Genital Lichen Sclerosus—The Role of the Vulvar Microbiome
Abstract
1. Introduction
2. Methods
2.1. Study Design and Population
2.2. Sample Preparation and 16S rRNA Variable Region Sequencing
2.3. Statistical Analysis
3. Ethics
4. Results
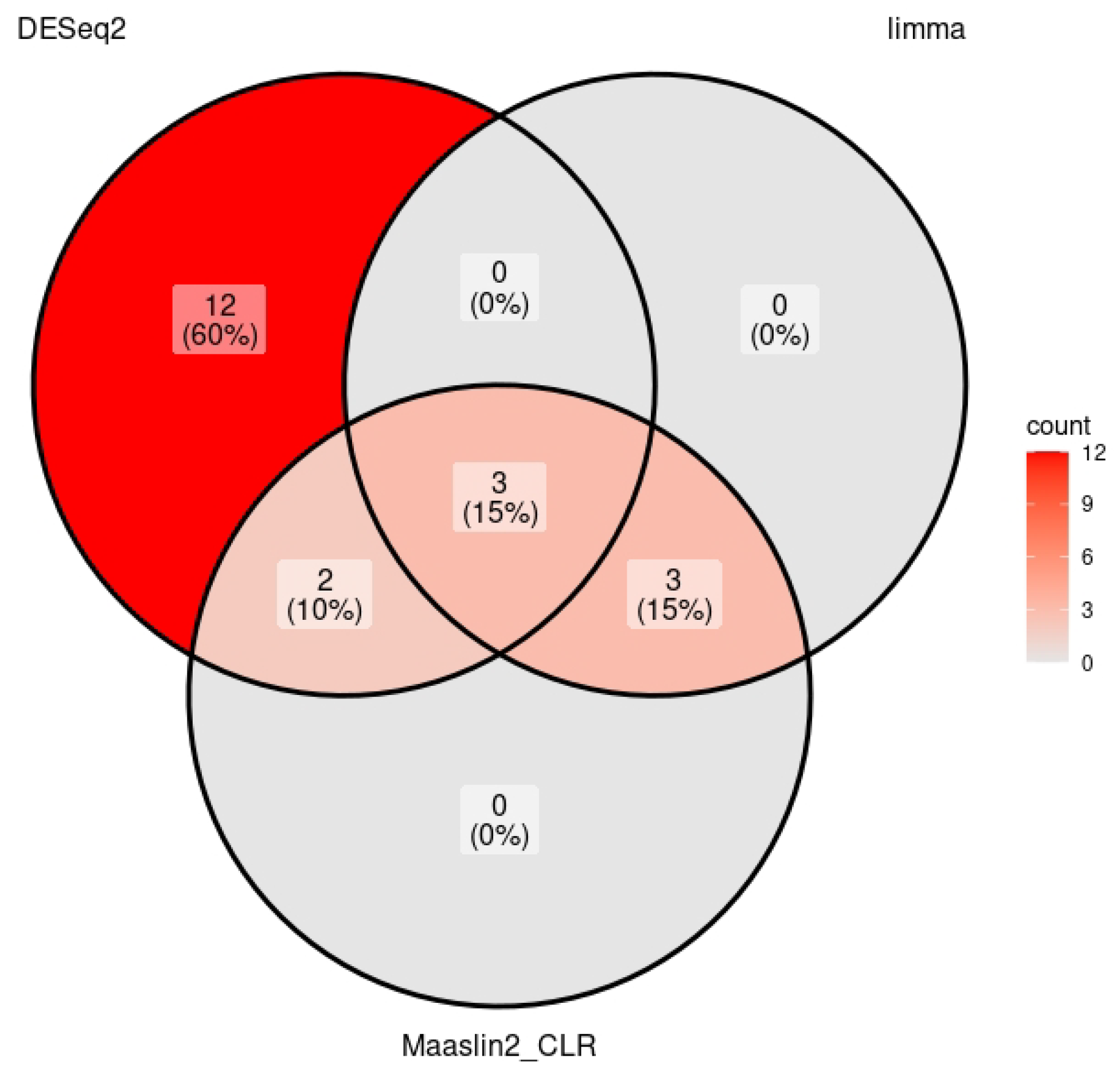
4.1. Deseq2
4.2. MaAslin2 CLR
4.3. Limma
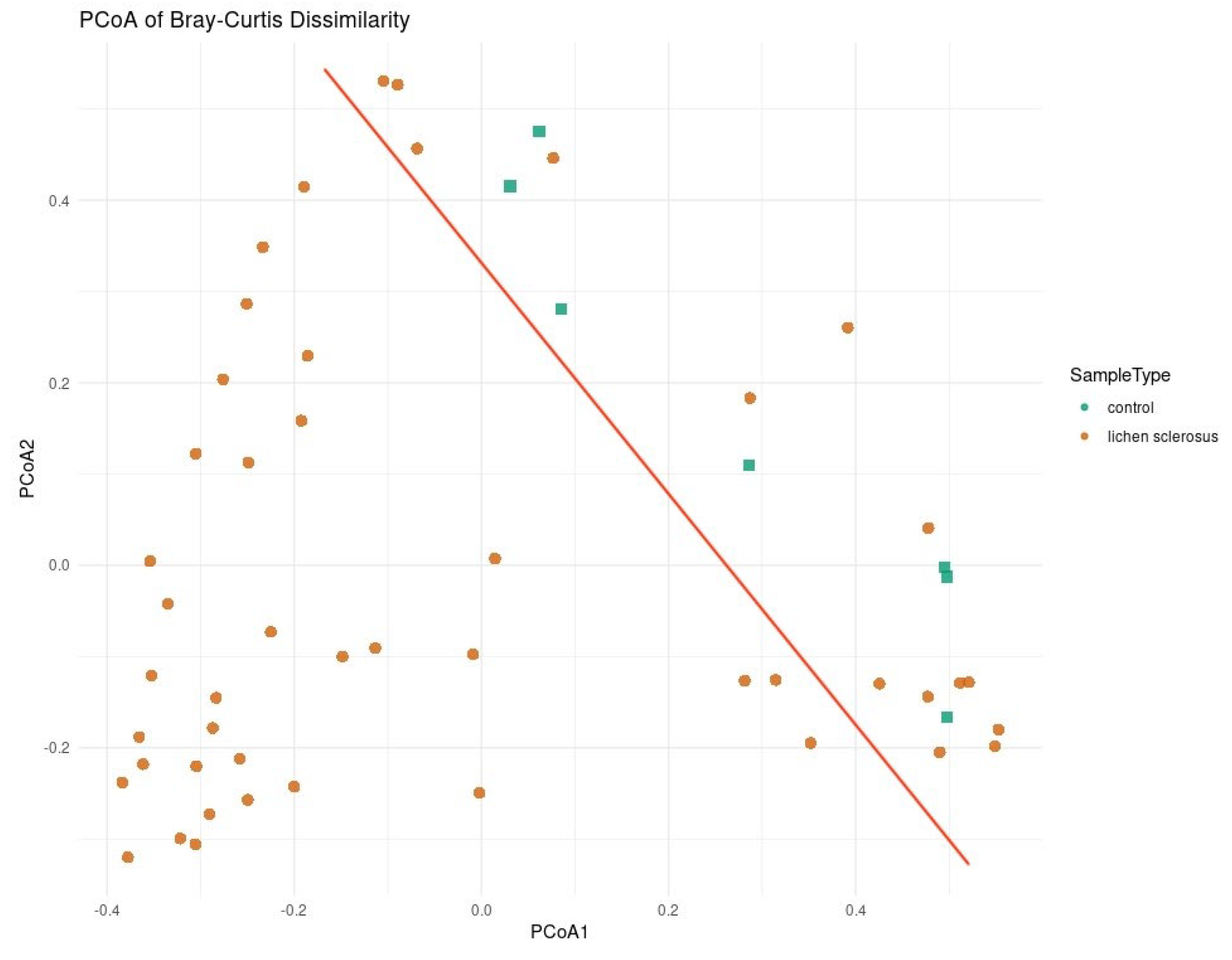
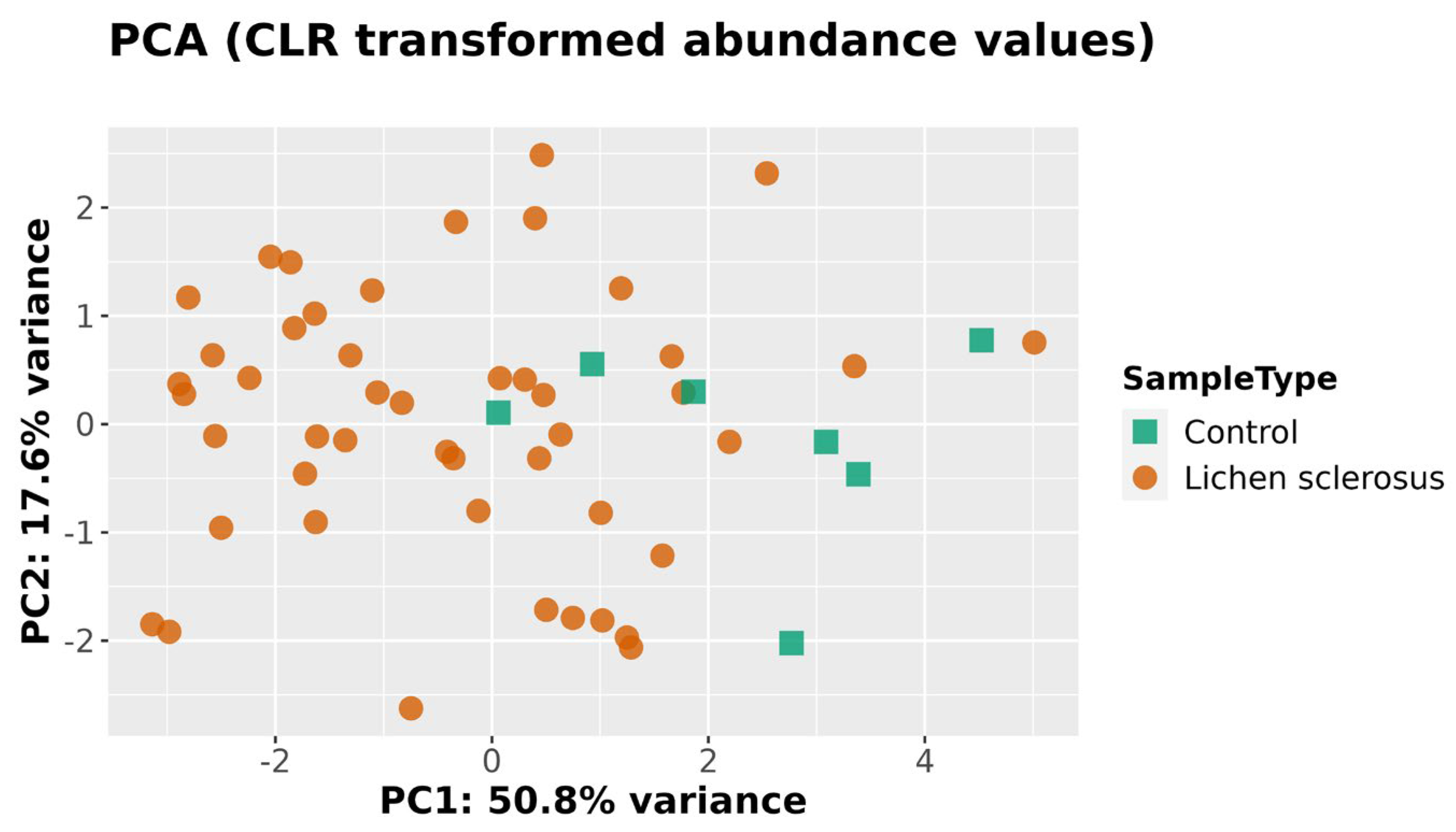
4.4. Beta Diversity
5. Discussion
Study Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Luca, D.A.; Papara, C.; Vorobyev, A.; Staiger, H.; Bieber, K.; Thaci, D.; Ludwig, R.J. Lichen sclerosus: The 2023 update. Front. Med. 2023, 10, 1106318. [Google Scholar] [CrossRef]
- Halonen, P.; Jakobsson, M.; Heikinheimo, O.; Gissler, M.; Pukkala, E. Incidence of lichen sclerosus and subsequent causes of death: A nationwide Finnish register study. BJOG Int. J. Obstet. Gynaecol. 2020, 127, 814–819. [Google Scholar] [CrossRef]
- Patterson, J.A.; Ackerman, A.B. Lichen sclerosus et atrophicus is not related to morphea. A clinical and histologic study of 24 patients in whom both conditions were reputed to be present simultaneously. Am. J. Dermatopathol. 1984, 6, 323–335. [Google Scholar] [CrossRef]
- Naswa, S.; Marfatia, Y.S. Physician-administered clinical score of vulvar lichen sclerosus: A study of 36 cases. Indian J. Sex. Transm. Dis. AIDS 2015, 36, 174–177. [Google Scholar] [CrossRef]
