Targeted Next-Generation Sequencing in the Molecular Diagnosis of Severe Combined Immunodeficiency
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Molecular Studies
3. Results
3.1. Genetic Analysis of Patient P1
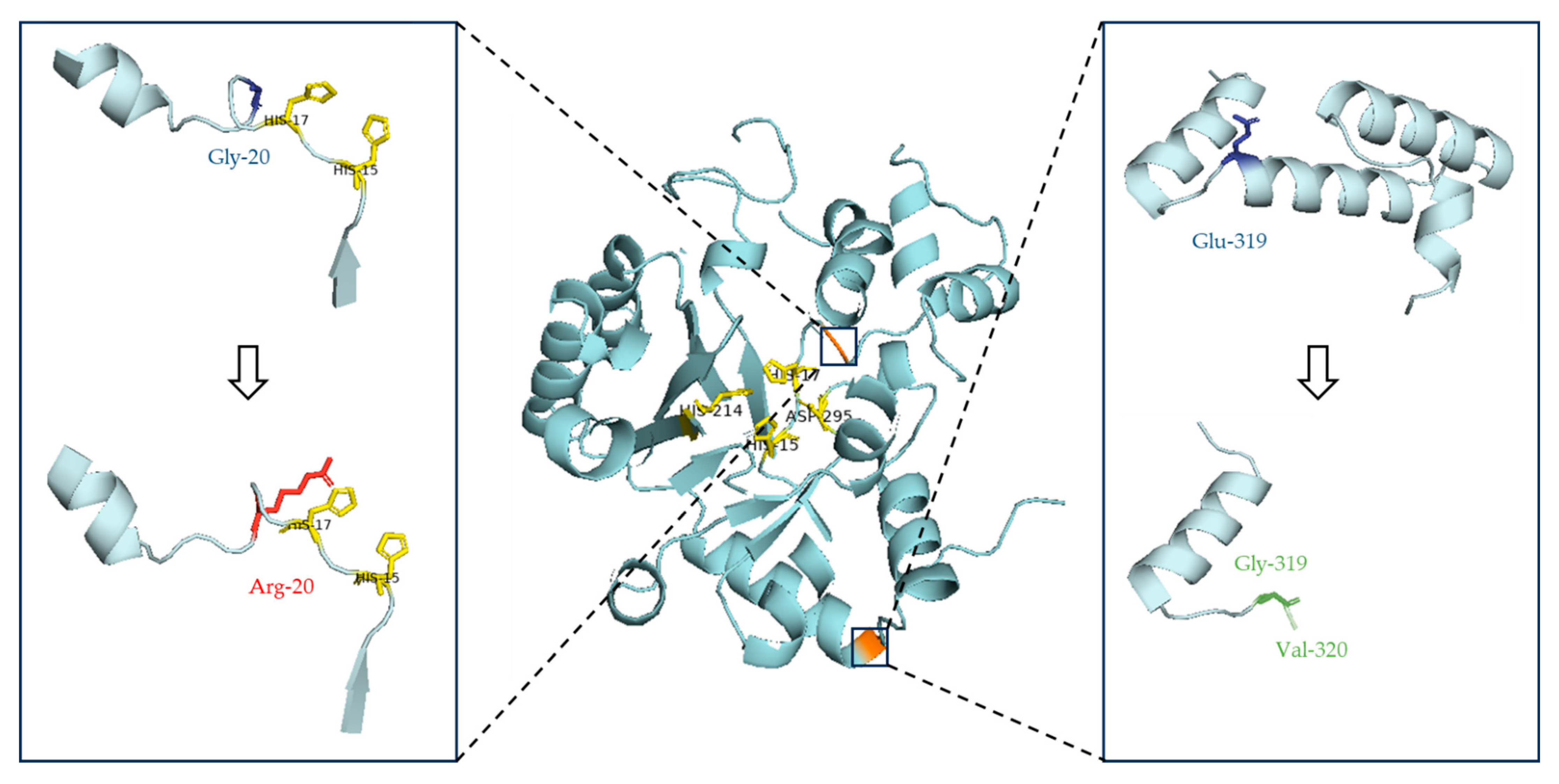
3.1.1. Pathogenicity Evaluation of the ADA:c.58G>A Variant
3.1.2. Pathogenicity Evaluation of the ADA:c.956_960del Variant
3.2. Genetic Variations in Patient P2
3.3. Genetic Variations in Patient P3
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Poli, M.C.; Aksentijevich, I.; Bousfiha, A.A.; Cunningham-Rundles, C.; Hambleton, S.; Klein, C.; Morio, T.; Picard, C.; Puel, A.; Rezaei, N.; et al. Human inborn errors of immunity: 2024 update on the classification from the International Union of Immunological Societies Expert Committee. J. Hum. Immun. 2025, 1, e20250003. [Google Scholar] [CrossRef]
- Barreiros, L.A.; Sousa, J.L.; Geier, C.; Leiss-Piller, A.; Kanegae, M.P.P.; França, T.T.; Boisson, B.; Lima, A.M.; Costa-Carvalho, B.T.; Aranda, C.S.; et al. SCID and Other Inborn Errors of Immunity with Low TRECs—The Brazilian Experience. J. Clin. Immunol. 2022, 42, 1171–1192. [Google Scholar] [CrossRef] [PubMed]
- Mikkers, H.; Pike-Overzet, K.; Staal, F.J.T. Induced pluripotent stem cells and severe combined immunodeficiency: Merely disease modeling or potentially a novel cure? Pediatr. Res. 2012, 71, 427–432. [Google Scholar] [CrossRef][Green Version]
- Puck, J.M. Population-based newborn screening for severe combined immunodeficiency: Steps toward implementation. J. Allergy Clin. Immunol. 2007, 120, 760–768. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.; Xu-Bayford, J.; Allwood, Z.; Slatter, M.; Cant, A.; Davies, E.G.; Veys, P.; Gennery, A.R.; Gaspar, H.B. Neonatal diagnosis of severe combined immunodeficiency leads to significantly improved survival outcome: The case for newborn screening. Blood 2011, 117, 3243–3246. [Google Scholar] [CrossRef] [PubMed]
