Assessment of the Bacteriocinogenic Potential of Marine Bacteria Reveals Lichenicidin Production by Seaweed-Derived Bacillus spp.
Abstract
:1. Introduction
2. Results
2.1. Isolation and Characterization of Bacteriocin-Producing Marine Bacteria
| Isolate no. (identified by 16S rRNA gene sequencing) | Origin | Isolation medium | Culture Supernatants (Well diffusion assay) a | Culture (deferred antagonism assay) a | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L. innocua WIT 361 | L. monocytogenes WIT 041 | B. subtilis ATCC 6633 | S. aureus DPC 5246 | MRSA W73365 | E. faecium ATCC 19434 | E. coli DSM 10720 | S. Typhimurium LT2 | E. faecalis ATCC 19433 | C. sakazakii ATCC 12868 | |||||
| B. licheniformis WIT 562 b | P. lanosa | LPMA | +/- | +/- | - | - | - | - | - | - | - | - | ||
| B. licheniformis WIT 564 b | Ulva spp. | LNA | - | - | + | + | - | - | - | - | - | - | ||
| B. licheniformis WIT 566 b | Ulva spp. | MA | + | - | + | - | - | - | - | - | - | - | ||
| B. pumilus WIT 560 c | F. vesiculosus | LNA | ++ | ++ | - | ++++ | +++ | - | ++++ | ++ | + | +++ | ||
| B. pumilus WIT 561 c | F. vesiculosus | LNA | ++ | ++ | - | +++ | +++ | - | ++++ | +++ | + | ++++ | ||
| B. pumilus WIT 563 | Ulva spp. | MA | + | +/- | - | +++ | ++ | - | ++++ | +/- | + | ++ | ||
| B. licheniformis WIT 565 | Ulva spp. | MA | ++ | +/- | + | + | - | - | ++++ | +/- | + | ++ | ||
| B. licheniformis WIT 567 | Sand | MA | - | ++ | + | ++ | + | ++++ | - | - | - | - | ||
| B. licheniformis WIT 568 | Seawater | MA | - | + | + | ++ | + | ++++ | - | - | - | - | ||
| B. licheniformis WIT 569 | U. lactuca | MA d | + | + | + | +++ | - | ++++ | - | - | + | - | ||
| B. licheniformis WIT 570 | U. lactuca | MA d | +++ | +++ | - | +++ | +++ | - | - | - | ++ | - | ||
| B. pumilus WIT 571 | P. lanosa | MA d | ++ | ++ | - | +++ | +++ | - | - | - | - | - | ||
| B. pumilus WIT 572 | P. lanosa | MA d | +++ | ++ | - | +++ | +++ | - | ++++ | +/- | + | +++ | ||
| B. pumilus WIT 573 | P. palmata | AIA | + | + | - | +++ | + | - | ++++ | +/- | ++ | +/- | ||
| B. pumilus WIT 574 | F. serratus | AIA | + | ++ | - | ++ | - | - | +/- | - | - | +/- | ||
| Lc. Lactis NZ 9700 e | NA f | NA | ++ | - | + | ++ | - | ++ | ND | ND | ND | ND | ||
2.2. Characterization of Antimicrobial Compounds Produced by Marine Bacteria
2.2.1. Effect of Growth Medium on Antimicrobial Production
2.2.2. Cross Sensitivity Assays
2.2.3. Physicochemical Characterization of Antimicrobial CFS
| Marine Isolate | Sensitive to b | Resistant to (temperature, °C) c | Resistant to(pH range) d |