- Spencer, A.; Uthayakumar, A.; Watchorn, R.E.; Palmer, E.L.; Akel, R.; Kravvas, G.; Muneer, A.; Fuller, L.C.; Lewis, F.; Bunker, C.B. Urinary incontinence and female genital lichen sclerosus. J. Eur. Acad. Dermatol. Venereol. 2025, in press. [Google Scholar] [CrossRef]
- Panou, E.; Panagou, E.; Foley, C.; Kravvas, G.; Watchorn, R.; Alnajjar, H.; Muneer, A.; Bunker, C.B. Male genital lichen sclerosus associated with urological interventions and microincontinence: A case series of 21 patients. Clin. Exp. Dermatol. 2022, 47, 107–109. [Google Scholar] [CrossRef]
- Bunker, C.B.; Patel, N.; Shim, T.N. Urinary voiding symptomatology (micro-incontinence) in male genital lichen sclerosus. Acta Derm. Venereol. 2013, 93, 246–248. [Google Scholar] [CrossRef] [PubMed]
- Åsbrink, E.; Hovmark, A.; Olsson, I. Clinical manifestations of acrodermatitis chronica atrophicans in 50 Swedish patients. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 1986, 263, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Kirtschig, G.; Kinberger, M.; Kreuter, A.; Simpson, R.; Gunthert, A.; van Hees, C.; Becker, K.; Ramakers, M.J.; Corazza, M.; Muller, S.; et al. EuroGuiderm guideline on lichen sclerosus—Treatment of lichen sclerosus. J. Eur. Acad. Dermatol. Venereol. 2024, 38, 1874–1909. [Google Scholar] [CrossRef] [PubMed]
- Kravvas, G.; Muneer, A.; Watchorn, R.E.; Castiglione, F.; Haider, A.; Freeman, A.; Hadway, P.; Alnajjar, H.; Lynch, M.; Bunker, C.B. Male genital lichen sclerosus, microincontinence and occlusion: Mapping the disease across the prepuce. Clin. Exp. Dermatol. 2022, 47, 1124–1130. [Google Scholar] [CrossRef]
