Proposed Comprehensive Methodology Integrated with Explainable Artificial Intelligence for Prediction of Possible Biomarkers in Metabolomics Panel of Plasma Samples for Breast Cancer Detection
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants, Dataset, and Power Analysis
2.2. Metabolomics Analysis
2.3. Modelling and Explainable Artificial Intelligence Methodology
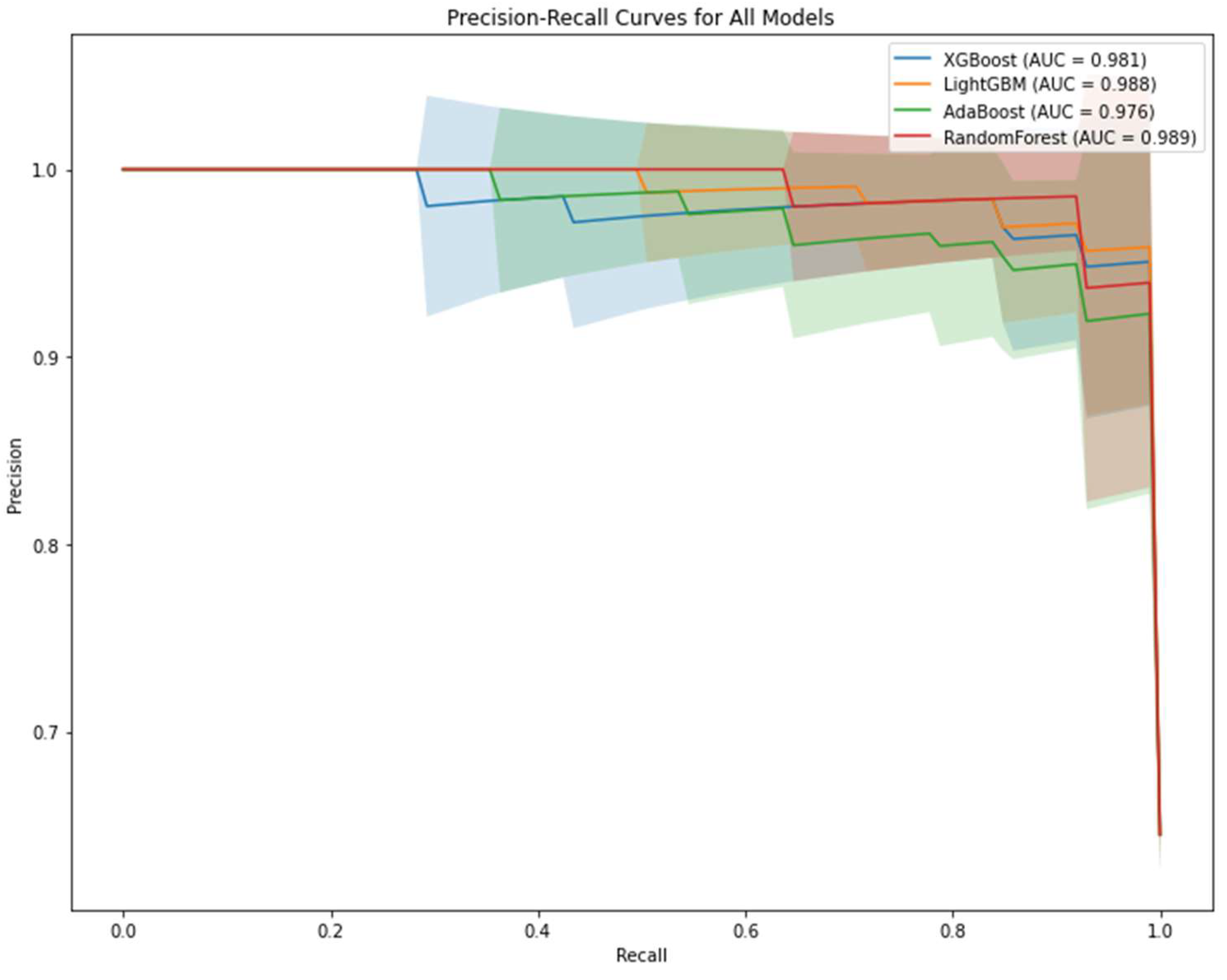
2.4. Performance Evaluation of Machine Learning Models
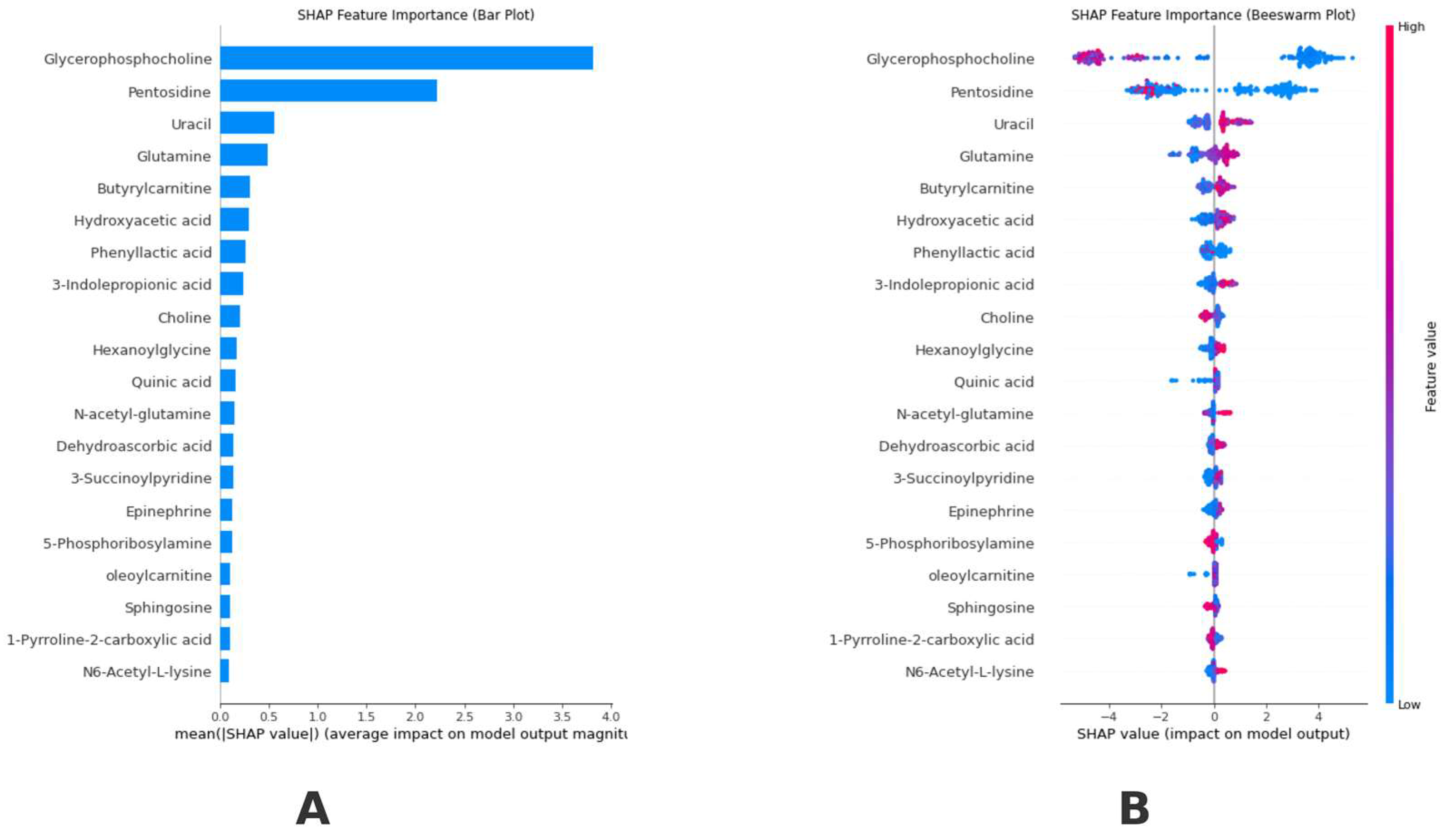
2.5. Explainable Artificial Intelligence and SHapley Additive exPlanations
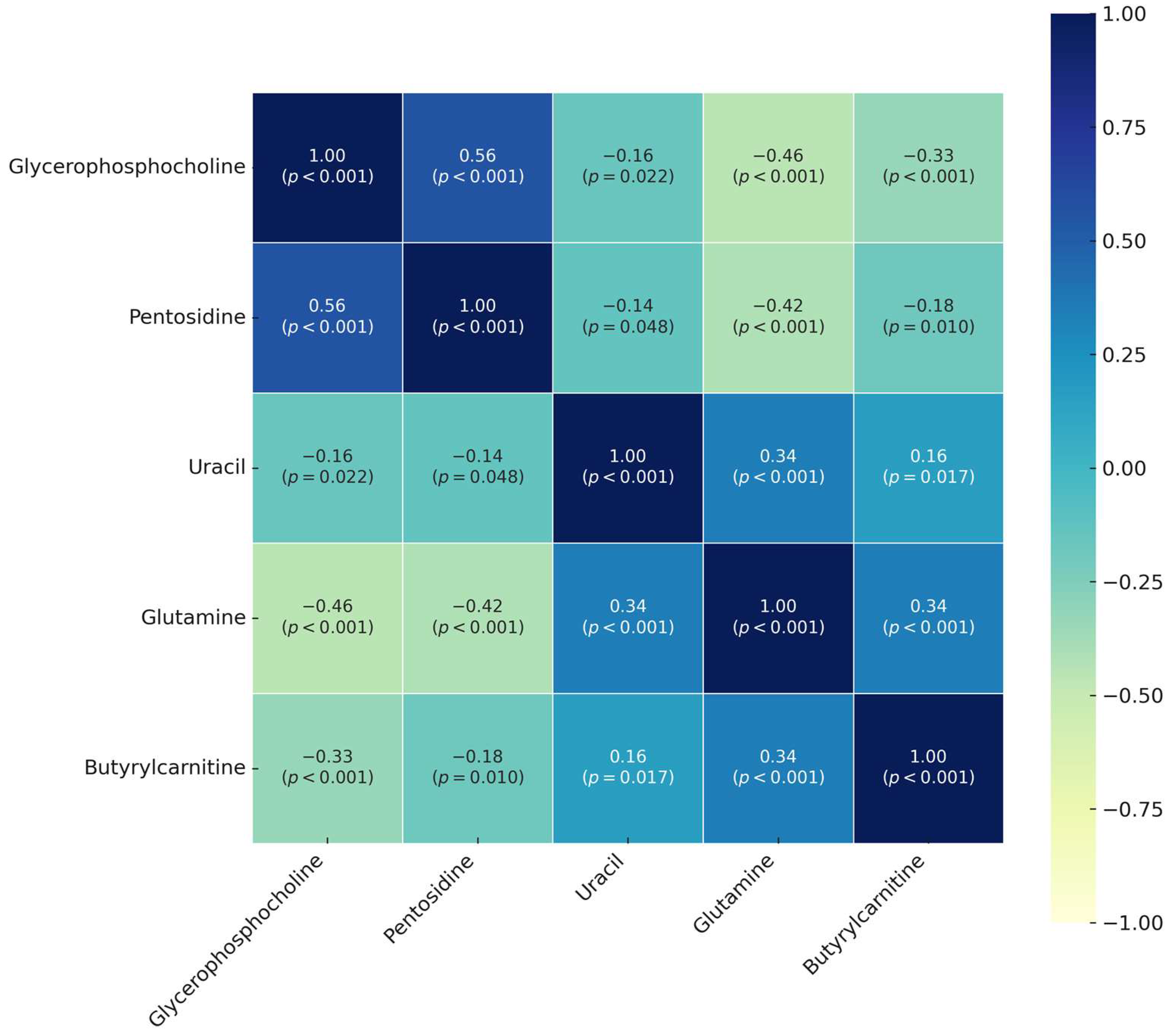
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations List
| AI | Artificial Intelligence |
| AUC | Area Under the Curve |
| PR-AUC | Precision-Recall Area Under the Curve |
| BC | Breast Cancer |
| CAD | Computer-Aided Diagnosis |
| CNN | Convolutional Neural Network |
| ER | Estrogen Receptor |
| GC-TOFMS | Gas Chromatography-Time of Flight Mass Spectrometry |
| HER-2 | Human Epidermal Growth Factor Receptor 2 |
| LC-TOFMS | Liquid Chromatography-Time of Flight Mass Spectrometry |
| LightGBM | Light Gradient Boosting Machine |
| mTOR | Mechanistic Target of Rapamycin |
| MRI | Magnetic Resonance Imaging |
| NMR | Nuclear Magnetic Resonance |
| PC | Phosphocholine |
| RF | Random Forest |
| ROC | Receiver Operating Characteristic |
| ROS | Reactive Oxygen Species |
| SHAP | SHapley Additive exPlanations |
| TNM | Tumor, Node, Metastasis |
| TOFMS | Time-of-Flight Mass Spectrometry |
| XAI | Explainable Artificial Intelligence |
| XGBoost | eXtreme Gradient Boosting |
References
- Smolarz, B.; Nowak, A.Z.; Romanowicz, H.J.C. Breast cancer—Epidemiology, classification, pathogenesis and treatment (review of literature). Cancers 2022, 14, 2569. [Google Scholar] [CrossRef]
- Skaane, P. Studies comparing screen-film mammography and full-field digital mammography in breast cancer screening: Updated review. Acta Radiol. 2009, 50, 3–14. [Google Scholar]
- Böhm, D.; Keller, K.; Wehrwein, N.; Lebrecht, A.; Schmidt, M.; Kölbl, H.; Grus, F.-H. Serum proteome profiling of primary breast cancer indicates a specific biomarker profile. Oncol. Rep. 2011, 26, 1051–1056. [Google Scholar] [PubMed]
