Association of Polymorphisms in PD-1 and LAG-3 Genes with Acute Myeloid Leukemia
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Healthy Individuals
2.2. DNA Extraction
2.3. SNP Selection and Genotyping Method
2.4. In Silico Analysis of Gene Expression Profiles and Their Correlation with Survival Prognosis
2.5. Statistical Analysis
3. Results
3.1. Association of PDCD1 (rs2227981A>G) Polymorphisms with AML
3.2. Association of LAG3 (rs12313899A>G) Polymorphisms with AML
3.3. Stratification Analysis by Gender and Age
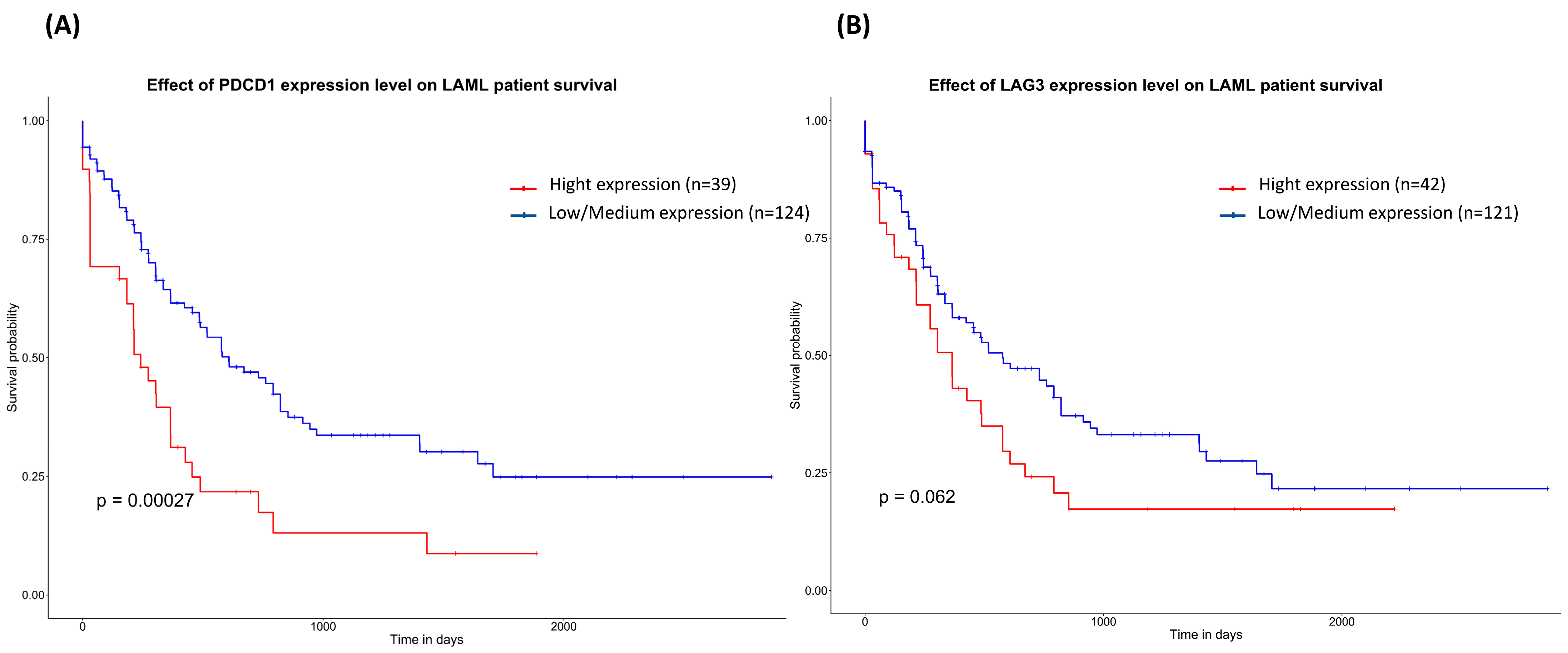
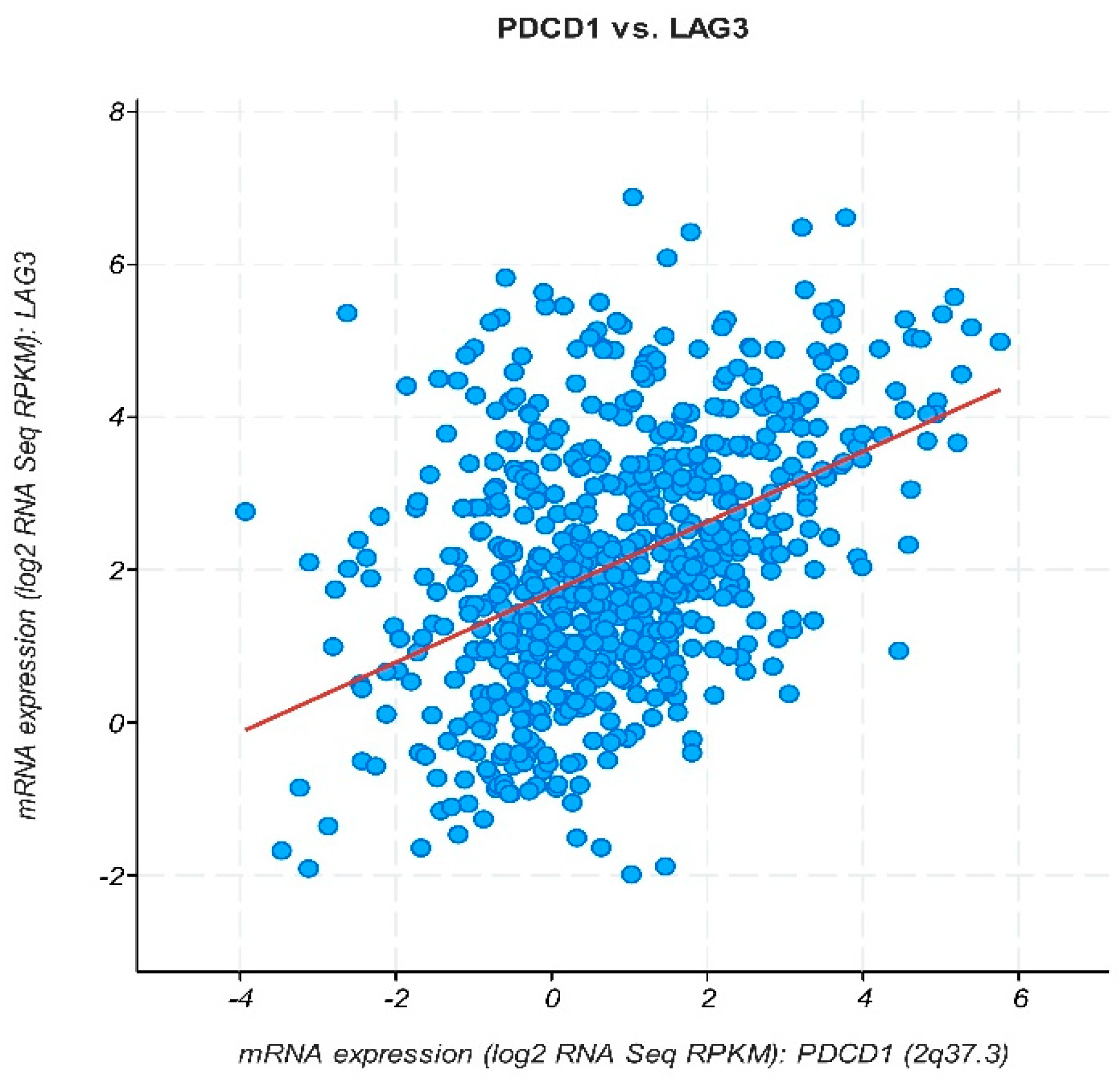
3.4. In Silico Analysis of mRNA Differential Expression and Prognosis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vardiman, J.W.; Harris, N.L.; Brunning, R.D. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood 2002, 100, 2292–2302. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Bawazir, A.; Al-Zamel, N.; Amen, A.; Akiel, M.A.; Alhawiti, N.M.; Alshehri, A. The burden of leukemia in the Kingdom of Saudi Arabia: 15 years period (1999–2013). BMC Cancer 2019, 19, 703. [Google Scholar] [CrossRef]
- Marrero, R.J.; Lamba, J.K. Current Landscape of Genome-Wide Association Studies in Acute Myeloid Leukemia: A Review. Cancers 2023, 15, 3583. [Google Scholar] [CrossRef]
- Lin, W.-Y.; Fordham, S.E.; Hungate, E.; Sunter, N.J.; Elstob, C.; Xu, Y.; Park, C.; Quante, A.; Strauch, K.; Gieger, C.; et al. Genome-wide association study identifies susceptibility loci for acute myeloid leukemia. Nat. Commun. 2021, 12, 6233. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.J.; Xie, L.; McCahan, S.M. Characterization of Pediatric Acute Lymphoblastic Leukemia Survival Patterns by Age at Diagnosis. J. Cancer Epidemiol. 2014, 2014, 865979. [Google Scholar] [CrossRef]
- Tebbi, C.K. Etiology of Acute Leukemia: A Review. Cancers 2021, 13, 2256. [Google Scholar] [CrossRef]
- Deschler, B.; Lübbert, M. Acute myeloid leukemia: Epidemiology and etiology. Cancer Interdiscip. Int. J. Am. Cancer Soc. 2006, 107, 2099–2107. [Google Scholar] [CrossRef] [PubMed]
- Kayser, S.; Schlenk, R.F. Genetic and immunophenotypical diagnostics for acute myeloid leukemia and their implication on treatment strategy. J. Lab. Med. 2012, 36. [Google Scholar] [CrossRef]