|---|---|---|---|
| WIT 562 e | Pronase E, proteinase K, pepsin, catalase, α-chymotrypsin, trypsin, protease type I | 50 (60) | 5–9 (2) |
| WIT 564 e | Pronase E, proteinase K, trypsin | 50 (60) | 3–9 (2) |
| WIT 566 e | Pronase E, proteinase K, catalase | 50 (60) | 5–9 (2, 11) |
| WIT 560 f | Pronase E, proteinase K, pepsin, α-chymotrypsin, trypsin, protease type I | 50 (70) | 5–9 (2, 11) |
| WIT 561 f | Pronase E, proteinase K, pepsin, α-chymotrypsin, protease type I | 50 (70) | 2–9 (12) |
| WIT 563 | Pronase E, proteinase K, pepsin, catalase, α-chymotrypsin, trypsin, protease type I | 70 (80) | 2–9 (12) |
| WIT 565 | Pronase E | 50 (60) | 3–9 (2, 11) |
| WIT 567 | Pronase E, proteinase K, pepsin, α-chymotrypsin, trypsin | 90 (121) | 2–9 (12) |
| WIT 568 | Pronase E, α-chymotrypsin, trypsin, protease type I | 50 (121) | 3–9 (2, 12) |
| WIT 569 | Pronase E, proteinase K, pepsin, α-chymotrypsin, trypsin | 90 (121) | 3–9 (2, 12) |
| WIT 570 | Proteinase K, pepsin, α-chymotrypsin, trypsin | 90 (121) | 5–9 (2, 12) |
| WIT 571 | Pronase E, proteinase K, trypsin | 40 (80) | 5–9 (2, 12) |
| WIT 572 | Pronase E, proteinase K, pepsin, α-chymotrypsin, trypsin, protease type I | 50 (60) | 3–9 (2, 12) |
| WIT 573 | Pronase E, proteinase K, pepsin, catalase, α-chymotrypsin, trypsin, protease type I | 50 (60) | 5–7 (2, 11) |
| WIT 574 | Proteinase K, α-chymotrypsin, protease type I | 50 (60) | 2–9 |
| NZ 9700 g | Pronase E, proteinase K, pepsin, α-chymotrypsin, trypsin, protease type I | 70 (100) | 2–9 (11) |
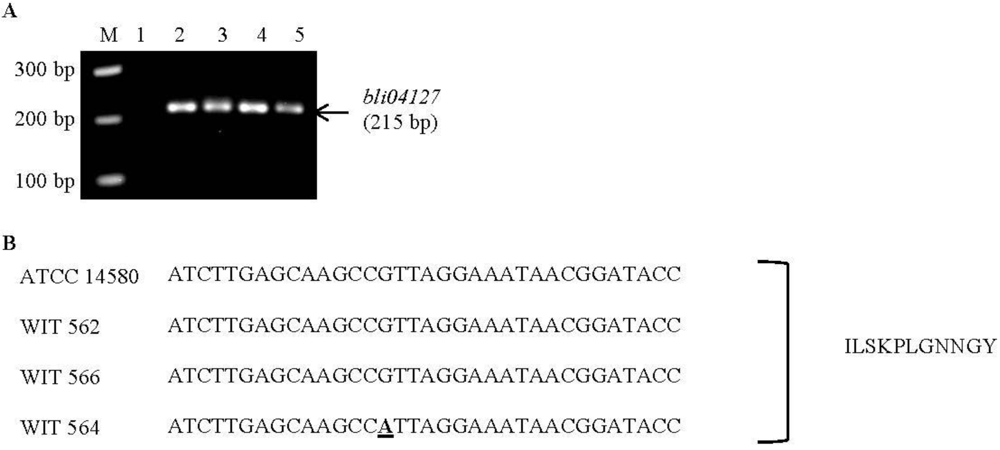
2.3. Screening for Known Bacteriocin-Associated Genes
| Bacteriocin | Gene | Sequence (5′-3′) | Size of expected product (bp) | Annealing temperature (°C) | Positive control strain a | Genbank accession number of bacteriocin sequence | Reference |
|---|---|---|---|---|---|---|---|
| Subtilin | Spa | ACTATGAATCAATGGAAGG | 370 | 50 | B. subtilis ATCC 6633 | M99263.1 | [8] |
| TTGCAGTTACAAGTTAGTG | |||||||
| Subtilosin | Sbo | GGTTGTGCAACATGCTCGAT | 300 | 58 | B. subtilis ATCC 6633 | AJ430547.1 | [8] |
| CTCAGGAAGCTGGTGAACTC | |||||||
| Sublancin | Sun | GTGTGCTGCGTTGTGGCTACAA | 230 | 62 | B. subtilis 168 | NC_000964.3 | [8] |
| TTGACGAGATACAAGCTAGTCC | |||||||
| Coagulin | CoaA | GGTGGTAAATACTACGGTAATGGGGT | ~600 | 66 | B. coagulans I4 | AF300457 | [9] |
| GTGTCTAAATTACTGGTTGATTCGT | |||||||
| Mersacidin | MrsA | CTTAATAAGGGGGTAATAC | 270 | 56 | B. subtilis HIL | Z47559.1 | This study |
| TAGGCTGTTCCTTCTGAAGG | Y-85,54728 | ||||||