- Jerkovic Gulin, S.; Liljeberg, L.; Seifert, O. The impact of genital lichen sclerosus in men and women on quality of life: A prospective cohort study. Int. J. Womens Dermatol. 2024, 10, e131. [Google Scholar] [CrossRef]
- Pagan, L.; Huisman, B.W.; van der Wurff, M.; Naafs, R.G.C.; Schuren, F.H.J.; Sanders, I.; Smits, W.K.; Zwittink, R.D.; Burggraaf, J.; Rissmann, R.; et al. The vulvar microbiome in lichen sclerosus and high-grade intraepithelial lesions. Front. Microbiol. 2023, 14, 1264768. [Google Scholar] [CrossRef]
- Pyle, H.J.; Evans, J.C.; Artami, M.; Raj, P.; Sridharan, S.; Arana, C.; Eckert, K.M.; McDonald, J.G.; Harris-Tryon, T.A.; Mauskar, M.M. Assessment of the cutaneous hormone landscapes and microbiomes in vulvar lichen sclerosus. J. Investig. Dermatol. 2024, 144, 1808–1816.e11. [Google Scholar] [CrossRef] [PubMed]
- Brunner, A.; Medvecz, M.; Makra, N.; Sárdy, M.; Komka, K.; Gugolya, M.; Szabó, D.; Gajdács, M.; Ostorházi, E. Human beta defensin levels and vaginal microbiome composition in post-menopausal women diagnosed with lichen sclerosus. Sci. Rep. 2021, 11, 15999. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhuo, Y.; Zhou, Y.; Hu, J.; Wen, H.; Xiao, C. Analysis of the vulvar skin microbiota in asymptomatic women and patients with vulvar lichen sclerosus based on 16S rRNA sequencing. Front. Cell Dev. Biol. 2022, 10, 842031. [Google Scholar] [CrossRef] [PubMed]