- Sree, S.V.; Ng, E.Y.-K.; Acharya U, R.; Tan, W. Breast imaging systems: A review and comparative study. J. Mech. Med. Biol. 2010, 10, 5–34. [Google Scholar]
- Zhu, J.; Thompson, C.B. Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell Biol. 2019, 20, 436–450. [Google Scholar] [CrossRef]
- Kalyanaraman, B. Teaching the basics of cancer metabolism: Developing antitumor strategies by exploiting the differences between normal and cancer cell metabolism. Redox Biol. 2017, 12, 833–842. [Google Scholar] [PubMed]
- Alwahsh, M.; Abumansour, H.; Althaher, A.R.; Hergenröder, R. Metabolic Profiling Techniques and their Application in Cancer Research. Curr. Pharm. Anal. 2024, 20, 485–499. [Google Scholar]
- Martínez-Reyes, I.; Chandel, N.S. Cancer metabolism: Looking forward. Nat. Rev. Cancer 2021, 21, 669–680. [Google Scholar]
- Muthubharathi, B.C.; Gowripriya, T.; Balamurugan, K. Metabolomics: Small molecules that matter more. Mol. Omics 2021, 17, 210–229. [Google Scholar]
- Sarhadi, V.K.; Armengol, G. Molecular biomarkers in cancer. Biomolecules 2022, 12, 1021. [Google Scholar] [CrossRef]
- Ren, S.; Hinzman, A.A.; Kang, E.L.; Szczesniak, R.D.; Lu, L.J. Computational and statistical analysis of metabolomics data. Metabolomics 2015, 11, 1492–1513. [Google Scholar]
- Guldogan, E.; Yagin, F.H.; Pinar, A.; Colak, C.; Kadry, S.; Kim, J. A proposed tree-based explainable artificial intelligence approach for the prediction of angina pectoris. Sci. Rep. 2023, 13, 22189. [Google Scholar]
- Cansel, N.; Hilal Yagin, F.; Akan, M.; Ilkay Aygul, B. Interpretable estimation of suicide risk and severity from complete blood count parameters with explainable artificial intelligence methods. Psychiatr. Danub. 2023, 35, 62–72. [Google Scholar]
- Bifarin, O.O.; Fernández, F.M. Automated machine learning and explainable AI (AutoML-XAI) for metabolomics: Improving cancer diagnostics. J. Am. Soc. Mass Spectrom. 2024, 35, 1089–1100. [Google Scholar] [PubMed]
- Novielli, P.; Romano, D.; Magarelli, M.; Bitonto, P.D.; Diacono, D.; Chiatante, A.; Lopalco, G.; Sabella, D.; Venerito, V.; Filannino, P. Explainable artificial intelligence for microbiome data analysis in colorectal cancer biomarker identification. Front. Microbiol. 2024, 15, 1348974. [Google Scholar] [CrossRef]