| Lichenicidin | Bli04127 | GGAAATGATTCTTTCATGG | 215 | 60 | B. licheniformis ATCC | CP000002.3 | This study |
| TTAGTTACAGCTTGGCATG | 14850 | ||||||
| Ericin A | EriSa | TGTCAAAGTTCGATGACTTC | 171 | 56 | B. subtilis A 1/3 | AF233755.1 | This study |
| TCAGCACTTAGCAAATGTTG | |||||||
| Haloduracin A1 | BH0454 | ATGGAAAATGCCTCTTGAG | 191 | 54 | B. halodurans ATCC | BA000004.3 | This study |
| TTAGTTGCAAGAAGGCATG | BAA-125D-5 | ||||||
| Haloduracin A2 | BH0453 | TTAGCACTGGCTTGTACACT | 180 | 58 | B. halodurans ATCC | BA000004.3 | This study |
| TTGCGTAATCCTGAATTCCG | BAA-125D-5 | ||||||
| Thuricin17 | TucA1, A2 & A3 | GTAGGTCAAATGGAAACAC | 589 | 52 | B. thuringiensis NEB17 | FJ159242.1 | This study |
| TTAACTTGCAGTACTAGCTC | |||||||
| Thurincin H | H1, H2 & H3 | ATGGAAACACCAGTAGTACA | 579 | 56 | B. thuringiensis SF361 | FJ977580.1 | This study |
| TTAACTTGCAGTACTAGCTC | |||||||
| Megacin A-216 | P293A | TTACATACCATGAGAAGCGCAT | 519 | 66 | B. megaterium 216 | EU014074.1 | This study |
| CATGTTAGTGCAGTTTACCTTC | |||||||
| Cerein 7B | Cer7B | ATAGCTGGGGTAAATGTGTTG | 153 | 62 | B. cereus CECT 5148 | AM087432.1 | This study |
| AAAGTAGCTGCACCTGTAAG | |||||||
| Class I-Type I | LanC | TAATTTAGGATWISYIMAYGG | ~250 | 40 | Lc. lactis NZ 9700 | NA b | [10] |
| Lantibiotic | ACCWGKIIIICCRTRRCACCA | ||||||
| LanB | TATGATCGAGAARYAKAWAGATATGG | ~400 | 44 | Lc. lactis NZ 9700 | NA | [10] | |
| TTATTAIRCAIATGIAYDAWACT | |||||||
| Class I-Type II | LanM | TTGCWAGWYWTGCWCATGG | 330 | 49 | Lc. lactis DPC 3147 | NA | [11] |
| Lantibiotic | CCTAATGAACCRTRRYAYCA |
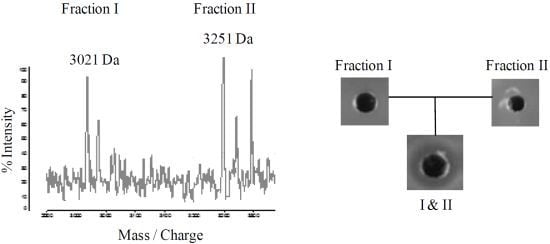
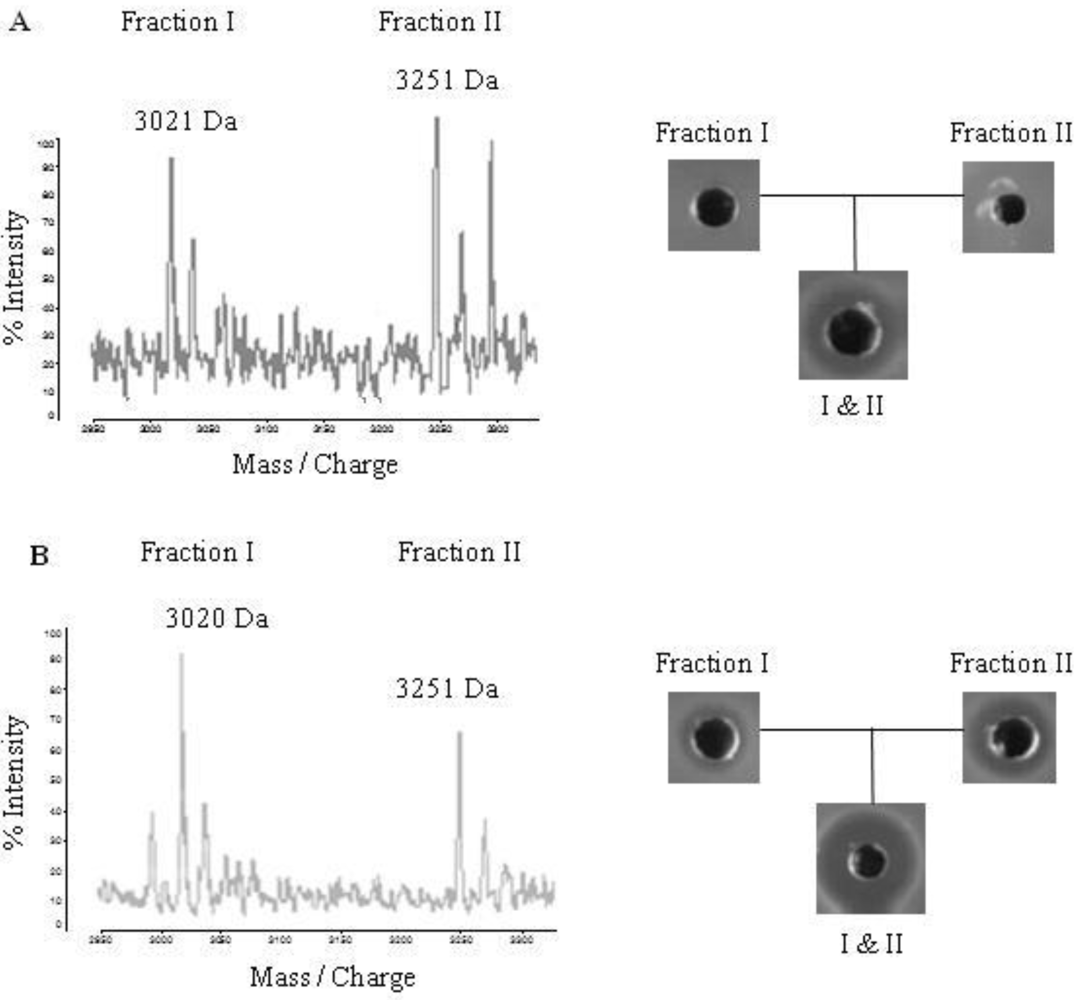
2.4. Purification of Lichenicidin
3. Discussion
4. Experimental Section
4.1. Bacterial Strains and Culture Conditions and Chemicals
4.2. Collection of Seaweed, Sand and Seawater Samples
4.3. Detection and Isolation of Bacteriocin-Producing Marine Bacteria
4.4. Deferred Antagonism Assays
4.5. Well Diffusion Assays and Determination of Spectra of Inhibition and Cross Sensitivity
4.6. Sensitivity of Antimicrobial Compounds to Enzymes, Heat and pH
4.7. Effect of Growth Medium on Antimicrobial Production
4.8. Identification and Differentiation of Bacteriocin-Producing Marine Isolates
4.9. Screening for Known Bacteriocin Genes and Sequencing of Lichenicidin Gene
4.10. Extraction and Purification of Lichenicidin
5. Conclusions
Supplementary Files
Acknowledgments
References
- Rahman, H.; Austin, B.; Mitchell, W.; Morris, P.; Jamieson, D.; Adams, D.; Spragg, A.M.; Schweizer, M. Novel anti-infective compounds from marine bacteria. Mar. Drugs 2010, 8, 498–518. [Google Scholar] [CrossRef]
