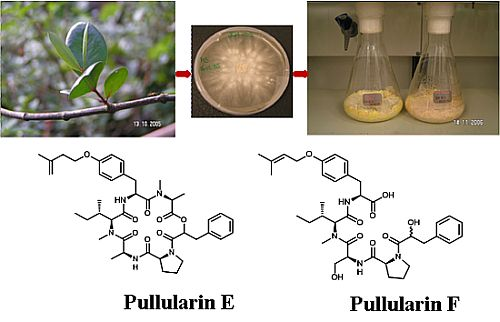
Pullularins E and F, Two New Peptides from the Endophytic Fungus Bionectria ochroleuca Isolated from the Mangrove Plant Sonneratia caseolaris
Abstract
:1. Introduction
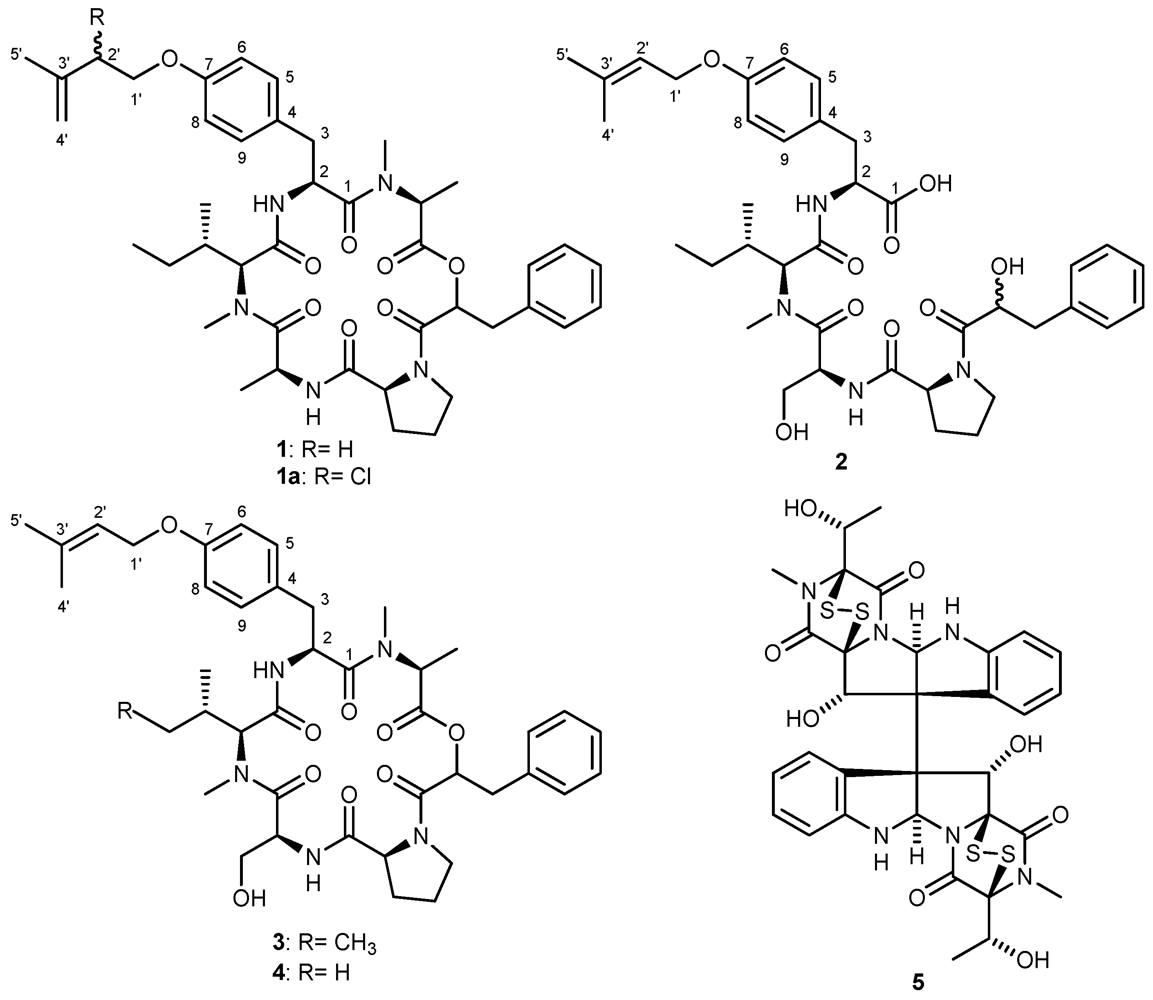
2. Results and Discussion
| Position | δC b | δH mult. (J Hz)c | COSY | HMBC (H→C) | ROESY |
|---|---|---|---|---|---|
| O-isoprenyl-Tyr | |||||
| 1 (C=O) | 168.1 C d | ||||
| 2 | 51.2 CH | 4.72 ddd (4.4, 8.8, 8.8) | 3, NH | 1, C=O (N-Me-lle) | N-CH3 (N-Me-Ala) |
| 3 | 36.9 CH2 | 2.95 dd (9.8, 13.8) | 2, 3b | 1, 2, 4, 5, 9 | |
| 2.41 m | 2, 3a | 1, 2, 4, 5, 9 | |||
| 4 | 130.7 C | ||||
| 5, 9 | 130.2 CH | 7.04 d (8.6) | 6, 8 | 3, 4, 6, 7 | |
| 6, 8 | 114.4 CH | 6.83 d (8.6) | 5, 9 | 5, 7 | |
| 7 | 156.2 C | ||||
| 1′ | 69.7 CH2 | 4.18 ddd (1.8, 6.0, 10.6) | 2′, 1′b | 2′, 3′, 7 | |
| 4.15 ddd (0.8, 7.3, 10.6) | 2′, 1′a | ||||
| 2′ | 63.0 CH | 4.86 ddd (2.1, 6.5, 6.5) | 1′ | 1′, 3′, 4′, 5′ | |
| 3′ | 141.3 C | ||||
| 4′ | 116.5 CH2 | 5.20 s | 4′b, 5′ (weak) | 2′, 3′, 5′ | |
| 5.04 s | 4′a, 5′ (weak) | 2′, 5′ | |||
| 5′ | 17.6 CH3 | 1.80 s | 4′ (weak) | 2′, 3′, 4′ | |
| NH | 9.53 d (8.7) | 2 | 1, 2, C=O (N-Me-lle) | 2 (N-Me-lle) | |
| N-Me-Ala | |||||
| 1 (C=O) | 169.5 C | ||||
| 2 | 58.4 CH | 3.69 q (6.7) | 3 | 1, 3, N-CH3, C=O (O-isoprenyl-Tyr) | N-CH3 |
| 3 | 13.0 CH3 | 1.05 d (6.6) | 2 | 1, 2 | N-CH3 |
| N-CH3 | 35.8 CH3 | 2.93 s | 2, C=O (O-isoprenyl-Tyr) | 2, 3, 2 (O-isoprenyl-Tyr) | |
| 3-Ph-Lac | |||||
| 1 (C=O) | 165.4 C | ||||
| 2 | 71.5 CH | 5.50 dd (5.1, 8.7) | 3 | 1, 3, 4, C=O (N-Me-Ala) | 5 (Pro) |
| 3 | 35.8 CH2 | 3.11 dd (8.8, 13.0) | 2, 3b | 1, 2, 4, 5,9 | |
| 2.81 dd (4.9, 13.0) | 2, 3a | 1, 2, 4, 5,9 | |||
| 4 | 136.4 C | ||||
| 5, 9 | 129.5 CH | 7.19 (overlapped) | 6, 8 | 3, 6, 7 | |
| 6, 8 | 128.4 CH | 7.26 t (7.5) | 5, 7, 9 | 4, 5, 7 | |
| 7 | 126.6 CH | 7.19 (overlapped) | 6, 8 | 5, 6 | |
| Pro | |||||
| 1 (C=O) | 171.8 C | ||||