- Khalturina, E.O.; Degtyareva, N.D.; Bairashevskaia, A.V.; Mulenkova, A.V.; Degtyareva, A.V. Modern diagnostic capabilities of neonatal screening for primary immunodeficiencies in newborns. Clin. Exp. Pediatr. 2021, 64, 504–510. [Google Scholar] [CrossRef]
- Chan, A.; Scalchunes, C.; Boyle, M.; Puck, J.M. Early vs. delayed diagnosis of severe combined immunodeficiency: A family perspective survey. Clin. Immunol. 2011, 138, 3–8. [Google Scholar] [CrossRef]
- Gennery, A.R.; Slatter, M.A.; Grandin, L.; Taupin, P.; Cant, A.J.; Veys, P.; Amrolia, P.J.; Gaspar, H.B.; Davies, E.G.; Friedrich, W.; et al. Transplantation of hematopoietic stem cells and long-term survival for primary immunodeficiencies in Europe: Entering a new century, do we do better? J. Allergy Clin. Immunol. 2010, 126, 602–610.e11. [Google Scholar] [CrossRef]
- Kuo, C.Y.; Kohn, D.B. Gene Therapy for the Treatment of Primary Immune Deficiencies. Curr. Allergy Asthma Rep. 2016, 16, 39. [Google Scholar] [CrossRef]
- Nijman, I.J.; van Montfrans, J.M.; Hoogstraat, M.; Boes, M.L.; van de Corput, L.; Renner, E.D.; van Zon, P.; van Lieshout, S.; Elferink, M.G.; van der Burg, M.; et al. Targeted next-generation sequencing: A novel diagnostic tool for primary immunodeficiencies. J. Allergy Clin. Immunol. 2014, 133, 529–534.e1. [Google Scholar] [CrossRef]
- Stoddard, J.L.; Niemela, J.E.; Fleisher, T.A.; Rosenzweig, S.D. Targeted NGS: A Cost-Effective Approach to Molecular Diagnosis of PIDs. Front. Immunol. 2014, 5, 531. [Google Scholar] [CrossRef]
- Moens, L.N.; Falk-Sörqvist, E.; Asplund, A.C.; Bernatowska, E.; Smith, C.I.E.; Nilsson, M. Diagnostics of Primary Immunodeficiency Diseases: A Sequencing Capture Approach. PLoS ONE 2014, 9, e114901. [Google Scholar] [CrossRef]
