Impact of NAD(P)H: Quinone Oxidoreductase 1 (NQO1) C609T Polymorphism on Lung Cancer Risk
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Data Collection
2.3. Statistical Analysis
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Cangır, A.K.; Yumuk, P.F.; Sak, S.D.; Akyürek, S.; Eralp, Y.; Yılmaz, Ü.; Selek, U.; Eroğlu, A.; Tatlı, A.M.; Dinçbaş, F.Ö.; et al. Lung Cancer in Turkey. J. Thorac. Oncol. 2022, 17, 1158–1170. [Google Scholar] [CrossRef]
- Turkish Statistical Institute. Death and Causes of Death Statistics. 2022. Available online: https://data.tuik.gov.tr/Bulten/Index?p=Olum-ve-Olum-Nedeni-Istatistikleri-2022-49679# (accessed on 15 September 2023).
- Luo, G.; Zhang, Y.; Rumgay, H.; Morgan, E.; Langselius, O.; Vignat, J.; Colombet, M.; Bray, F. Estimated worldwide variation and trends in incidence of lung cancer by histological subtype in 2022 and over time: A population-based study. Lancet Respir. Med. 2025, 13, 348–363. [Google Scholar] [CrossRef]
- Ensarioğlu, K.; Özyürek, B.A.; Dinçer, M.; Gümüşler, H.A. Reimbursement of Lung Cancer in Turkey: A 10-Year Single Center Study. Cancer Med. 2025, 14, e71014. [Google Scholar] [CrossRef]
- Kaghazchi, B.; Um, I.H.; Elshani, M.; Read, O.J.; Harrison, D.J. Spatial Analysis of NQO1 in Non-Small Cell Lung Cancer Shows Its Expression Is Independent of NRF1 and NRF2 in the Tumor Microenvironment. Biomolecules 2022, 12, 1652. [Google Scholar] [CrossRef]
- Soerjomataram, I.; Bray, F. Planning for tomorrow: Global cancer incidence and the role of prevention 2020–2070. Nat. Rev. Clin. Oncol. 2021, 18, 663–672. [Google Scholar] [CrossRef]
- Siegel, D.; Ross, D. İnsan dokularında NAD(P)H:kinon oksidoredüktaz 1’in (NQO1) immünodeksiyonu. Free Radic. Biol. Med. 2000, 29, 246–253. [Google Scholar] [CrossRef]
- Tossetta, G.; Fantone, S.; Goteri, G.; Giannubilo, S.R.; Ciavattini, A.; Marzioni, D. The Role of NQO1 in Ovarian Cancer. Int. J. Mol. Sci. 2023, 24, 7839. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhong, B.; Zhao, L.; Hou, Y.; Ai, N.; Lu, J.J.; Ge, W.; Chen, X. Fighting drug-resistant LC by induction of NAD(P)H:quinone oxidoreductase 1 (NQO1)-mediated ferroptosis. Drug Resist. Updat. 2023, 70, 100977. [Google Scholar] [CrossRef]
- Madajewski, B.; Boatman, M.A.; Martinez, I.; Carter, J.H.; Bey, E.A. NAD(P)H Quinone Oxidoreductase-1 Expression Promotes Self-Renewal and Therapeutic Resistance in Non-Small Cell Lung Cancer. Genes 2023, 14, 607. [Google Scholar] [CrossRef] [PubMed]