| 2 | 58.4 CH | 4.31 dd (1.8, 8.6) | 3 | 3, 4, 5 | NH (Ala) |
| 3 | 29.1 CH2 | 2.03 m | 2, 3b, 4 | 1 | |
| 1.68 m | 2, 3a, 4 | 4 | |||
| 4 | 23.8 CH2 | 1.82 m | 3, 5 | ||
| 5 | 45.8 CH2 | 3.75 ddd (3.0, 8.7, 8.7) | 4, 5b | 3 | 2 (3-Ph-Lac) |
| 3.21 q (8.5) | 4, 5a | 4 | 2 (3-Ph-Lac) | ||
| Ala | |||||
| 1 (C=O) | 173.8 C | ||||
| 2 | 44.4 CH | 4.65 m | 3, NH | 1, 3, C=O (Pro) | |
| 3 | 17.2 CH3 | 1.21 d (6.6) | 2 | 1, 2 | N-CH3 (N-Me-lle) |
| NH | 8.89 d (4.3) | 2 | 1, 1 (Pro), 3 | 3, 2 (Pro), 3b (Pro) | |
| N-Me-lle | |||||
| 1 (C=O) | 168.0 C d | ||||
| 2 | 64.8 CH | 4.53 d (10.8) | 3 | 1, 3, 3-CH3, N-CH3, C=O (Ala) | NH (O-isoprenyl-Tyr) |
| 3 | 32.0 CH | 2.00 m | 2, 4, 3-CH3 | ||
| 4 | 24.3 CH2 | 1.22 m | 3, 5 | ||
| 0.90 m | |||||
| 5 | 11.8 CH3 | 0.89 t (6.7) | 4 | 3, 4 | |
| 3-CH3 | 15.9 CH3 | 0.91 d (6.4) | 3 | 2, 3, 4 | |
| N-CH3 | 28.4 CH3 | 2.40 s | 2, C=O (Ala) | 3 (Ala) |
| Compound | L5178Y Survival Rate in % (10 µg/mL) | EC50 (µg/mL) |
|---|---|---|
| Chloro-derivative of pullularin E (1a) | 15.6 | 5.60 |
| Pullularin F (2) | 114.3 | >10 |
| Pullularin A (3) | 1.7 | 2.60 |
| Pullularin C (4) | 21.7 | 6.70 |
| Verticillin D (5) | 0.5 | <0.1 |
| Kahalalide F (positive control) | 6.40 |
3. Experimental Section
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Identification of Fungal Cultures
3.4. Cultivation
3.5. Extraction and Fractionation
3.6. Preparation and HPLC Analysis of Marfey Derivatives [21,22]
| Position | δC | δH mult. (J Hz) b | COSY | HMBC (H→C) |
|---|---|---|---|---|
| O-prenyl-Tyr | ||||
| 1 (C=O) | 172.0 C | |||
| 2 | 55.4 CH | 4.72 ddd (4.4, 8.8, 8.8) | 1, 3, 4, C=O (N-Me-lle) | |
| 3 | 35.8 CH2 | 2.96 dd (4.0, 12.8) | 2, 5 | 1, 4, 5 |
| 2.59 br d (4.3) | 2, 5 | 1, 4, 5 | ||
| 4 | 131.0 C | |||
| 5, 9 | 130.5 CH | 7.04 d (8.6) | 6, 8 | 3, 5, 6, 7 |
| 6, 8 | 116.0 CH | 6.82 d (8.6) | 5, 9 | 4 |
| 7 | 156.5 C | |||
| 1′ | 64.0 CH2 | 4.49 d (6.4) | 2′ | 2′, 3′, 7 |
| 2′ | 120.5 CH | 5.42 br m | 1′ | 4′, 5′ |