- Al-Mousa, H.; Abouelhoda, M.; Monies, D.M.; Al-Tassan, N.; Al-Ghonaium, A.; Al-Saud, B.; Al-Dhekri, H.; Arnaout, R.; Al-Muhsen, S.; Ades, N.; et al. Unbiased targeted next-generation sequencing molecular approach for primary immunodeficiency diseases. J. Allergy Clin. Immunol. 2016, 137, 1780–1787. [Google Scholar] [CrossRef] [PubMed]
- Beck, T.F.; Mullikin, J.C. the NISC Comparative Sequencing Program. Systematic Evaluation of Sanger Validation of Next-Generation Sequencing Variants. Clin. Chem. 2016, 62, 647–654. [Google Scholar] [CrossRef] [PubMed]
- About OMIM—OMIM—(OMIM.ORG). Available online: https://omim.org/about (accessed on 11 July 2025).
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–423. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Hogner, S.; Lundman, E.; Strand, J.; Ytre-Arne, M.E.; Tangeraas, T.; Stray-Pedersen, A. Newborn Genetic Screening—Still a Role for Sanger Sequencing in the Era of NGS. Int. J. Neonatal Screen. 2023, 9, 67. [Google Scholar] [CrossRef]
- Puck, J.M.; Pepper, A.E.; Henthorn, P.S.; Candotti, F.; Isakov, J.; Whitwam, T.; Conley, M.E.; Fischer, R.E.; Rosenblatt, H.M.; Small, T.N.; et al. Mutation Analysis of IL2RG in Human X-Linked Severe Combined Immunodeficiency. Blood 1997, 89, 1968–1977. [Google Scholar] [PubMed]
- Yang, D.R.; Huie, M.L.; Hirschhorn, R. Homozygosity for a missense mutation (G20R) associated with neonatal onset adenosine deaminase-deficient severe combined immunodeficiency (ADA-SCID). Clin. Immunol. Immunopathol. 1994, 70, 171–175. [Google Scholar] [CrossRef]
- Firtina, S.; Ng, Y.Y.; Ng, O.H.; Kiykim, A.; Aydiner, E.; Nepesov, S.; Camcioglu, Y.; Sayar, E.H.; Reisli, I.; Torun, S.H.; et al. Mutational landscape of severe combined immunodeficiency patients from Turkey. Int. J. Immunogenet. 2020, 47, 529–538. [Google Scholar] [CrossRef]