- Takakuwa, O.; Oguri, T.; Ozasa, H.; Uemura, T.; Kunii, E.; Kasai, D.; Miyazaki, M.; Maeno, K.; Sato, S. C609T polymorphism of NAD(P)H quinone oxidoreductase 1 as a predictive biomarker for response to amrubicin. J. Thorac. Oncol. 2011, 6, 1826–1832. [Google Scholar] [CrossRef]
- Beaver, S.K.; Mesa-Torres, N.; Pey, A.L.; Timson, D.J. NQO1: A target for the treatment of cancer and neurological diseases, and a model to understand loss of function disease mechanisms. Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 663–676. [Google Scholar] [CrossRef]
- Nebert, D.W.; Roe, A.L.; Vandale, S.E.; Bingham, E.; Oakley, G.G. NAD(P)H:quinone oxidoreductase (NQO1) polymorphism, exposure to benzene, and predisposition to disease: A HuGE review. Genet. Med. 2002, 4, 62–70. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, D. The NQO1 C609T polymorphism and risk of lung cancer: A meta-analysis. Asian Pac. J. Cancer Prev. 2011, 12, 3091–3095. [Google Scholar] [PubMed]
- Schabath, M.B.; Cote, M.L. Cancer Progress and Priorities: LC. Cancer Epidemiol. Biomark. Prev. 2019, 28, 1563–1579. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Lin, H.; Wu, X.; Jin, W.; Zhang, Z. NQO1 C609T polymorphism and lung cancer susceptibility: Evidence from a comprehensive meta-analysis. Oncotarget 2017, 8, 102301–102309. [Google Scholar] [CrossRef]
- Chao, C.; Zhang, Z.F.; Berthiller, J.; Boffetta, P.; Hashibe, M. NAD(P)H:quinone oxidoreductase 1 (NQO1) Pro187Ser polymorphism and the risk of lung, bladder, and colorectal cancers: A meta-analysis. Cancer Epidemiol. Biomark. Prev. 2006, 15, 979–987. [Google Scholar] [CrossRef]
- Akkiz, H.; Bayram, S.; Bekar, A.; Akgöllü, E.; Ülger, Y.; Kaya, B.Y.; Sandikçi, M.; Özdil, B. No association of NAD(P)H: Quinone oxidoreductase 1 (NQO1) C609T polymorphism and risk of hepatocellular carcinoma development in Turkish subjects. Asian Pac. J. Cancer Prev. 2010, 11, 1051–1058. [Google Scholar] [PubMed]
- Lin, P.; Hsueh, Y.M.; Ko, J.L.; Liang, Y.F.; Tsai, K.J.; Chen, C.Y. Analysis of NQO1, GSTP1, and MnSOD genetic polymorphisms on lung cancer risk in Taiwan. Lung Cancer 2003, 40, 123–129. [Google Scholar] [CrossRef]