- Irajizad, E.; Wu, R.; Vykoukal, J.; Murage, E.; Spencer, R.; Dennison, J.B.; Moulder, S.; Ravenberg, E.; Lim, B.; Litton, J.; et al. Application of artificial intelligence to plasma metabolomics profiles to predict response to neoadjuvant chemotherapy in triple-negative breast cancer. Front. Artif. Intell. 2022, 5, 876100. [Google Scholar]
- Li, N.; Yang, C.; Zhou, S.; Song, S.; Jin, Y.; Wang, D.; Liu, J.; Gao, Y.; Yang, H.; Mao, W.; et al. Combination of plasma-based metabolomics and machine learning algorithm provides a novel diagnostic strategy for malignant mesothelioma. Diagnostics 2021, 11, 1281. [Google Scholar] [CrossRef]
- Xie, G.; Zhou, B.; Zhao, A.; Qiu, Y.; Zhao, X.; Garmire, L.; Shvetsov, Y.B.; Yu, H.; Yen, Y.; Jia, W. Lowered circulating aspartate is a metabolic feature of human breast cancer. Oncotarget 2015, 6, 33369. [Google Scholar]
- Arslan, A.K.; Yaşar, Ş.; Çolak, C.; Yoloğlu, S. WSSPAS: An interactive web application for sample size and power analysis with R using shiny. Türkiye Klin. Biyoistatistik 2018, 10, 224–246. [Google Scholar]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Freund, Y.; Schapire, R.E. A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci. 1997, 55, 119–139. [Google Scholar]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. Lightgbm: A highly efficient gradient boosting decision tree. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Huang, Y.; Li, W.; Macheret, F.; Gabriel, R.A.; Ohno-Machado, L. A tutorial on calibration measurements and calibration models for clinical prediction models. J. Am. Med. Inform. Assoc. 2020, 27, 621–633. [Google Scholar] [CrossRef]
- Liu, J.; Wang, C.; Yan, R.; Lu, Y.; Bai, J.; Wang, H.; Li, R. Machine learning-based prediction of postpartum hemorrhage after vaginal delivery: Combining bleeding high risk factors and uterine contraction curve. Arch. Gynecol. Obstet. 2022, 306, 1015–1025. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Rehmsmeier, M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS ONE 2015, 10, e0118432. [Google Scholar] [CrossRef]