- Rawat, R.S.; Kumar, S. Understanding the mode of inhibition and molecular interaction of taxifolin with human adenosine deaminase. J. Biomol. Struct. Dyn. 2023, 41, 377–385. [Google Scholar] [CrossRef]
- Adams, S.P.; Wilson, M.; Harb, E.; Fairbanks, L.; Xu-Bayford, J.; Brown, L.; Kearney, L.; Madkaikar, M.; Gaspar, H.B. Spectrum of mutations in a cohort of UK patients with ADA deficient SCID: Segregation of genotypes with specific ethnicities. Clin. Immunol. 2015, 161, 174–179. [Google Scholar] [CrossRef]
- Baffelli, R.; Notarangelo, L.D.; Imberti, L.; Hershfield, M.S.; Serana, F.; Santisteban, I.; Bolda, F.; Porta, F.; Lanfranchi, A. Diagnosis, Treatment and Long-Term Follow Up of Patients with ADA Deficiency: A Single-Center Experience. J. Clin. Immunol. 2015, 35, 624–637. [Google Scholar] [CrossRef]
- Geelen, J.; Pfundt, R.; Meijer, J.; Verheijen, F.W.; van Kuilenburg, A.B.; Warris, A.; Marcelis, C. Severe phenotype of severe combined immunodeficiency caused by adenosine deaminase deficiency in a patient with a homozygous mutation due to uniparental disomy. J. Allergy Clin. Immunol. 2013, 132, 222–223. [Google Scholar] [CrossRef]
- Cagdas, D.; Cetinkaya, P.G.; Karaatmaca, B.; Esenboga, S.; Tan, C.; Yılmaz, T.; Gümüş, E.; Barış, S.; Kuşkonmaz, B.; Ozgur, T.T.; et al. ADA Deficiency: Evaluation of the Clinical and Laboratory Features and the Outcome. J. Clin. Immunol. 2018, 38, 484–493. [Google Scholar] [CrossRef]
- Felgentreff, K.; Lee, Y.N.; Frugoni, F.; Du, L.; van der Burg, M.; Giliani, S.; Tezcan, I.; Reisli, I.; Mejstrikova, E.; de Villartay, J.-P.; et al. Functional analysis of naturally occurring DCLRE1C mutations and correlation with the clinical phenotype of ARTEMIS deficiency. J. Allergy Clin. Immunol. 2015, 136, 140–150.e7. [Google Scholar] [CrossRef]