| 3′ | 137.0 C | |||
| 4′ | 25.0 CH3 | 1.72 s | 2′, 3′, 5′ | |
| 5′ | 18.0 CH3 | 1.76 s | 2′, 3′, 4′ | |
| NH | 8.45 d (8.7) | 2 (O-isoprenyl-Tyr) | 1 | |
| N-Me-lle | ||||
| 1 (C=O) | 168.5 C | |||
| 2 | 61.0 CH | 4.52 d (11.0) | 1, C=O (Ser) | |
| 3 | 31.0 CH | 1.90 m * | 2, 4, 3-CH3 | 3-CH3 |
| 4 | 24.0 CH2 | 1.25 m * | 4b, 5 | 3, 3-CH3 |
| 0.92 m * | 4a, 5 | |||
| 5 | 10.0 CH3 | 0.75 m * | 4 | 3, 4 |
| 3-CH3 | 15.5 CH3 | 0.80 m * | 3 | 2, 3, 4 |
| N-CH3 | 28.4 CH3 | 2.85 s | 2, C=O (Ser) | |
| Ser | ||||
| 1 (C=O) | 171.8 C | |||
| 2 | 44.4 CH | 4.80 m | 3, NH | 3, 1 (Pro) |
| 3 | 62.0 CH2 | 3.65 m | 2 | 1 |
| 3.48 m | 2 | 1 | ||
| NH | 8.18 d (4.3) | 2 | 1 | |
| Pro | ||||
| 1 (C=O) | 171.5 C | |||
| 2 | 52.0 CH | 4.70 m | 3 | 3, 4 |
| 3 | 32.0 CH2 | 3.48 m * | 2, 4 | 2 |
| 3.20 m * | 2, 4 | |||
| 4 | 22.0 CH2 | 1.80 m * | 3, 5 | |
| 1.75 m * | 3, 5 | |||
| 5 | 32.0 CH2 | 1.78 m * | 4, 5b | |
| 3.20 m * | 4, 5a | |||
| 3-Ph-Lac | ||||
| 1 (C=O) | 165.4 C | |||
| 2 | 71.5 CH | 4.10 m | 3 | |
| 3 | 41.0 CH2 | 2.85 m * | 2 | 1, 4, 5 |
| 2.70 m * | 1, 4, 5 | |||
| 4 | 138.0 C | |||
| 5, 9 | 129.0 CH | 7.19 dd (2.0, 8.8) * | 6, 8 | 4, 7 |
| 6, 8 | 126.0 CH | 7.25 d (7.2) * | 5, 9 | 4, 5, 6, 8 |
| 7 | 128.0 CH | 7.26 m * |
3.7. Cell Proliferation Assay
4. Conclusions
Acknowledgments
References
- Aly, A.H.; Debbab, A.; Kjer, J.; Proksch, P. Fungal endophytes from higher plants: A prolific source of phytochemicals and other bioactive natural products. Fungal Divers. 2010, 41, 1–16. [Google Scholar] [CrossRef]
- Aly, A.H.; Debbab, A.; Proksch, P. Fifty years of drug discovery from fungi. Fungal Divers. 2011, 50, 3–19. [Google Scholar] [CrossRef]
- Bigelis, R.; He, H.; Yang, H.Y.; Chang, L.P.; Greenstein, M. Production of fungal antibiotics using polymeric solid supports in solid-state and liquid fermentation. J. Ind. Microbiol. Biotechnol. 2006, 33, 815–826. [Google Scholar] [CrossRef]