- Penesyan, A.; Kjelleberg, S.; Egan, S. Development of novel drugs from marine surface-associated microorganisms. Mar. Drugs 2010, 8, 438–459. [Google Scholar] [CrossRef]
- Lee, H.; Kim, H.Y. Lantibiotics, class I bacteriocins from the genus Bacillus. J. Microbiol. Biotechnol. 2011, 21, 229–235. [Google Scholar]
- Cotter, P.D.; Hill, C.; Ross, R.P. Bacteriocins: Developing innate immunity for food. Nat. Rev. Microb. 2005, 3, 777–788. [Google Scholar] [CrossRef]
- Riley, M. Bacteriocins, Biology, Ecology, and Evolution. In Encyclopedia of Microbiology, 3rd; Schaechter, M., Ed.; Academic Press: Oxford, UK, 2009; pp. 32–44. [Google Scholar]
- Abriouel, H.; Franz, C.; Omar, N.; Gálvez, A. Diversity and applications of Bacillus bacteriocins. FEMS Microbiol. Rev. 2011, 35, 201–232. [Google Scholar] [CrossRef]
- Desriac, F.; Defer, D.; Bourgougnon, N.; Brillet, B.; Le Chevalier, P.; Fleury, Y. Bacteriocin as weapons in the marine animal-associated bacteria warfare: Inventory and potential applications as an aquaculture probiotic. Mar. Drugs 2010, 8, 1153–1177. [Google Scholar] [CrossRef]
- Barbosa, T.M. University College Cork: Cork, Ireland, Unpublished work. 2010.
- Riazi, S.; Wirawan, R.; Badmaev, V.; Chikindas, M. Characterization of lactosporin, a novel antimicrobial protein produced by Bacillus coagulans ATCC 7050. J. Appl. Microbiol. 2009, 106, 1370–1377. [Google Scholar] [CrossRef]
- Wirawan, R.; Klesse, N.; Jack, R.; Tagg, J. Molecular and genetic characterization of a novel nisin variant produced by Streptococcus uberis. Appl. Environ. Microb. 2006, 72, 1148–1156. [Google Scholar] [CrossRef]
- Hyink, O.; Balakrishnan, M.; Tagg, J.R. Streptococcus rattus strain BHT produces both a class I two component lantibiotic and a class II bacteriocin. FEMS Microbiol. Lett. 2005, 252, 235–241. [Google Scholar] [CrossRef]
- Begley, M.; Cotter, P.; Hill, C.; Ross, R. Identification of a novel two-peptide lantibiotic, lichenicidin, following rational genome mining for LanM proteins. Appl. Environ. Microb. 2009, 75, 5451–5460. [Google Scholar] [CrossRef]
- Lemos, M.L.; Toranzo, A.E.; Barja, J.L. Antibiotic activity of epiphytic bacteria isolated from intertidal seaweeds. Microb. Ecol. 1985, 11, 149–1693. [Google Scholar] [CrossRef]
- Penesyan, A.; Marshall-Jones, Z.; Holmstrom, C.; Kjelleberg, S.; Egan, S. Antimicrobial activity observed among cultured marine epiphytic bacteria reflects their potential as a source of new drugs. FEMS Microbiol. Ecol. 2009, 69, 113–124. [Google Scholar] [CrossRef]