- Moshous, D.; Callebaut, I.; de Chasseval, R.; Corneo, B.; Cavazzana-Calvo, M.; Le Deist, F.; Tezcan, I.; Sanal, O.; Bertrand, Y.; Philippe, N.; et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 2001, 105, 177–186. [Google Scholar] [CrossRef]
- Moshous, D.; Callebaut, I.; De Chasseval, R.; Poinsignon, C.; Villey, I.; Fischer, A.; DE Villartay, J. The V(D)J Recombination/DNA Repair Factor Artemis Belongs to the Metallo-β-Lactamase Family and Constitutes a Critical Developmental Checkpoint of the Lymphoid System. Ann. N. Y. Acad. Sci. 2003, 987, 150–157. [Google Scholar] [CrossRef]
- Niemela, J.E.; Puck, J.M.; Fischer, R.E.; Fleisher, T.A.; Hsu, A.P. Efficient detection of thirty-seven new IL2RG mutations in human X-linked severe combined immunodeficiency. Clin. Immunol. 2000, 95 Pt 1, 33–38. [Google Scholar] [CrossRef]
- Michos, A.; Tzanoudaki, M.; Villa, A.; Giliani, S.; Chrousos, G.; Kanariou, M. Severe combined immunodeficiency in Greek children over a 20-year period: Rarity of γc-chain deficiency (X-linked) type. J. Clin. Immunol. 2011, 31, 778–783. [Google Scholar] [CrossRef]
| Patient | Sex | Age on Diagnosis | Clinical Manifestations | Immune Phenotype | Outcome |
|---|---|---|---|---|---|
| P1 | M | 37 days | Failure to thrive, severe leucopenia, neutropenia and lymphopenia, absence of thymic shadow | Τ−Β−ΝΚ− SCID | Alive, under enzyme replacement therapy and chemoprotective regimen until gene therapy |
| P2 | F | 3.5 months | Persistent CMV infection of the CNS, hypogammaglobulinemia, severe lymphopenia from 50th day of life, normal levels of NK cells | Τ−Β−ΝΚ+ SCID | Death due to HSCT complications and CMV treatment toxicities |
| P3 | M | 8.5 months | Persistent respiratory infection, leucopenia, severe lymphopenia, hypogammaglobulinemia, reduced CD3+, CD4+, CD8+ T and NK cells | Τ−Β+ΝΚ− SCID | Death prior to HSCT due to respiratory failure |