- Taylor, O.A.; Birse, K.D.; Hill, D.J.; Knodel, S.; Noel-Romas, L.; Myers, A.; Marino, J.; Burgener, A.D.; Pope, R.; Farr Zuend, C. The relationship between the vaginal and vulvar microbiomes and lichen sclerosus symptoms in post-menopausal women. Sci. Rep. 2024, 14, 27094. [Google Scholar] [CrossRef]
- Nygaard, S.; Gerlif, K.; Bundgaard-Nielsen, C.; Saleh Media, J.; Leutscher, P.; Sørensen, S.; Brusen Villadsen, A.; Thomsen Schmidt Arenholt, L. The urinary, vaginal and gut microbiota in women with genital lichen sclerosus—A case-control study. Eur. J. Obstet. Gynecol. Reprod. Biol. 2023, 289, 1–8. [Google Scholar] [CrossRef]
- Watchorn, R.E.; van den Munckhof, E.H.A.; Quint, K.D.; Eliahoo, J.; de Koning, M.N.C.; Quint, W.G.V.; Bunker, C.B. Balanopreputial sac and urine microbiota in patients with male genital lichen sclerosus. Int. J. Dermatol. 2021, 60, 201–207. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Croatti, V.; Parolin, C.; Giordani, B.; Foschi, C.; Fedi, S.; Vitali, B. Lactobacilli extracellular vesicles: Potential postbiotics to support the vaginal microbiota homeostasis. Microb. Cell Fact. 2022, 21, 237. [Google Scholar] [CrossRef]
- Chattopadhyay, S.; Arnold, J.D.; Malayil, L.; Hittle, L.; Mongodin, E.F.; Marathe, K.S.; Gomez-Lobo, V.; Sapkota, A.R. Potential role of the skin and gut microbiota in premenarchal vulvar lichen sclerosus: A pilot case-control study. PLoS ONE 2021, 16, e0245243. [Google Scholar] [CrossRef]