- Shallis, R.M.; Wang, R.; Davidoff, A.; Ma, X.; Zeidan, A.M. Epidemiology of acute myeloid leukemia: Recent progress and enduring challenges. Blood Rev. 2019, 36, 70–87. [Google Scholar] [CrossRef]
- Newell, L.F.; Cook, R.J. Advances in acute myeloid leukemia. BMJ 2021, 375, n2026. [Google Scholar] [CrossRef] [PubMed]
- Im, A.; Pavletic, S.Z. Immunotherapy in hematologic malignancies: Past, present, and future. J. Hematol. Oncol. 2017, 10, 94. [Google Scholar] [CrossRef] [PubMed]
- Moradbeygi, F.; Ghasemi, Y.; Farmani, A.R.; Hemmati, S. Glucarpidase (carboxypeptidase G2): Biotechnological production, clinical application as a methotrexate antidote, and placement in targeted cancer therapy. Biomed. Pharmacother. 2023, 166, 115292. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Wu, T.; Zhou, X.; Xie, S.; Sun, H.; Sun, Y.; Li, Y. Progress of research on PD-1/PD-L1 in leukemia. Front. Immunol. 2023, 14, 1265299. [Google Scholar] [CrossRef] [PubMed]
- Estey, E.H. Acute myeloid leukemia: 2019 update on risk-stratification and management. Am. J. Hematol. 2018, 93, 1267–1291. [Google Scholar] [CrossRef] [PubMed]
- Kansal, R. Classification of acute myeloid leukemia by the revised fourth edition World Health Organization criteria: A retrospective single-institution study with appraisal of the new entities of acute myeloid leukemia with gene mutations in NPM1 and biallelic CEBPA. Hum. Pathol. 2019, 90, 80–96. [Google Scholar] [CrossRef]
- Li, Z.; Philip, M.; Ferrell, P.B. Alterations of T-cell-mediated immunity in acute myeloid leukemia. Oncogene 2020, 39, 3611–3619. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, H.; Hagerling, C.; Werb, Z. Roles of the immune system in cancer: From tumor initiation to metastatic progression. Genes. Dev. 2018, 32, 1267–1284. [Google Scholar] [CrossRef] [PubMed]
- Rovatti, P.E.; Gambacorta, V.; Lorentino, F.; Ciceri, F.; Vago, L. Mechanisms of leukemia immune evasion and their role in relapse after haploidentical hematopoietic cell transplantation. Front. Immunol. 2020, 11, 502733. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Chen, S.; Yang, L.; Li, Y. The role of PD-1 and PD-L1 in T-cell immune suppression in patients with hematological malignancies. J. Hematol. Oncol. 2013, 6, 74. [Google Scholar] [CrossRef]