| No | Implicated Gene/OMIM | Associated IEI | Clinical Manifestations | Inheritance |
|---|---|---|---|---|
| 1 | IL2RG/308380 | γc deficiency | CID moderate, SCID (T−B+NK−) | XLR |
| 2 | JAK3/600173 | JAK3 deficiency | SCID (T−B+NK−) | AR |
| 3 | IL7R/146661 | IL7Rα deficiency | IMD 104, SCID (T−B+NK+) | AR |
| 4 | PTPRC/151460 | CD45 deficiency | IMD 105, SCID (T−B+NK+) | AR |
| 5 | CD3D/186790 | CD3δ deficiency | IMD19, SCID (T−B+NK+) | AR |
| 6 | CD3E/186830 | CD3ε deficiency | IMD18, SCID (T−B+NK+) | AR |
| 7 | CD247/186780 | CD3ζ deficiency | IMD25 (provisional, T−B+NK+) | AR |
| 8 | RAG1/179615 | RAG deficiency | Combined cellular and humoral immune defects with granulomas, Omenn syndrome, SCID (T−B−NK+) | AR |
| 9 | RAG2/179616 | |||
| 10 | DCLRE1C/605988 | Artemis deficiency | Ommen syndrome, Athabascan-type SCID (T−B−NK+) | AR |
| 11 | PRKDC/600899 | DNA-PKcs deficiency | IMD26, with or without neurologic abnormalities, SCID (T−B−NK+) | AR |
| 12 | LIG4/601837 | DNA ligase IV deficiency | LIG4 syndrome, SCID (T−B−NK+) | AR |
| 13 | NHEJ1/611290 | Cernunnos/XLF deficiency | IMD124, SCID (T−B−NK+), Microphthalmia/coloboma 13 | AR |
| 14 | AK2/103020 | AK2 defect | Reticular dysgenesis (T−B−NK−) | AR |
| 15 | ADA/608958 | ADA deficiency | SCID due to ADA deficiency (T−B−NK−) | AR |
| 16 | RAC2/602049 | Activated RAC2 defect | IMD73A with defective neutrophil chemotaxis and leukocytosis, IMD73B with defective neutrophil chemotaxis and lymphopenia, SCID (T−B−NK−) | AD GoF |
| 17 | ZAP70/176947 | ZAP-70 deficiency | Autoimmune disease, multisystem, infantile-onset, 2, IMD48 | AR |
| 18 | RFX5/601863 | MHC class II deficiency | MHC class II deficiency 3, MHC class II deficiency 5 (provisional) | AR |
| 19 | RFXAP/601861 | MHC class II deficiency 4 | ||
| 20 | CIITA/600005 | MHC class II deficiency 1 | ||
| 21 | RFXANK/603200 | MHC class II deficiency 2 | ||