- Li, X.; Chen, S.; Chen, S.; Cheng, S.; Lan, H.; Wu, Y.; Qiu, G.; Zhang, L. Skin microbiome and causal relationships in three dermatological diseases: Evidence from Mendelian randomization and Bayesian weighting. Skin Res. Technol. 2024, 30, e70035. [Google Scholar] [CrossRef]
- Cheung, M.K.; Leung, T.F.; Tam, W.H.; Leung, A.S.Y.; Chan, O.M.; Ng, R.W.Y.; Yau, J.W.K.; Yuen, L.Y.; Tong, S.L.Y.; Ho, W.C.S.; et al. Development of the early-life gut microbiome and associations with eczema in a prospective Chinese cohort. mSystems 2023, 8, e0052123. [Google Scholar] [CrossRef]
- McCarthy, S.; Barrett, M.; Kirthi, S.; Pellanda, P.; Vlckova, K.; Tobin, A.M.; Murphy, M.; Shanahan, F.; O’Toole, P.W. Altered skin and gut microbiome in hidradenitis suppurativa. J. Investig. Dermatol. 2022, 142, 459–468.e415. [Google Scholar] [CrossRef] [PubMed]
- Neumann, A.; Björck, L.; Frick, I.M. Finegoldia magna, an anaerobic Gram-positive bacterium of the normal human microbiota, induces inflammation by activating neutrophils. Front. Microbiol. 2020, 11, 65. [Google Scholar] [CrossRef] [PubMed]
- Constantinou, A.; Polak-Witka, K.; Tomazou, M.; Oulas, A.; Kanti, V.; Schwarzer, R.; Helmuth, J.; Edelmann, A.; Blume-Peytavi, U.; Spyrou, G.M.; et al. Dysbiosis and enhanced beta-defensin production in hair follicles of patients with lichen planopilaris and frontal fibrosing alopecia. Biomedicines 2021, 9, 266. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef]


| DeSeq2 | MaAslin CLR | Limma | |
|---|---|---|---|
| Lactobacillus | −3.47 | −4.66 | −7.16 |