- Ozenne, B.; Subtil, F.; Maucort-Boulch, D. The precision–recall curve overcame the optimism of the receiver operating characteristic curve in rare diseases. J. Clin. Epidemiol. 2015, 68, 855–859. [Google Scholar] [CrossRef] [PubMed]
- Petch, J.; Di, S.; Nelson, W. Opening the black box: The promise and limitations of explainable machine learning in cardiology. Can. J. Cardiol. 2022, 38, 204–213. [Google Scholar] [CrossRef]
- Saranya, A.; Subhashini, R. A systematic review of Explainable Artificial Intelligence models and applications: Recent developments and future trends. Decis. Anal. J. 2023, 7, 100230. [Google Scholar]
- Nasir, Y.; Kadian, K.; Sharma, A.; Dwivedi, V. Interpretable machine learning for dermatological disease detection: Bridging the gap between accuracy and explainability. Comput. Biol. Med. 2024, 179, 108919. [Google Scholar] [CrossRef]
- Scott, M.; Su-In, L.J.A. A unified approach to interpreting model predictions. Neural Inf. Process. Syst. 2017, 30, 4765–4774. [Google Scholar]
- Aas, K.; Jullum, M.; Løland, A. Explaining individual predictions when features are dependent: More accurate approximations to Shapley values. Artif. Intell. 2021, 298, 103502. [Google Scholar] [CrossRef]
- Hassija, V.; Chamola, V.; Mahapatra, A.; Singal, A.; Goel, D.; Huang, K.; Scardapane, S.; Spinelli, I.; Mahmud, M.; Hussain, A. Interpreting black-box models: A review on explainable artificial intelligence. Cogn. Comput. 2024, 16, 45–74. [Google Scholar] [CrossRef]
- Vimbi, V.; Shaffi, N.; Mahmud, M. Interpreting artificial intelligence models: A systematic review on the application of LIME and SHAP in Alzheimer’s disease detection. Brain Inform. 2024, 11, 10. [Google Scholar]
- Tasnim, N.; Al Mamun, S.; Shahidul Islam, M.; Kaiser, M.S.; Mahmud, M. Explainable mortality prediction model for congestive heart failure with nature-based feature selection method. Appl. Sci. 2023, 13, 6138. [Google Scholar] [CrossRef]
- Yang, L.; Wang, Y.; Cai, H.; Wang, S.; Shen, Y.; Ke, C. Application of metabolomics in the diagnosis of breast cancer: A systematic review. J. Cancer 2020, 11, 2540. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Jasbi, P.; Shi, X.; Turner, C.; Hrovat, J.; Liu, L.; Rabena, Y.; Porter, P.; Gu, H. Early breast cancer detection using untargeted and targeted metabolomics. J. Proteome Res. 2021, 20, 3124–3133. [Google Scholar] [CrossRef]