- O’Shea, E.F.; Gardiner, G.E.; O’Connor, P.M.; Mills, S.; Ross, R.P.; Hill, C. Characterization of enterocin- and salivaricin-producing lactic acid bacteria from the mammalian gastrointestinal tract. FEMS Microbiol. Lett. 2009, 291, 24–34. [Google Scholar] [CrossRef]
- Phelan, R.W.; O’Halloran, J.A.; Kennedy, J.; Morrissey, J.P.; Dobson, A.D.; O’Gara, F.; Barbosa, T.M. Diversity and bioactive potential of endospore-forming bacteria cultured from the marine sponge Haliclona simulans. J. Appl. Microbiol. 2012, 112, 65–78. [Google Scholar] [CrossRef]
- Wiese, J.; Thiel, V.; Nagel, K.; Staufenberger, T.; Imhoff, J. Diversity of antibiotic-active bacteria associated with the brown alga Laminaria saccharina from the Baltic Sea. Mar. Biotechnol. 2009, 11, 287–300. [Google Scholar] [CrossRef] [Green Version]
- Dischinger, J.; Josten, M.; Szekat, C.; Sahl, H.; Bierbaum, G. Production of the novel two-peptide lantibiotic lichenicidin by Bacillus licheniformis DSM 13. PLoS One 2009, 4, e6788. [Google Scholar] [CrossRef]
- Shenkarev, Z.O.; Finkina, E.I.; Nurmukhamedova, E.K.; Balandin, S.V.; Mineev, K.S.; Nadezhdin, K.D.; Yakimenko, Z.A.; Tagaev, A.A.; Temirov, Y.V.; Arseniev, A.S.; et al. Isolation, structure elucidation, and synergistic antibacterial activity of a novel two-component lantibiotic lichenicidin from Bacillus licheniformis VK21. Biochemistry 2010, 3, 6462–6472. [Google Scholar]
- Mendo, S.; Faustino, N.A.; Sarmento, A.C.; Amado, F.; Moir, A.J. Purification and characterization of a new peptide antibiotic produced by a thermotolerant Bacillus licheniformis strain. Biotechnol. Lett. 2004, 26, 115–119. [Google Scholar] [CrossRef]
- Caetano, T.; Krawczyk, J.M.; Mosker, E.; Sussmuth, R.D.; Mendo, S. Heterologous expression, biosynthesis, and mutagenesis of type II lantibiotics from Bacillus licheniformis in Escherichia coli. Chem. Biol. 2011, 18, 90–100. [Google Scholar] [CrossRef]
- Kwaadsteniet, M.; Doeschate, K.T.; Dicks, L.M.T. Characterization of the structural gene encoding nisin F, a new lantibiotic produced by a Lactococcus lactis subsp. lactis isolate from freshwater catfish (Clarias gariepinus). Appl. Environ. Microbiol. 2008, 74, 547–549. [Google Scholar] [CrossRef]
- Ghanbari, M.; Rezaei, M.; Soltani, M.; Shah-Hosseini, G. Production of bacteriocin by a novel Bacillus sp. strain RF 140, an intestinal bacterium of Caspian Frisian Roach (Rutilus frisii kutum). Iran J. Vet. Res. 2009, 10, 267–272. [Google Scholar]