- Cragg, G.; Newman, D. Nature: A vital source of leads for anticancer drug development. Phytochem. Rev. 2009, 8, 313–331. [Google Scholar] [CrossRef]
- Lugo, A.E.; Snedaker, S.C. The ecology of mangroves. Annu. Rev. Ecol. Syst. 1974, 5, 39–64. [Google Scholar]
- Wen, L.; Chen, G.; She, Z.G.; Yan, C.Y.; Cai, J.N.; Mu, L. Two new paeciloxocins from a mangrove endophytic fungus Paecilomyces sp. Russ. Chem. Bull. 2010, 59, 1656–1659. [Google Scholar] [CrossRef]
- Shao, C.L.; Wang, C.Y.; Gu, Y.C.; Wei, M.Y.; Pan, J.H.; Deng, D.S.; She, Z.G.; Lin, Y.C. Penicinoline, a new pyrrolyl 4-quinolinone alkaloid with an unprecedented ring system from an endophytic fungus Penicillium sp. Bioorg. Med. Chem. Lett. 2010, 20, 3284–3286. [Google Scholar]
- Liu, F.; Cai, X.L.; Yang, H.; Xia, X.K.; Guo, Z.Y.; Yuan, J.; Li, M.F.; She, Z.G.; Lin, Y.C. The bioactive metabolites of the mangrove endophytic Fungus Talaromyces sp. ZH-154 isolated from Kandelia candel (L.) Druce. Planta Med. 2010, 76, 185–186. [Google Scholar] [CrossRef]
- Aly, A.H.; Debbab, A.; Proksch, P. Fungal endophytes: Unique plant inhabitants with great promises. Appl. Microbiol. Biotechnol. 2011, 90, 1829–1845. [Google Scholar] [CrossRef]
- Xu, J.; Aly, A.; Wray, V.; Proksch, P. Polyketide derivatives of endophytic fungus Pestalotiopsis sp. isolated from the Chinese mangrove plant Rhizophora mucronata. Tetrahedron Lett. 2011, 52, 21–25. [Google Scholar]
- Debbab, A.; Aly, A.H.; Edrada-Ebel, R.; Wray, V.; Mueller, W.E.; Totzke, F.; Zirrgiebel, U.; Schaechtele, C.; Kubbutat, M.H.; Lin, W.H.; et al. Bioactive metabolites from the endophytic fungus Stemphylium globuliferum isolated from Mentha pulegium. J. Nat. Prod. 2009, 72, 626–631. [Google Scholar]
- Freinkman, E.; Oh, D.C.; Scott, J.J.; Currie, C.R.; Clardy, J. Bionectriol A, a polyketide glycoside from the fungus Bionectria sp. associated with the fungus-growing ant, Apterostigma dentigerum. Tetrahedron Lett. 2009, 50, 6834–6837. [Google Scholar]