- Piffoux, M.; Jacquemin, J.; Pétéra, M.; Durand, S.; Abila, A.; Centeno, D.; Joly, C.; Lyan, B.; Martin, A.-L.; Everhard, S.; et al. Metabolomic Prediction of Breast Cancer Treatment–Induced Neurologic and Metabolic Toxicities. Clin. Cancer Res. 2024, 30, 4654–4666. [Google Scholar] [CrossRef]
- Díaz-Beltrán, L.; González-Olmedo, C.; Luque-Caro, N.; Díaz, C.; Martín-Blázquez, A.; Fernández-Navarro, M.; Ortega-Granados, A.L.; Gálvez-Montosa, F.; Vicente, F.; Pérez del Palacio, J.P.; et al. Human plasma metabolomics for biomarker discovery: Targeting the molecular subtypes in breast cancer. Cancers 2021, 13, 147. [Google Scholar] [CrossRef]
- Haince, J.-F.; Zhang, L.; Bux, R.A.; Tappia, P.S.; Ramjiawan, B.; Wishart, D.; Maksymiuk, A. Abstract PO5-13-03: Early Detection of Breast Cancer using Targeted Plasma Metabolomic Profiling. Cancer Res. 2024, 84, PO5-13-03. [Google Scholar] [CrossRef]
- Sterin, M.; Cohen, J.S.; Mardor, Y.; Berman, E.; Ringel, I. Levels of phospholipid metabolites in breast cancer cells treated with antimitotic drugs: A 31P-magnetic resonance spectroscopy study. Cancer Res. 2001, 61, 7536–7543. [Google Scholar]
- Stoica, C.; Ferreira, A.; Hannan, K.; Bakovic, M. Bilayer Forming Phospholipids as Targets for Cancer Therapy. Int. J. Mol. Sci. 2022, 23, 5266. [Google Scholar] [CrossRef] [PubMed]
- Wijnen, J.; Jiang, L.; Greenwood, T.; Cheng, M.; Döpkens, M.; Cao, M.; Bhujwalla, Z.; Krishnamachary, B.; Klomp, D.; Glunde, K. Silencing of the glycerophosphocholine phosphodiesterase GDPD5 alters the phospholipid metabolite profile in a breast cancer model in vivo as monitored by 31P MRS. NMR Biomed. 2014, 27, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Scheijen, J.L.; van de Waarenburg, M.P.; Stehouwer, C.D.; Schalkwijk, C.G. Measurement of pentosidine in human plasma protein by a single-column high-performance liquid chromatography method with fluorescence detection. J. Chromatogr. B 2009, 877, 610–614. [Google Scholar]