- Cladera Olivera, F.; Caron, G.; Brandelli, A. Bacteriocin like substance production by Bacillus licheniformis strain P40. Lett. Appl. Microbiol. 2004, 38, 251–256. [Google Scholar] [CrossRef]
- Jamal, M.; Morris, P.; Hansen, R.; Jamieson, D.; Burgess, J.; Austin, B. Recovery and characterization of a 30.7-kDa protein from Bacillus licheniformis associated with inhibitory activity against methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococci, and Listeria monocytogenes. Mar. Biotechnol. 2006, 8, 587–592. [Google Scholar] [CrossRef]
- Martirani, L.; Varcamonti, M.; Naclerio, G.; De Felice, M. Purification and partial characterization of bacillocin 490, a novel bacteriocin produced by a thermophilic strain of Bacillus licheniformis. Microb. Cell Fact. 2002, 1, 1–5. [Google Scholar] [CrossRef]
- Pattnaik, P.; Kaushik, J.; Grover, S.; Batish, V. Purification and characterization of a bacteriocin like compound (Lichenin) produced anaerobically by Bacillus licheniformis isolated from water buffalo. J. Appl. Microbiol. 2001, 91, 636–645. [Google Scholar] [CrossRef]
- Kayalvizhi, N.; Gunasekaran, P. Purification and characterization of a novel broad-spectrum bacteriocin from Bacillus licheniformis MKU3. Biotechnol. Bioprocess. Eng. 2010, 15, 365–370. [Google Scholar] [CrossRef]
- He, L.; Chen, W.; Liu, Y. Production and partial characterization of bacteriocin-like peptides by Bacillus licheniformis ZJU12. Microbiol. Res. 2006, 161, 321–326. [Google Scholar] [CrossRef]
- Aunpad, R.; Na-Bangchang, K. Pumilicin 4, a novel bacteriocin with anti-MRSA and anti-VRE activity produced by newly isolated bacteria Bacillus pumilus strain WAPB4. Curr. Microbiol. 2007, 55, 308–313. [Google Scholar] [CrossRef]
- Korenblum, E.; von Der Weid, I.; Santos, A.L.S.; Rosado, A.S.; Sebastián, G.V.; Coutinho, C.M.L.M.; Magalhães, F.C.M.; de Paiva, M.M.; Seldin, L. Production of antimicrobial substances by Bacillus subtilis LFE-1, B. firmus H2O-1 and B. licheniformis T6-5 isolated from an oil reservoir in Brazil. J. Appl. Microbiol. 2005, 98, 667–675. [Google Scholar] [CrossRef]
- Kennedy, J.; Baker, P.; Piper, C.; Cotter, P.D.; Rea, M.C.; O’Connor, P.M.; Ross, R.P.; Hill, C.; O’Gara, F.; Marchesi, J.R.; et al. Isolation and analysis of bacteria with antimicrobial activities from the marine sponge Haliclona simulans collected from Irish waters. Mar. Biotechnol. 2009, 11, 384–396. [Google Scholar] [CrossRef]