- Masroor, M.; Jain, A.; Javid, J.; Mir, R.; Prashant, Y.; Imtiyaz, A.; Mariyam, Z.; Mohan, A.; Ray, P.; Saxena, A. Clinical Significance of the NQO1 C609T Polymorphism in Non Small Cell Lung Adenocarcinoma Patients. Asian Pac. J. Cancer Prev. 2015, 16, 7653–7658. [Google Scholar] [CrossRef]
- Banerjee, S. Evaluation of the risk of lung cancer associated with NAD(P)H: Quinone oxidoreductase 1 (NQO1) C609T polymorphism in male current cigarette smokers from the Eastern India. Mol. Biol. Res. Commun. 2020, 9, 111–115. [Google Scholar]
- Ergen, H.A.; Gormus, U.; Narter, F.; Zeybek, U.; Bulgurcuoglu, S.; Isbir, T. Investigation of NAD(P)H:quinone oxidoreductase 1 (NQO1) C609T polymorphism in prostate cancer. Anticancer Res. 2007, 27, 4107–4110. [Google Scholar]
- Sirma, S.; Agaoglu, L.; Yildiz, I.; Cayli, D.; Horgusluoglu, E.; Anak, S.; Yuksel, L.; Unuvar, A.; Celkan, T.; Apak, H.; et al. NAD(P)H: Quinone oxidoreductase 1 null genotype is not associated with pediatric de novo acute leukemia. Pediatr. Blood Cancer 2004, 43, 568–570. [Google Scholar] [CrossRef]
- Ghorbani, F.; Mazidimoradi, A.; Biyabani, A.; Allahqoli, L.; Salehiniya, H. Role of NADPH Quinone Reductase 1 (NQO1) Polymorphism in Prevention, Diagnosis, and Treatment of Gastrointestinal Cancers. Curr. Cancer Drug Targets 2024, 24, 1213–1221. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, H.; Ishikawa, T.; Sugai, M.; Muneoka, Y.; Kano, Y.; Ueki, H.; Abe, S.; Moro, K.; Hirose, Y.; Miura, K.; et al. Prognostic Significance of NQO1 Expression in Non-neoplastic Esophageal Squamous Epithelium for Patients With Esophageal Cancer. Anticancer Res. 2024, 44, 1915–1924. [Google Scholar] [CrossRef]
- Yamamoto, S.; Koyanagi, Y.N.; Iwashita, Y.; Shinozaki, T.; Fujiwara, Y.; Sakakura, N.; Hara, M.; Nishida, Y.; Otonari, J.; Ikezaki, H.; et al. Smoking behavior-related genetic variants and lung cancer risk in Japanese: An assessment by mediation analysis. Carcinogenesis 2025, 46, bgaf011. [Google Scholar] [CrossRef] [PubMed]
- Cheng, E.S.; Weber, M.; Steinberg, J.; Yu, X.Q. Lung cancer risk in never-smokers: An overview of environmental and genetic factors. Chin. J. Cancer Res. 2021, 33, 548–562. [Google Scholar] [CrossRef]
- Katoh, T. Application of molecular biology to occupational health field--the frequency of gene polymorphism of cytochrome P450 1A1 and glutathione S-transferase M1 in patients with lung, oral and urothelial cancer. J. UOEH 1995, 17, 271–278. [Google Scholar] [CrossRef]