- Günther, U.L. Metabolomics biomarkers for breast cancer. Pathobiology 2015, 82, 153–165. [Google Scholar]
- Demas, D.M.; Demo, S.; Fallah, Y.; Clarke, R.; Nephew, K.P.; Althouse, S.; Sandusky, G.; He, W.; Shajahan-Haq, A.N. Glutamine metabolism drives growth in advanced hormone receptor positive breast cancer. Front. Oncol. 2019, 9, 686. [Google Scholar]
- Wang, Y.; Zhang, H.; Chu, Y. Advances in Research on the Relationship between Glutamine Metabolism and Breast Cancer. Int. J. Biol. Life Sci. 2023, 3, 1–3. [Google Scholar]
- Liu, Y.; Qi, C.; Zheng, L.; Li, J.; Wang, L.; Yang, Y. 1 H-NMR based metabolic study of MMTV-PyMT mice along with pathological progress to screen biomarkers for the early diagnosis of breast cancer. Mol. Omics 2022, 18, 167–177. [Google Scholar]
- Yu, R.; Peng, M.; Zhao, S.; Wang, Z.; Ma, Y.; Zhang, X.; Lv, X.; Wang, S.; Ju, S.; Zhao, R.; et al. Comprehensive Characterization of the Function of Metabolic Genes and Establishment of a Prediction Model in Breast Cancer. Dis. Markers 2022, 2022, 3846010. [Google Scholar] [CrossRef] [PubMed]
- Mirshekaran, R.; Ahmadi, K.; Shahbazi, B.; Farshidfar, G.; Eftekhar, E.; Kavousipour, S. Cancer Therapy Potential Unveiled: FDA-Approved Drugs Targeting Glutaminase for Breast Cancer Treatment. ChemistrySelect 2024, 9, e202401043. [Google Scholar]
- Sari, L.; Romadloni, A.; Lityaningrum, R.; Hastuti, H.D. Implementation of LightGBM and Random Forest in Potential Customer Classification. TIERS Inf. Technol. J. 2023, 4, 43–55. [Google Scholar] [CrossRef]
- Duran, F.; Wijaya, F.; Hulu, Y.R.; Harahap, M.; Prabowo, A. Perbandingan Kinerja Algoritma Random Forest Classifier Dan Lightgbm Classifier Untuk Prediksi Penyakit Jantung. Data Sci. Indones. (DSI) 2023, 3, 98–103. [Google Scholar]
- Mohanty, F.; Rup, S.; Dash, B.; Majhi, B.; Swamy, M.N.S. A computer-aided diagnosis system using Tchebichef features and improved grey wolf optimized extreme learning machine. Appl. Intell. 2019, 49, 983–1001. [Google Scholar]
- Hamid, M.A.; Mondher, H.M.; Ayoub, B. Deep Learning CNNs for Breast Cancer Classification and Detection” Enhancing Diagnostic Accuracy in Medical Practice. In Proceedings of the 2024 2nd International Conference on Electrical Engineering and Automatic Control (ICEEAC), Setif, Algeria, 12–14 May 2024; pp. 1–6. [Google Scholar]