| Finegoldia | −1.85 | −1.43 | −2.36 |
| Lawsonella | −1.62 | −1.55 | −2.49 |
| Satphylococcus | Not significant | −2.79 | −3.52 |
| Cutibacterium | Not significant | −1.29 | −3.12 |
| Unknown Genus Belonging to Actinomycetaceae | Not significant | −2.28 | −3.58 |
| Porphyromonas | 2.07 | 2.06 | Not significant |
| Ezekiella | 4.18 | 2.42 | Not significant |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the Lithuanian University of Health Sciences. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jerkovic Gulin, S.; Almlöf, O.; Seifert, O.; Kravvas, G.; Lundin, F.; Bergman Jungeström, M.; Söderman, J. Genital Lichen Sclerosus—The Role of the Vulvar Microbiome. Medicina 2025, 61, 1632. https://doi.org/10.3390/medicina61091632
Jerkovic Gulin S, Almlöf O, Seifert O, Kravvas G, Lundin F, Bergman Jungeström M, Söderman J. Genital Lichen Sclerosus—The Role of the Vulvar Microbiome. Medicina. 2025; 61(9):1632. https://doi.org/10.3390/medicina61091632
Chicago/Turabian StyleJerkovic Gulin, Sandra, Olivia Almlöf, Oliver Seifert, Georgios Kravvas, Filippa Lundin, Malin Bergman Jungeström, and Jan Söderman. 2025. "Genital Lichen Sclerosus—The Role of the Vulvar Microbiome" Medicina 61, no. 9: 1632. https://doi.org/10.3390/medicina61091632
APA StyleJerkovic Gulin, S., Almlöf, O., Seifert, O., Kravvas, G., Lundin, F., Bergman Jungeström, M., & Söderman, J. (2025). Genital Lichen Sclerosus—The Role of the Vulvar Microbiome. Medicina, 61(9), 1632. https://doi.org/10.3390/medicina61091632