- Teague, R.M.; Kline, J. Immune evasion in acute myeloid leukemia: Current concepts and future directions. J. Immunother. Cancer 2013, 1, 13. [Google Scholar] [CrossRef]
- Niu, B.; Zhou, F.; Su, Y.; Wang, L.; Yi, Z.; Du, H.; Ren, G. Different expression characteristics of LAG3 and PD-1 in sepsis and their synergistic effect on T cell exhaustion: A new strategy for immune checkpoint blockade. Front. Immunol. 2019, 10, 467887. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, M.; Meng, G.; Ma, Y.; Ye, J.; Sun, T.; Ji, C. Immune checkpoint-related gene polymorphisms are associated with acute myeloid leukemia. Cancer Med. 2023, 12, 18588–18596. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.; Jasek, M.; Karabon, L. Immune checkpoint molecules—Inherited variations as markers for cancer risk. Front. Immunol. 2021, 11, 606721. [Google Scholar] [CrossRef] [PubMed]
- Al-Harbi, N.; Vaali-Mohammed, M.-A.; Al-Omar, S.; Zubaidi, A.; Al-Obeed, O.; Abdulla, M.-H.; Mansour, L. Rs10204525 polymorphism of the programmed death (PD-1) gene is associated with increased risk in a Saudi Arabian population with colorectal cancer. Medicina 2022, 58, 1439. [Google Scholar] [CrossRef] [PubMed]
- Ok, C.Y.; Young, K.H. Checkpoint inhibitors in hematological malignancies. J. Hematol. Oncol. 2017, 10, 103. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Solé, X.; Guinó, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef]
- Yin, L.; Guo, H.; Zhao, L.; Wang, J. The programmed death-1 gene polymorphism (PD-1.5 C/T) is associated with non-small cell lung cancer risk in a Chinese Han population. Int. J. Clin. Exp. Med. 2014, 7, 5832. [Google Scholar]
- Li, X.F.; Jiang, X.Q.; Zhang, J.W.; Jia, Y.J. Association of the programmed cell death-1 PD1.5 C > T polymorphism with cervical cancer risk in a Chinese population. Genet. Mol. Res. 2016, 15, 10-4238. [Google Scholar] [CrossRef]