- Ju, Y.-M.; Juang, S.-H.; Chen, K.-J.; Lee, T.-H. TMC-151 a monoacetate, a new polyketide from Bionectria ochroleuca. Z. Naturforsch. B 2007, 62b, 561–564. [Google Scholar]
- Zheng, C.J.; Kim, C.J.; Bae, K.S.; Kim, Y.H.; Kim, W.G. Bionectins A–C, epidithiodioxopiperazines with anti-MRSA activity, from Bionectra byssicola F120. J. Nat. Prod. 2006, 69, 1816–1819. [Google Scholar]
- Zheng, C.J.; Park, S.H.; Koshino, H.; Kim, Y.H.; Kim, W.G. Verticillin G, a new antibacterial compound from Bionectra byssicola. J. Antibiot. 2007, 60, 61–64. [Google Scholar] [CrossRef]
- Zheng, C.J.; Kim, Y.H.; Kim, W.G. Glioperazine B, as a new antimicrobial agent against Staphylococcus aureus, and glioperazine C: Two new dioxopiperazines from Bionectra byssicola. Biosci. Biotechnol. Biochem. 2007, 71, 1979–1983. [Google Scholar] [CrossRef]
- Isaka, M.; Berkaew, P.; Intereya, K.; Komwijit, S.; Sathitkunanon, T. Antiplasmodial and antiviral cyclohexadepsipeptides from the endophytic fungus Pullularia sp. BCC 8613. Tetrahedron 2007, 63, 6855–6860. [Google Scholar]
- Joshi, B.K.; Gloer, G.B.; Wicklow, D.T. New verticillin and glisoprenin analogues from Gliocladium catenulatum, a mycoparasite of Aspergillus flavus sclerotia. J. Nat. Prod. 1999, 62, 730–733. [Google Scholar] [CrossRef]
- Harada, K.; Kiyonga, F.; Mayuni, T.; Hibino, Y.; Suzuki, M.; Ikai, Y.; Oka, H. A method using LCMS for determination of absolute configuration of constituent amino acids in peptide-advanced method. Tetrahedron Lett. 1995, 35, 1515–1518. [Google Scholar]
- Wang, S.; Li, X.M.; Teuscher, F.; Li, D.L.; Diesel, A.; Ebel, R.; Proksch, P.; Wang, B.G. Chaetopyranin, a benzaldehyde derivative, and other related metabolites from Chaetomium globosum, an endophytic fungus derived from the marine red alga Polysiphonia urceolata. J. Nat. Prod. 2006, 69, 1622–1625. [Google Scholar] [CrossRef]
- Marfey, P. Determination of D-amino acids. II: Use of a bifunctional reagent 1,5-difluoro-2,4-dinitrobenzene. Carlsberg Res. Commun. 1984, 49, 591–596. [Google Scholar] [CrossRef]
