaxanthin biosynthetic pathway proposed at the gene level. J. Bacteriol. 1995, 177, 6575–6584. [Google Scholar]
- Eskling, M.; Arvidsson, P.O.; Åkerlund, H.E. The xanthophyll cycle, its regulation and components. Physiol. Plant 1997, 100, 806–816. [Google Scholar]
- Sandmann, G.; Albrecht, M.; Schnurr, G.; Knorzer, O.; Böger, P. The biotechnological potential and design of novel carotenoids by gene combination in Escherichia coli. Trends Biotechnol. 1999, 17, 233–237. [Google Scholar] [CrossRef]
- Takaichi, S. Carotenoids in algae: Distributions, biosyntheses and functions. Mar. Drugs 2011, 9, 1101–1118. [Google Scholar] [CrossRef]
- Frey, A.; Audran, C.; Marin, E.; Sotta, B.; Marion-Poll, A. Engineeringseed dormancy by the modification of zeaxanthin epoxidase gene expression. Plant Mol. Biol. 1999, 39, 1267–1274. [Google Scholar] [CrossRef]
- Wang, N.; Fang, W.; Han, H.; Sui, N.; Li, B.; Meng, Q.W. Overexpression of zeaxanthin epoxidase gene enhances thesensitivity of tomato PSII photoinhibition to high light andchilling stress. Physiol. Plant. 2008, 132, 384–396. [Google Scholar] [CrossRef]
- Ruban, A.V.; Young, A.J.; Pascal, A.A.; Horton, P. The effects of illumination on the xanthophyll composition of PSII light harvesting complexesof spinach thylakoid membranes. Plant Physiol. 1994, 104, 227–234. [Google Scholar]
- Lee, A.J.; Thornber, J.P. Analysis of the pigment-protein complexes from barley (Hordeum vulgare): The xanthophyll cycle intermediates occur mainly in the light-harvesting complexes of photosystem I and II. Plant Physiol. 1995, 107, 565–574. [Google Scholar]
- Goss, R.; Lepetit, B.; Wilhelm, C. Evidence for a rebinding of antheraxanthin to the light-harvesting complex during the epoxidation reaction of the violaxanthin cycle. J. Plant Physiol. 2006, 163, 585–590. [Google Scholar] [CrossRef]
- Kalituho, L.; Rech, J.; Jahns, P. The roles of specific xanthophylls in light utilization. Planta 2007, 225, 423–439. [Google Scholar]
- Dall’Osto, L.; Caffarri, S.; Bassi, R. A mechanism of non photochemical energy dissipation, independent from PsbS, revealed by a conformational change in the antenna protein CP26. Plant Cell 2005, 17, 1217–1232. [Google Scholar] [CrossRef]
- Kalituho, L.; Beran, K.C.; Jahns, P. The transiently generated nonphotochemical quenching of excitation energy in arabidopsis leaves is modulated by zeaxanthin. Plant Physiol. 2007, 143, 1861–1870. [Google Scholar] [CrossRef]
- Arnon, D.I.; McSwain, B.D.; Tsujimoto, H.Y.; Wada, K. Photochemical activity and components of membrane preparations from blue-green algae. I. Coexistence of two photosystems in relation to chlorophyll a and removal of phycocyanin. Biochim. Biophys. Acta 1974, 357, 231–245. [Google Scholar] [CrossRef]
- Gorman, D.S.; Levine, R.P. Cytochrome f and plastocyanin: Their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 1965, 54, 1665–1669. [Google Scholar] [CrossRef]
- Anwaruzzaman, N.; Chin, B.L.; Li, X.P.; Lohr, M.; Martínez, D.A.; Niyogi, K.K. Genomic analysis of mutants affecting xanthophyll biosynthesis and regulation of photosynthetic light harvesting in Chlamydomonas reinhardtii. Photosynth. Res. 2004, 82, 265–276. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J., Walker, M., Eds.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Nielsen, H.; von Hejne, G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999, 8, 978–984. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Von Heijne, G. Membrane protein structure prediction: hydrophobicity analysis and the “Positive inside”rule. J. Mol. Biol. 1992, 225, 487–494. [Google Scholar] [CrossRef]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L.; et al. InterPro: The integrative protein signature database. Nucleic Acids Res. 2009, 37, D211–D215. [Google Scholar] [CrossRef]
- León, R.; Couso, I.; Fernández, E. Metabolic engineering of ketocarotenoids biosynthesis in the unicelullar microalga Chlamydomonas reinhardtii. J. Biotechnol. 2007, 15, 143–152. [Google Scholar]
- Sizova, I.; Fuhrman, M.; Hegemann, P. A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 2001, 277, 221–229. [Google Scholar] [CrossRef]
- Von Kampen, J.; Nieländer, U.; von Wettern, M. Stress-dependent transcription of a gene encoding a Gb-like polypeptide from Chlamydomonas reinhardtii. J. Plant Physiol. 1994, 143, 756–758. [Google Scholar] [CrossRef]
- Sun, N.; Wang, Y.; Huang, J.C.; Chen, F. Sugar-based growth, astaxanthin accumulation and carotenogenic transcription of heterotrophic Chlorella zofingiensis (Chlorophyta). Process Biochem. 2008, 43, 1288–1292. [Google Scholar] [CrossRef]
- Vidhyavathi, R.; Venkatachalam, L.; Sarada, R.; Ravishankar, G.A. Regulation of carotenoid biosynthetic genes expression and carotenoid accumulation in the green alga Haematococcus pluvialis under nutrient stress conditions. J. Exp. Bot. 2008, 59, 1409–1418. [Google Scholar] [CrossRef]
- León, R.; Vila, M.; Hernánz, D.; Vilches, C. Production of phytoene by herbicide-treated microalgae Dunaliella bardawil in two-phase systems. Biotechnol. Bioeng. 2005, 92, 695–701. [Google Scholar] [CrossRef]
- Samples Availability: Available from the authors.
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Couso, I.; Cordero, B.F.; Vargas, M.Á.; Rodríguez, H. Efficient Heterologous Transformation of Chlamydomonas reinhardtii npq2 Mutant with the Zeaxanthin Epoxidase Gene Isolated and Characterized from Chlorella zofingiensis. Mar. Drugs 2012, 10, 1955-1976. https://doi.org/10.3390/md10091955
Couso I, Cordero BF, Vargas MÁ, Rodríguez H. Efficient Heterologous Transformation of Chlamydomonas reinhardtii npq2 Mutant with the Zeaxanthin Epoxidase Gene Isolated and Characterized from Chlorella zofingiensis. Marine Drugs. 2012; 10(9):1955-1976. https://doi.org/10.3390/md10091955
Chicago/Turabian StyleCouso, Inmaculada, Baldo F. Cordero, María Ángeles Vargas, and Herminia Rodríguez. 2012. "Efficient Heterologous Transformation of Chlamydomonas reinhardtii npq2 Mutant with the Zeaxanthin Epoxidase Gene Isolated and Characterized from Chlorella zofingiensis" Marine Drugs 10, no. 9: 1955-1976. https://doi.org/10.3390/md10091955
APA StyleCouso, I., Cordero, B. F., Vargas, M. Á., & Rodríguez, H. (2012). Efficient Heterologous Transformation of Chlamydomonas reinhardtii npq2 Mutant with the Zeaxanthin Epoxidase Gene Isolated and Characterized from Chlorella zofingiensis. Marine Drugs, 10(9), 1955-1976. https://doi.org/10.3390/md10091955