- Dong, W.; Gong, M.; Shi, Z.; Xiao, J.; Zhang, J.; Peng, J. Programmed cell death-1 polymorphisms decrease the cancer risk: A meta-analysis involving twelve case-control studies. PLoS ONE 2016, 11, e0152448. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Wang, Y.; Jiang, H.; Liu, P.; Liu, C.; Gu, H.; Chen, S.; Kang, M. Programmed death-1 (PD-1) rs2227981 C > T polymorphism is associated with cancer susceptibility: A meta-analysis. Int. J. Clin. Exp. Med. 2015, 8, 22278. [Google Scholar] [PubMed]
- Mamat, U.; Arkinjan, M. Association of programmed death-1 gene polymorphism rs2227981 with tumor: Evidence from a meta analysis. Int. J. Clin. Exp. Med. 2015, 8, 13282. [Google Scholar] [PubMed]
- Hashemi, M.; Karami, S.; Sarabandi, S.; Moazeni-Roodi, A.; Małecki, A.; Ghavami, S.; Wiechec, E. Association between PD-1 and PD-L1 Polymorphisms and the Risk of Cancer: A Meta-Analysis of Case-Control Studies. Cancers 2019, 11, 1150. [Google Scholar] [CrossRef] [PubMed]
- Kasamatsu, T.; Awata, M.; Ishihara, R.; Murakami, Y.; Gotoh, N.; Matsumoto, M.; Sawamura, M.; Yokohama, A.; Handa, H.; Tsukamoto, N. PDCD1 and PDCD1LG1 polymorphisms affect the susceptibility to multiple myeloma. Clin. Exp. Med. 2020, 20, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Grzywnowicz, M.; Karabon, L.; Karczmarczyk, A.; Zajac, M.; Skorka, K.; Zaleska, J.; Wlasiuk, P.; Chocholska, S.; Tomczak, W.; Bojarska-Junak, A. The function of a novel immunophenotype candidate molecule PD-1 in chronic lymphocytic leukemia. Leuk. Lymphoma 2015, 56, 2908–2913. [Google Scholar] [CrossRef] [PubMed]
- Chocarro, L.; Blanco, E.; Zuazo, M.; Arasanz, H.; Bocanegra, A.; Fernández-Rubio, L.; Morente, P.; Fernández-Hinojal, G.; Echaide, M.; Garnica, M. Understanding LAG-3 signaling. Int. J. Mol. Sci. 2021, 22, 5282. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.C.; Joller, N.; Kuchroo, V.K. Lag-3, Tim-3, and TIGIT: Co-inhibitory receptors with specialized functions in immune regulation. Immunity 2016, 44, 989–1004. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ye, Y.; Yu, H.; Lin, S.H.; Tu, H.; Liang, D.; Chang, D.W.; Huang, M.; Wu, X. Immune checkpoint-related serum proteins and genetic variants predict outcomes of localized prostate cancer, a cohort study. Cancer Immunol. Immunother. 2021, 70, 701–712. [Google Scholar] [CrossRef]