- Tang, D.L.; Rundle, A.; Warburton, D.; Santella, R.M.; Tsai, W.Y.; Chiamprasert, S.; Hsu, Y.Z.; Perera, F.P. Associations between both genetic and environmental biomarkers and lung cancer: Evidence of a greater risk of lung cancer in women smokers. Carcinogenesis 1998, 19, 1949–1953. [Google Scholar] [CrossRef] [PubMed]

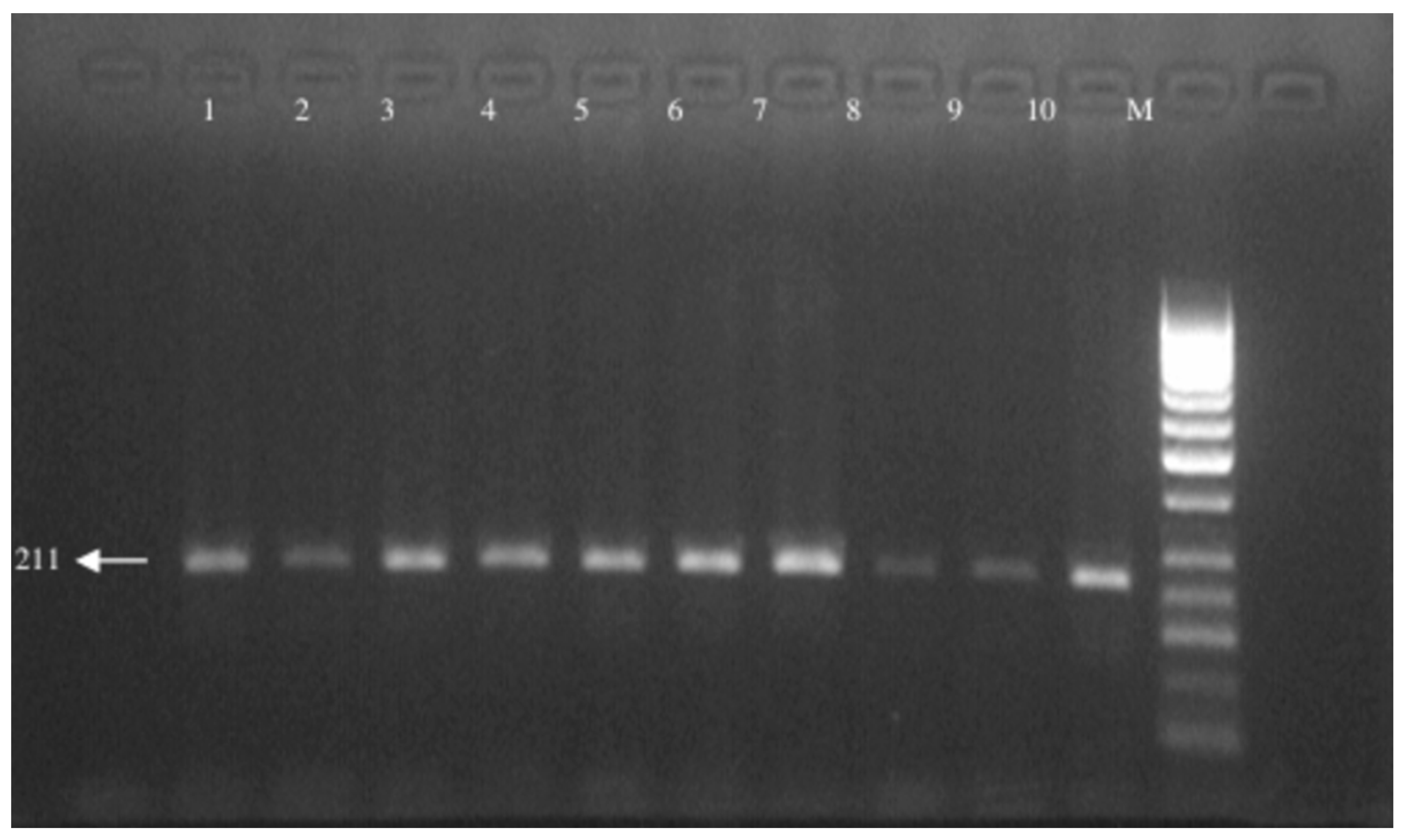
| F: 5′-TCC TCA GAG TGG CAT TCT GC -3′ |
| R: 5′- TCT CCT CAT CCT GTA CCT CT -3′ |
| Patients | Controls | |
|---|---|---|
| Gender | ||
| Female, n (%) | 3 (4.0%) | 4 (6.2%) |
| Male, n (%) | 72 (96.0%) | 61 (93.8%) |
| Smoking | 75 (100%) | 65 (100%) |
| Histopathology | ||
| Adenocarcinoma | 9 (12.0%) | |
| Squamous-cell carcinoma | 19 (25.3%) | |
| Small-cell carcinoma | 14 (18.7%) | |
| Non-small-cell carcinoma | 27 (36.0%) | |
| Unknown | 6 (8.0%) |
| Genotype | Patient | Control | p-Value |
|---|---|---|---|
| CC | 50 (66.7%) | 39 (60.0%) | |
| CT | 24 (32.0%) | 23 (35.4%) | 0.433 |
| TT | 1 (1.3%) | 3 (4.6%) |
| Histopathology | CC | CT | TT |
|---|---|---|---|
| Adenocarcinoma | 7 (14.0%) | 2 (8.3%) | |
| Squamous-cell carcinoma | 13 (26.0%) | 6 (25.0%) | |
| Small-cell carcinoma | 7 (14.0%) | 7 (29.2%) | |
| Non-small-cell carcinoma | 19 (38.0%) | 7 (29.2%) | 1 (100%) |
| Unknown | 4 (8%) | 2 (8.3%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the Lithuanian University of Health Sciences. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gumuskaya, P.O.; Ozkan, K.; Ay, A.; Sipahi, T.; Uzun, H. Impact of NAD(P)H: Quinone Oxidoreductase 1 (NQO1) C609T Polymorphism on Lung Cancer Risk. Medicina 2025, 61, 1504. https://doi.org/10.3390/medicina61091504
Gumuskaya PO, Ozkan K, Ay A, Sipahi T, Uzun H. Impact of NAD(P)H: Quinone Oxidoreductase 1 (NQO1) C609T Polymorphism on Lung Cancer Risk. Medicina. 2025; 61(9):1504. https://doi.org/10.3390/medicina61091504
Chicago/Turabian StyleGumuskaya, Perihan Ozkan, Kamile Ozkan, Arzu Ay, Tammam Sipahi, and Hafize Uzun. 2025. "Impact of NAD(P)H: Quinone Oxidoreductase 1 (NQO1) C609T Polymorphism on Lung Cancer Risk" Medicina 61, no. 9: 1504. https://doi.org/10.3390/medicina61091504
APA StyleGumuskaya, P. O., Ozkan, K., Ay, A., Sipahi, T., & Uzun, H. (2025). Impact of NAD(P)H: Quinone Oxidoreductase 1 (NQO1) C609T Polymorphism on Lung Cancer Risk. Medicina, 61(9), 1504. https://doi.org/10.3390/medicina61091504