- Long, N.P.; Nghi, T.D.; Kang, Y.P.; Anh, N.H.; Kim, H.M.; Park, S.K.; Kwon, S.W. Toward a standardized strategy of clinical metabolomics for the advancement of precision medicine. Metabolites 2020, 10, 51. [Google Scholar] [CrossRef] [PubMed]
- Marchand, C.R.; Farshidfar, F.; Rattner, J.; Bathe, O.F. A framework for development of useful metabolomic biomarkers and their effective knowledge translation. Metabolites 2018, 8, 59. [Google Scholar] [CrossRef]

| Characteristic | Breast Cancer | Healthy Control |
|---|---|---|
| Number of Participants | 138 | 76 |
| Age (median, range) | 49.0 (31–73) | 34.0 (21–40) |
| TNM Stage I | 19 | |
| TNM Stage II | 50 | |
| TNM Stage III | 49 | |
| TNM Stage IV | 20 | |
| ER (Positive/Negative/Unknown) | 77/54/7 | |
| PR (Positive/Negative/Unknown) | 64/67/7 | |
| HER-2 (Positive/Negative/Unknown) | 50/80/8 |
| Model | Type | Description |
|---|---|---|
| RF | Ensemble ML; Bagging | Random Forest is an ensemble technique that generates several decision trees during the training phase and produces the mode of the classes for classification purposes. By averaging several deep decision trees trained on different parts of the same training dataset, it improves the accuracy of predictions and reduces overfitting. |
| AdaBoost | Ensemble ML; Boosting | AdaBoost is an enhancement method that integrates weak learners to form a robust classifier. It assigns weights to misclassified instances and modifies them in subsequent iterations to emphasize difficult-to-classify occurrences, thereby enhancing the model’s accuracy. |
| LightGBM | Ensemble ML; Boosting | LightGBM is a gradient-boosting framework that emphasizes the efficiency of model construction. It employs methods such as histogram-based learning and exclusive feature bundling to accelerate training and enhance performance, particularly on extensive datasets. |
| XGBoost | Ensemble ML; Boosting | XGboost is a sophisticated version of gradient boosting that employs a regularized model structure to mitigate overfitting. It has sophisticated capabilities like parallel processing, tree pruning, and the management of missing information, rendering it rapid and effective for structured data analysis. |