- Lee, K.M.; Baris, D.; Zhang, Y.; Hosgood III, H.D.; Menashe, I.; Yeager, M.; Zahm, S.H.; Wang, S.S.; Purdue, M.P.; Chanock, S. Common single nucleotide polymorphisms in immunoregulatory genes and multiple myeloma risk among women in Connecticut. Am. J. Hematol. 2010, 85, 560–563. [Google Scholar] [CrossRef]
- Cha, J.H.; Chan, L.C.; Li, C.W.; Hsu, J.L.; Hung, M.C. Mechanisms Controlling PD-L1 Expression in Cancer. Mol. Cell 2019, 76, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.C.; Latchman, Y.E.; Buhlmann, J.E.; Tomczak, M.F.; Horwitz, B.H.; Freeman, G.J.; Sharpe, A.H. Regulation of PD-1, PD-L1, and PD-L2 expression during normal and autoimmune responses. Eur. J. Immunol. 2003, 33, 2706–2716. [Google Scholar] [CrossRef] [PubMed]
- Armand, P. Immune checkpoint blockade in hematologic malignancies. Blood J. Am. Soc. Hematol. 2015, 125, 3393–3400. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Liang, C.; Wang, S.; Chio, C.L.; Zhang, Y.; Zeng, C.; Chen, S.; Wang, C.; Li, Y. Expression patterns of immune checkpoints in acute myeloid leukemia. J. Hematol. Oncol. 2020, 13, 28. [Google Scholar] [CrossRef] [PubMed]
- Radwan, S.M.; Elleboudy, N.S.; Nabih, N.A.; Kamal, A.M. The immune checkpoints Cytotoxic T lymphocyte antigen-4 and Lymphocyte activation gene-3 expression is up-regulated in acute myeloid leukemia. HLA 2020, 96, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Keane, C.; Law, S.C.; Gould, C.; Birch, S.; Sabdia, M.B.; Merida de Long, L.; Thillaiyampalam, G.; Abro, E.; Tobin, J.W.; Tan, X. LAG3: A novel immune checkpoint expressed by multiple lymphocyte subsets in diffuse large B-cell lymphoma. Blood Adv. 2020, 4, 1367–1377. [Google Scholar] [CrossRef]
 ) significant difference. Data were obtained from the GEPIA database (http://gepia.cancer-pku.cn, accessed on 14 February 2024).
) significant difference. Data were obtained from the GEPIA database (http://gepia.cancer-pku.cn, accessed on 14 February 2024).
 ) significant difference. Data were obtained from the GEPIA database (http://gepia.cancer-pku.cn, accessed on 14 February 2024).