- Ashour, M.; Edrada-Ebel, R.A.; Ebel, R.; Wray, V.; Waetjen, W.; Padmakumar, K.; Mueller, W.E.G.; Lin, W.; Proksch, P. Kahalalide derivatives from the indian sacoglossan mollusk Elysia grandifolia. J. Nat. Prod. 2006, 69, 1547–1553. [Google Scholar] [CrossRef]
- Carmichael, J.; DeGraff, W.G.; Gazdar, A.F.; Minna, J.D.; Mitchell, J.B. Evaluation of a tetrazolium-based semiautomated colorimetric assay: Assessment of radiosensitivity. Cancer Res. 1987, 47, 943–946. [Google Scholar]
- Samples Availability: Available from the authors.
Supplementary Files
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ebrahim, W.; Kjer, J.; El Amrani, M.; Wray, V.; Lin, W.; Ebel, R.; Lai, D.; Proksch, P. Pullularins E and F, Two New Peptides from the Endophytic Fungus Bionectria ochroleuca Isolated from the Mangrove Plant Sonneratia caseolaris. Mar. Drugs 2012, 10, 1081-1091. https://doi.org/10.3390/md10051081
Ebrahim W, Kjer J, El Amrani M, Wray V, Lin W, Ebel R, Lai D, Proksch P. Pullularins E and F, Two New Peptides from the Endophytic Fungus Bionectria ochroleuca Isolated from the Mangrove Plant Sonneratia caseolaris. Marine Drugs. 2012; 10(5):1081-1091. https://doi.org/10.3390/md10051081
Chicago/Turabian StyleEbrahim, Weaam, Julia Kjer, Mustapha El Amrani, Victor Wray, Wenhan Lin, Rainer Ebel, Daowan Lai, and Peter Proksch. 2012. "Pullularins E and F, Two New Peptides from the Endophytic Fungus Bionectria ochroleuca Isolated from the Mangrove Plant Sonneratia caseolaris" Marine Drugs 10, no. 5: 1081-1091. https://doi.org/10.3390/md10051081
APA StyleEbrahim, W., Kjer, J., El Amrani, M., Wray, V., Lin, W., Ebel, R., Lai, D., & Proksch, P. (2012). Pullularins E and F, Two New Peptides from the Endophytic Fungus Bionectria ochroleuca Isolated from the Mangrove Plant Sonneratia caseolaris. Marine Drugs, 10(5), 1081-1091. https://doi.org/10.3390/md10051081