| Metric | XGBoost | LightGBM | AdaBoost | Random Forest |
|---|---|---|---|---|
| Accuracy | 0.949 ± 0.056 | 0.949 ± 0.046 | 0.939 ± 0.071 | 0.963 ± 0.043 |
| Sensitivity | 0.948 ± 0.080 | 0.956 ± 0.062 | 0.949 ± 0.084 | 0.977 ± 0.051 |
| Specificity | 0.948 ± 0.067 | 0.934 ± 0.070 | 0.920 ± 0.097 | 0.934 ± 0.091 |
| F1 | 0.958 ± 0.048 | 0.959 ± 0.038 | 0.951 ± 0.056 | 0.971 ± 0.034 |
| AUC | 0.974 ± 0.045 | 0.983 ± 0.028 | 0.963 ± 0.043 | 0.982 ± 0.030 |
| Brier | 0.044 ± 0.044 | 0.034 ± 0.036 | 0.134 ± 0.024 | 0.069 ± 0.022 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the Lithuanian University of Health Sciences. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colak, C.; Yagin, F.H.; Algarni, A.; Algarni, A.; Al-Hashem, F.; Ardigò, L.P. Proposed Comprehensive Methodology Integrated with Explainable Artificial Intelligence for Prediction of Possible Biomarkers in Metabolomics Panel of Plasma Samples for Breast Cancer Detection. Medicina 2025, 61, 581. https://doi.org/10.3390/medicina61040581
Colak C, Yagin FH, Algarni A, Algarni A, Al-Hashem F, Ardigò LP. Proposed Comprehensive Methodology Integrated with Explainable Artificial Intelligence for Prediction of Possible Biomarkers in Metabolomics Panel of Plasma Samples for Breast Cancer Detection. Medicina. 2025; 61(4):581. https://doi.org/10.3390/medicina61040581
Chicago/Turabian StyleColak, Cemil, Fatma Hilal Yagin, Abdulmohsen Algarni, Ali Algarni, Fahaid Al-Hashem, and Luca Paolo Ardigò. 2025. "Proposed Comprehensive Methodology Integrated with Explainable Artificial Intelligence for Prediction of Possible Biomarkers in Metabolomics Panel of Plasma Samples for Breast Cancer Detection" Medicina 61, no. 4: 581. https://doi.org/10.3390/medicina61040581
APA StyleColak, C., Yagin, F. H., Algarni, A., Algarni, A., Al-Hashem, F., & Ardigò, L. P. (2025). Proposed Comprehensive Methodology Integrated with Explainable Artificial Intelligence for Prediction of Possible Biomarkers in Metabolomics Panel of Plasma Samples for Breast Cancer Detection. Medicina, 61(4), 581. https://doi.org/10.3390/medicina61040581