) significant difference. Data were obtained from the GEPIA database (http://gepia.cancer-pku.cn, accessed on 14 February 2024).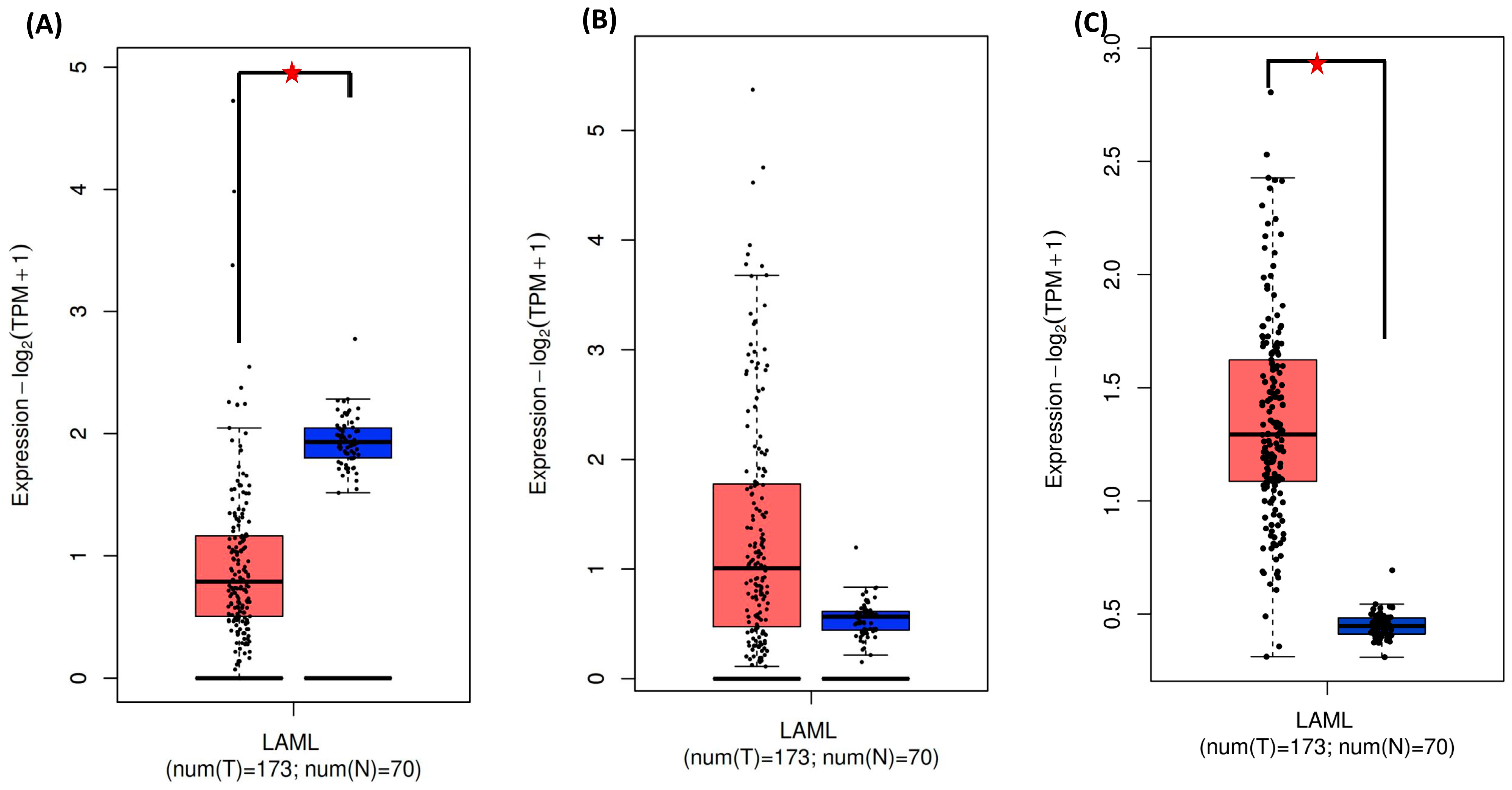
| Gene | SNP ID/Assay ID | Chromosome Position | Nucleotide Change | Region | MAF in Human Populations | ||||
|---|---|---|---|---|---|---|---|---|---|
| Global | European | East Asian | American | Qatari | |||||
| PDCD1 | rs2227981 | Chr2/241851121 | A>G | Exon5 | 0.35 | 0.40 | 0.27 | 0.44 | 0.33 |
| LAG3 | rs12313899 | Chr 12/6768692 | A>G | Intro | 0.47 | 0.39 | 0.43 | 0.42 | 0.40 |
| Locus | Model | Genotype | Control (%) n = 132 | AML n = 98 | OR (95% CI) | p-Value | AIC |
|---|---|---|---|---|---|---|---|
| PDCD1 | Allele | A | 0.65 | 0.48 | 1 | 0.0008 | |
| G | 0.35 | 0.52 | 1.93 (1.31–2.81) | ||||
| Codominant | AA | 50 (38.2%) | 4 (4.1%) | 1 | <0.0001 | 274.2 | |
| AG | 69 (52.7%)) | 87 (88.8%) | 15.76 (5.43–45.78) | ||||
| GG | 12 (9.2%) | 7 (7.1%) | 7.29 (1.83–29.00) | ||||
| Dominant | AA | 50 (38.2%) | 4 (4.1%) | 1 | <0.0001 | 284 | |
| AG + GG | 81 (61.8%) | 94 (95.9%) | 14.51 (5.02–41.91) | ||||
| Recessive | AA + AG | 119 (90.8%) | 91 (92.9%) | 1 | 0.58 | 316.4 | |
| GG | 12 (9.2%) | 7 (7.1%) | 0.76 (0.29–2.01) | ||||
| Overdominant | G/G + A/A | 62 (47.3%) | 11 (11.2%) | 1 | <0.0001 | 280.1 | |
| A/G | 69 (52.7%) | 87 (88.8%) | 7.11 (3.48–14.52) | ||||
| Log-Additive | 3.35 (1.90–5.91) | <0.0001 | 296.1 |
| Locus | Model | Genotype | Control (%) n = 132 | AML n = 98 | OR (95% CI) | p-Value | AIC |
|---|---|---|---|---|---|---|---|
| LAG3 | Allele | A | 0.54 | 0.63 | 1 | 0.23 | |
| G | 0.46 | 0.37 | 1.163 (0.907–1.492) | ||||
| Codominant | AA | 44 (34.1%) | 41 (41.8%) | 1 | 0.2 | 323.5 | |
| AG | 52 (40.3%) | 41 (41.8%) | 0.85 (0.47–1.53) | ||||
| GG | 33 (25.6%) | 16 (16.3%) | 0.52 (0.25–1.08) | ||||
| Dominant | AA | 44 (34.1%) | 41 (41.8%) | 1 | 0.23 | 319.9 | |
| AG + GG | 85 (65.9%) | 57 (58.2%) | 0.72 (0.42–1.24) | ||||
| Recessive | AA + AG | 96 (74.4%) | 82 (83.7%) | 1 | 0.09 | 318.4 | |
| GG | 33 (25.6%) | 16 (16.3%) | 0.57 (0.29–1.10) | ||||
| Overdominant | AA + GG | 77 (59.7%) | 57 (58.2%) | 1 | 0.82 | 321.2 | |