| 22 | TAP1/170260 | MHC class I deficiency | MHC class I deficiency 1 | AR |
| 23 | TAP2/170261 | MHC class I deficiency 2 | ||
| 24 | TAPBP/601962 | MHC class I deficiency 3 (provisional) | ||
| 25 | B2M/109700 | IMD43 | ||
| 26 | CD3G/186740 | CD3γ deficiency | IMD 17, CD3 gamma deficient | AR |
| 27 | DOCK2/603122 | DOCK2 deficiency | IMD40 | AR |
| 28 | CARD11/607210 | CARD11 deficiency | IMD11A | AR LoF |
| 29 | BCL10/616098 | BCL10 deficiency | IMD37 (provisional) | AR |
| 30 | MALT1/604860 | MALT1 deficiency | IMD12 | AR |
| Gene | Targeted Region | Transcript | Primer Sequence | PCR Product (bp) | PCR Conditions | Reference |
|---|---|---|---|---|---|---|
| ADA | Exon 2 | NM_000022.4 | F: 5′-AACATTAAGCTCTGAAAGGTCCTTCG-3′ R: 5′-GCTTGATTCCCACAGGGAGAC-3′ | 272 | 94 °C 5 min, 35 cycles (94 °C 30 s, 62 °C 45 s, 72 °C 45 s), 72 °C 10 min | [18] |
| Exon 10 | F: 5′-GGCTGCCATTCTGCCTGGTT-3′ R: 5′-CCTCTCTCCAAAGATTCCAGGC-3′ | 495 | 94 °C 5 min, 40 cycles (94 °C 30 s, 60 °C 45 s, 72 °C 45 s), 72 °C 10 min | |||
| DCLRE1C | Exon 3 | NM_001033855.3 | F: 5′-TCTAACAGATTTTGTGCCAGCG-3′ R: 5′-CTGAAGTATGTTACAAACTGAGGC-3′ | 413 | 94 °C 5 min, 32 cycles (94 °C 30 s, 62 °C 1 min, 72 °C 45 s), 72 °C 3 min | In house design of primers |
| IL2RG | Exons 3–4 | NM_000206.3 | F: 5′-TGCAGTACCCAGATTGGCC-3′ R: 5′-GGCCTTAGCTGCTACATTCACG-3′ | 625 | 94 °C 5 min, 36 cycles (94 °C 45 s, 60 °C 1 min, 72 °C 30 s), 72 °C 2 min | [19] |
| Patient | Sex | Immune Phenotype | Gene | Pathogenic Variants | Zygosity | Mode of Inheritance |
|---|---|---|---|---|---|---|
| P1 | M | Τ−Β−ΝΚ− SCID | ADA | c.58G>A (p.Gly20Arg) | Het | Not performed (Maternal inheritance or de novo) |
| c.956_960del (p.Glu319Glyfs) | Het | Paternal inheritance | ||||
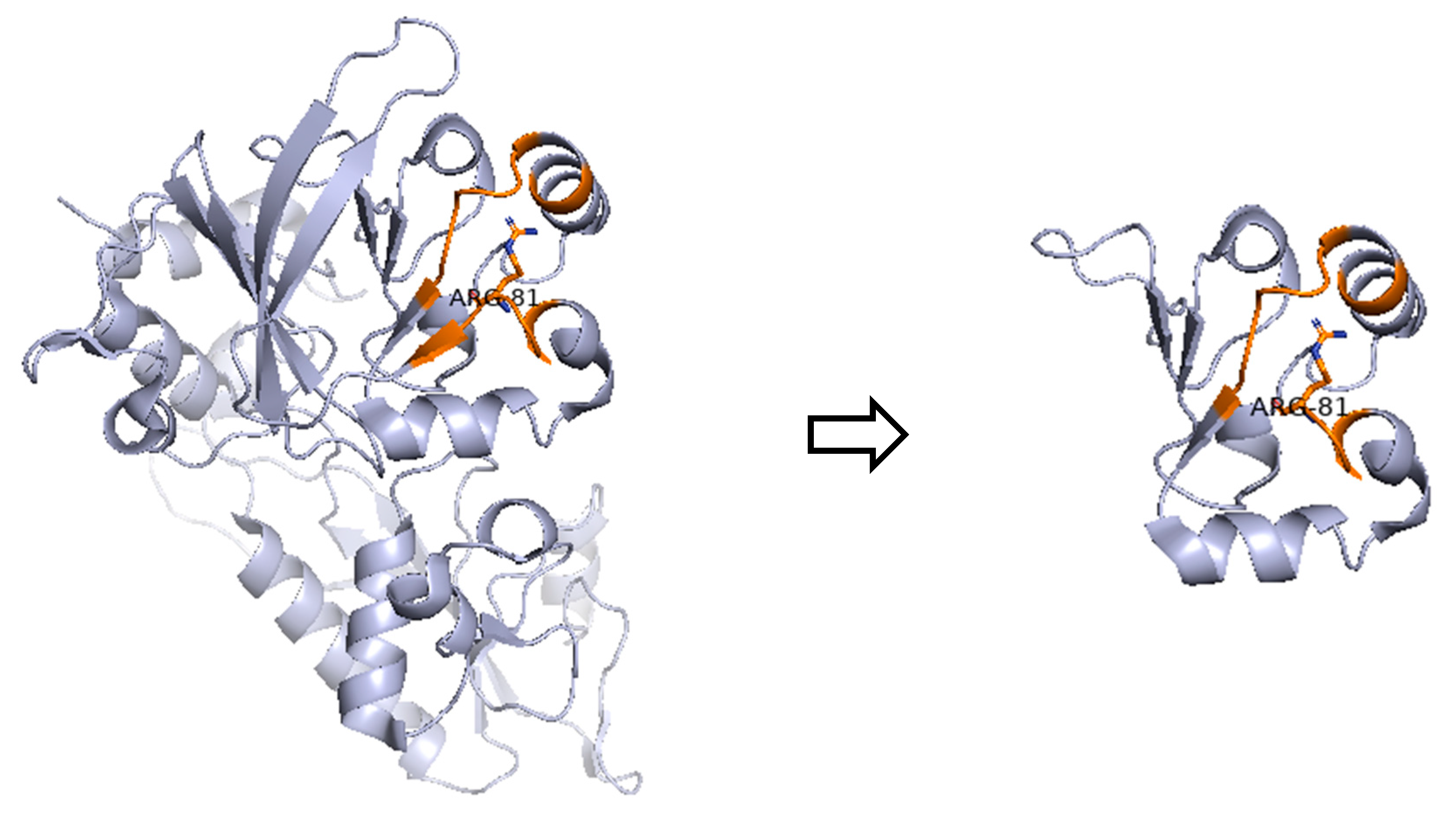
| P2 | F | Τ−Β−ΝΚ+ SCID | DCLRE1C | c.241C>T (p.Arg81 *) | Het | Paternal inheritance |