- Ryan, M.P.; Rea, M.C.; Hill, C.; Ross, R.P. An application in cheddar cheese manufacture for a strain of Lactococcus lactis producing a novel broad-spectrum bacteriocin, lacticin 3147. Appl. Environ. Microbiol. 1996, 62, 612–619. [Google Scholar]
- Bierbaum, G.; Brötz, H.; Koller, K.; Sahl, H. Cloning, sequencing and production of the lantibiotic mersacidin. FEMS Microbiol. Lett. 1995, 127, 121–126. [Google Scholar] [CrossRef]
- Coffey, L.; Owens, E.; Tambling, K.; O’Neill, D.; O’Connor, L.; O’Reilly, C. Real-time PCR detection of Fe-type nitrile hydratase genes from environmental isolates suggests horizontal gene transfer between multiple genera. Antonie Van Leeuwenhoek 2010, 98, 455–463. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar]
- National Center for Biotechnology Information. Basic Local Alignment Search Tool. Available online: http://www.ncbi.nlm.nih.gov/BLAST/ (accessed on 10 September 2010).
- Roderic, D.M. TreeView. Available online: http://taxonomy.zoology.gla.ac.uk/rod/treeview.html (accessed on 20 September 2010).
- Science Foundation Ireland, University College Dublin. Clustal: Multiple Sequence Alignment. Available online: ftp://ftp-gbmc.ustrasbg.fr/pub/ClustalX/ (accessed on 25 September 2010).
- Ouoba, L.; Diawara, B.; Amoa-Awua, W.; Traoré, A.; Møller, P. Genotyping of starter cultures of Bacillus subtilis and Bacillus pumilus for fermentation of African locust bean (Parkia biglobosa) to produce Soumbala. Int. J. Food Microbiol. 2004, 90, 197–205. [Google Scholar] [CrossRef]
- Samples Availability: Available from the authors.
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Prieto, M.L.; O’Sullivan, L.; Tan, S.P.; McLoughlin, P.; Hughes, H.; O’Connor, P.M.; Cotter, P.D.; Lawlor, P.G.; Gardiner, G.E. Assessment of the Bacteriocinogenic Potential of Marine Bacteria Reveals Lichenicidin Production by Seaweed-Derived Bacillus spp. Mar. Drugs 2012, 10, 2280-2299. https://doi.org/10.3390/md10102280
Prieto ML, O’Sullivan L, Tan SP, McLoughlin P, Hughes H, O’Connor PM, Cotter PD, Lawlor PG, Gardiner GE. Assessment of the Bacteriocinogenic Potential of Marine Bacteria Reveals Lichenicidin Production by Seaweed-Derived Bacillus spp. Marine Drugs. 2012; 10(10):2280-2299. https://doi.org/10.3390/md10102280
Chicago/Turabian StylePrieto, Maria Luz, Laurie O’Sullivan, Shiau Pin Tan, Peter McLoughlin, Helen Hughes, Paula M. O’Connor, Paul D. Cotter, Peadar G. Lawlor, and Gillian E. Gardiner. 2012. "Assessment of the Bacteriocinogenic Potential of Marine Bacteria Reveals Lichenicidin Production by Seaweed-Derived Bacillus spp." Marine Drugs 10, no. 10: 2280-2299. https://doi.org/10.3390/md10102280