| AG | 52 (40.3%) | 41 (41.8%) | 1.07 (0.62–1.82) | ||||
| Log-Additive | --- | --- | 0.74 (0.52–1.05) | 0.091 | 311.6 |
| Locus | Model | Genotype | AML Female n = 44 | AML Male n = 54 | OR (95% CI) | p-Value | AIC |
|---|---|---|---|---|---|---|---|
| PDCD1 rs2227981A>G | Allele | A | 0.5 | 0.57 | 0.77 | ||
| G | 0.5 | 0.47 | 0.89 (0.50–1.57) | ||||
| Codominant | AA | 2 (4.5%) | 5 (9.3%) | 1 | |||
| AG | 40 (90.9%) | 47 (87%) | 0.47 (0.09–2.56) | 0.352 | 139.9 | ||
| GG | 2 (4.5%) | 2 (3.7%) | 0.40 (0.03–5.15) | ||||
| Dominant | AA | 2 (4.5%) | 5 (9.3%) | 1 | 0.36 | 138.1 | |
| AG + GG | 42 (95.5%) | 49 (90.7%) | 0.47 (0.09–2.53) | ||||
| AA + AG | 42 (95.5%) | 52 (96.3%) | 1 | 0.83 | 138.8 | ||
| Recessive | GG | 2 (4.5%) | 2 (3.7%) | 0.81 (0.11–5.98) | |||
| Overdominant | AA + GG | 4 (9.1%) | 7 (13%) | 1 | 0.54 | 138.5 | |
| AG | 40 (90.9%) | 47 (87%) | 0.67 (0.18–2.46) | ||||
| Log-Additive | 40 (90.9%) | 47 (87%) | 0.67 (0.18–2.46) | 0.51 | 138.4 | ||
| LAG3 rs12313899A>G | A | 0.63 | 0.62 | 1 | |||
| Alleles | G | 0.37 | 0.38 | 1.02 (0.56–182) | |||
| AA | 17 (38.6%) | 24 (44.4%) | 1 | ||||
| Codominant | AG | 21 (47.7%) | 20 (37%) | 0.67 (0.28–1.61) | 0.55 | 139.6 | |
| GG | 6 (13.6%) | 10 (18.5%) | 1.18 (0.36–3.87) | ||||
| AA | 17 (38.6%) | 24 (44.4%) | 1 | 0.56 | 138.5 | ||
| Dominant | AG + GG | 27 (61.4%) | 30 (55.6%) | 0.79 (0.35–1.77) | |||
| AA + AG | 38 (86.4%) | 44 (81.5%) | 1 | 0.51 | 138.4 | ||
| Recessive | GG | 6 (13.6%) | 10 (18.5%) | 1.44 (0.48–4.33) | |||
| Overdominant | AA + GG | 23 (52.3%) | 34 (63%) | 1 | 0.29 | 137.7 | |
| AG | 21 (47.7%) | 20 (37%) | 0.64 (0.29–1.45) | ||||
| Log-Additive | 0.98 (0.56–1.71) | 0.95 | 138.8 |
| Locus | Model | Genotype | AML Age < 29 n = 52 | AML Age ≥ 29 n = 42 | OR (95% CI) | p-Value | AIC |
|---|---|---|---|---|---|---|---|
| PDCD1 rs2227981A>G | Allele | A | 54 (52%) | 47 (51%) | 1 | 1 | |
| G | 50 (48%) | 45 (49%) | 0.96 (0.55–1.69) | ||||
| Codominant | AA | 2 (4.5%) | 5 (9.3%) | 1 | |||
| AG | 40 (90.9%) | 47 (87%) | 0.47 (0.09–2.56) | 0.352 | 139.9 | ||
| GG | 2 (4.5%) | 2 (3.7%) | 0.40 (0.03–5.15) | ||||
| Dominant | AA | 2 (4.5%) | 5 (9.3%) | 1 | 0.36 | 138.1 | |