| Deletion of exons 1–3 | Het | Maternal inheritance | ||||
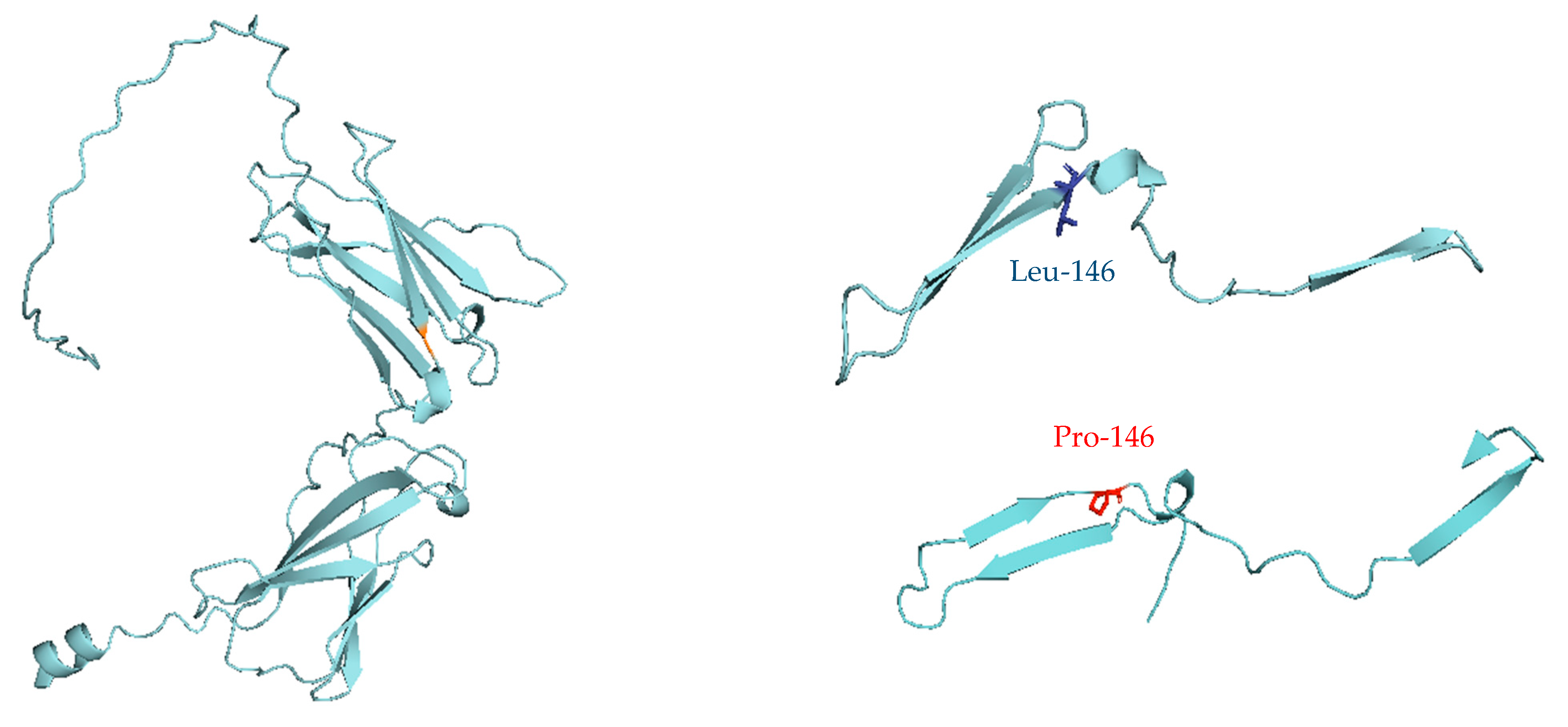
| P3 | M | Τ−Β+ΝΚ− SCID | IL2RG | c.437T>C (p.Leu146Pro) | Hmz | De novo (Mother did not harbor the pathogenic variant) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the Lithuanian University of Health Sciences. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakaros, E.; Sarrou, S.; Gkantaras, A.; Matziri, A.; Galanopoulos, A.P.; Charisi, K.; Bangeas, A.; Taparkou, A.; Papadimitriou, E.; Mouchtouri, V.A.; et al. Targeted Next-Generation Sequencing in the Molecular Diagnosis of Severe Combined Immunodeficiency. Medicina 2025, 61, 1644. https://doi.org/10.3390/medicina61091644
Bakaros E, Sarrou S, Gkantaras A, Matziri A, Galanopoulos AP, Charisi K, Bangeas A, Taparkou A, Papadimitriou E, Mouchtouri VA, et al. Targeted Next-Generation Sequencing in the Molecular Diagnosis of Severe Combined Immunodeficiency. Medicina. 2025; 61(9):1644. https://doi.org/10.3390/medicina61091644
Chicago/Turabian StyleBakaros, Evangelos, Styliani Sarrou, Antonios Gkantaras, Alexia Matziri, Achilleas P. Galanopoulos, Konstantina Charisi, Athanasios Bangeas, Anna Taparkou, Eleni Papadimitriou, Varvara A. Mouchtouri, and et al. 2025. "Targeted Next-Generation Sequencing in the Molecular Diagnosis of Severe Combined Immunodeficiency" Medicina 61, no. 9: 1644. https://doi.org/10.3390/medicina61091644
APA StyleBakaros, E., Sarrou, S., Gkantaras, A., Matziri, A., Galanopoulos, A. P., Charisi, K., Bangeas, A., Taparkou, A., Papadimitriou, E., Mouchtouri, V. A., Kalala, F., Hadjichristodoulou, C., Speletas, M., & Farmaki, E. (2025). Targeted Next-Generation Sequencing in the Molecular Diagnosis of Severe Combined Immunodeficiency. Medicina, 61(9), 1644. https://doi.org/10.3390/medicina61091644