| AG + GG | 42 (95.5%) | 49 (90.7%) | 0.47 (0.09–2.53) | ||||
| Recessive | AA + AG | 42 (95.5%) | 52 (96.3%) | 1 | 0.83 | 138.8 | |
| GG | 2 (4.5%) | 2 (3.7%) | 0.81 (0.11–5.98) | ||||
| Overdominant | AA + GG | 4 (9.1%) | 7 (13%) | 1 | 0.54 | 138.5 | |
| AG | 40 (90.9%) | 47 (87%) | 0.67 (0.18–2.46) | ||||
| Log-Additive | 40 (90.9%) | 47 (87%) | 0.67 (0.18–2.46) | 0.51 | 138.4 | ||
| LAG3 rs12313899A>G | A | 0.66 (69) | 0.59 (54) | 1 | 0.30 | ||
| Alleles | G | 0.34 (35) | 0.41 (38) | 0.72 (0.40–1.28) | |||
| Codominant | AA | 4 (7.7%) | 3 (6.5%) | 1 | 0.97 | 141.4 | |
| AG | 46 (88.5%) | 41 (89.1%) | 1.19 (0.25–5.63) | ||||
| GG | 2 (3.8%) | 2 (4.3%) | 1.33 (0.11–15.70) | ||||
| Dominant | AA | 4 (7.7%) | 3 (6.5%) | 1 | 0.82 | 139.4 | |
| AG + GG | 48 (92.3%) | 43 (93.5%) | 1.19 (0.25–5.64) | ||||
| Recessive | AA + AG | 50 (96.2%) | 44 (95.7%) | 1 | 0.9 | 139.5 | |
| GG | 2 (3.8%) | 2 (4.3%) | 1.14 (0.15–8.41) | ||||
| Overdominant | AA + GG | 6 (11.5%) | 5 (10.9%) | 1 | 0.92 | 139.5 | |
| AG | 46 (88.5%) | 41 (89.1%) | 1.07 (0.30–3.77) | ||||
| Log-Additive | --- | --- | 1.16 (0.35–3.83) | 0.8 | 139.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mansour, L.; Alqahtani, M.; Aljuaimlani, A.; Al-Tamimi, J.; Al-Harbi, N.; Alomar, S. Association of Polymorphisms in PD-1 and LAG-3 Genes with Acute Myeloid Leukemia. Medicina 2024, 60, 721. https://doi.org/10.3390/medicina60050721
Mansour L, Alqahtani M, Aljuaimlani A, Al-Tamimi J, Al-Harbi N, Alomar S. Association of Polymorphisms in PD-1 and LAG-3 Genes with Acute Myeloid Leukemia. Medicina. 2024; 60(5):721. https://doi.org/10.3390/medicina60050721
Chicago/Turabian StyleMansour, Lamjed, Mashael Alqahtani, Ali Aljuaimlani, Jameel Al-Tamimi, Nouf Al-Harbi, and Suliman Alomar. 2024. "Association of Polymorphisms in PD-1 and LAG-3 Genes with Acute Myeloid Leukemia" Medicina 60, no. 5: 721. https://doi.org/10.3390/medicina60050721
APA StyleMansour, L., Alqahtani, M., Aljuaimlani, A., Al-Tamimi, J., Al-Harbi, N., & Alomar, S. (2024). Association of Polymorphisms in PD-1 and LAG-3 Genes with Acute Myeloid Leukemia. Medicina, 60(5), 721. https://doi.org/10.3390/medicina60050721






