Abstract
Background and Objectives: An extracellular vesicle is part of a class of submicron particles derived from cells, mediating cellular crosstalk through microRNA (miRNA). MiRNA is a group of RNA molecules, each of which consists of 15–22 nucleotides and post-transcriptionally modulates gene expression. The complementary mRNAs—onto which the miRNAs hybridize—are involved in processes such as implantation, tumor suppression, proliferation, angiogenesis, and metastasis that define the entire tumor microenvironment. The endometrial biopsy is a standard technique used to recognize cellular atypia, but other non-invasive markers may reduce patient discomfort during the use of invasive methods. The present study aims to examine the distribution and the regulation of the differentially expressed miRNAs (DEMs) and EV-derived substances in women with endometrial cancer. Materials and Methods: We systematically searched the PubMed, EMBASE, Scopus, Cochrane Library, and ScienceDirect databases in April 2023, adopted the string “Endometrial Neoplasms AND Exosomes”, and followed the recommendations in the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement. We selected all the studies that included patients with endometrial cancer and that described the regulation of miRNA molecules in that context. The differences in molecule expression between patients and controls were evaluated as significant when the proteins had a fold change of ±1.5. Results: Seventeen records fulfilled the inclusion criteria: a total of 371 patients and 273 controls were analyzed. The upregulated molecules that had the widest delta between endometrial cancer patients and controls—relative expression ≥ 1 > 3 log2(ratio)—were miR-20b-5p, miR-204-5p, miR-15a-5p, and miR-320a. In particular, miR-20b-5p and miR-204-5p were extracted from both serum and endometrial specimens, whereas miR-15a-5p was only isolated from plasma, and miR-320a was only extracted from the endometrial specimens. In parallel, the most downregulated miRNA in the endometrial cancer patients compared to the healthy subjects was miR-320a, which was found in the endometrial specimens. Conclusions: Although their epigenetic regulation remains unknown, these upregulated molecules derived from EVs are feasible markers for the early detection of endometrial cancer. The modulation of these miRNA molecules should be assessed during different treatments or if recurrence develops in response to a targeted treatment modality.
1. Introduction
Endometrial cancer (EC) is a malignancy of the inner epithelial lining of the uterus, with an increasing worldwide incidence and disease-associated mortality [1]. The global incidence of EC in 2020 was 417,336, and EC is the sixth most commonly occurring female cancer [2]. Most cases occur between 65 and 75 years of age [3]. Among perimenopausal and postmenopausal women, postmenopausal bleeding (PMB) accounts for approximately two-thirds of all gynecological visits and is a common symptom of EC [4]. For early-stage disease, the main treatment is surgery. Depending on the stage of the disease and other risk factors, adjuvant radiotherapy and/or chemotherapy can be added as an adjuvant treatment option to decrease the risk of relapse [5]. Surgical staging is used for prognostication and identification of women who might benefit from adjuvant treatment, according to the current stratification of patients according to risk [6]. Total hysterectomy with bilateral salpingo-oophorectomy (BSO) is the standard of care and can be performed using an open or minimally invasive approach [7,8,9]. Moreover, lymph node status must be investigated to address adjuvant treatment [10]. EC management remains challenging, and a deeper understanding of the genetic diversity as well as the drivers of the various pathogenic states of this disease has led to the development of divergent management approaches in an effort to improve therapeutic precision in this complex malignancy [11]. Recently, greater emphasis has been given to the molecular profile of the disease, but it still neglects to totally explain the differences in the evolution of EC subtypes [12]. A substantial difference in EC management and treatment may be caused by extracellular vesicles (EVs), which are a class of submicron particles derived from cells that mediate cellular crosstalk through microRNA. MicroRNA (miRNA) is a small non-coding RNA that binds to target messenger ribonucleic acid (mRNA) to inhibit post-transcriptional gene expression and plays an essential role in regulating gene expression, the cell cycle, the timing of biological development, etc. [13]. Complementary mRNA—onto which miRNAs hybridize—are involved in the implantation, tumor suppression, apoptosis, proliferation, angiogenesis, and metastasis that define the tumor microenvironment [14]. EVs have a role as carriers of molecular pathways. In that context, miRNAs are the most investigated substances in EVs [13]. Their sources are blood samples (to be intended as serum and/or plasma), biopsy specimens, urine samples, and cellular compartments. Many studies have revealed the role of miRNAs in the biological processes of various cancers and other conditions: these molecules can help clinicians detect the existence of cancer as early as possible and screen out those with undiagnosed suspicious cases and healthy people [15,16]. On the other hand, it may reproduce the concrete risk for disease recurrence in patients, which is still a major oncological issue [17]. The wide variation in the recurrence patterns of endometrial carcinoma, to date, shows no gold standard in the timing and modalities of investigation. Although major international guidelines suggest checks every four months during the first two years of disease, the most suitable method to prevent recurrence is unknown. Moreover, this scenario is complicated by the possibility of the disease recurring locally or at a distant site because of metastasis. Deepening our knowledge of miRNA expression in endometrial carcinoma may facilitate the diagnosis of recurrences and the prediction of ways in which the disease may recur, thus enabling clinicians to choose the most appropriate follow-up method. Several studies have shown that specific miRNAs and EV-derived molecules can be used as high-precision biomarkers for EC detection [18]. In light of this, circulating molecules show great potential for use as cancer biomarkers because of their stability in peripheral serum or plasma. Because blood samples are easy to obtain and blood testing is cheap and convenient, circulating biomarkers can be used alone or in combination with other traditional screening methods for preliminary screening before invasive pathological and imaging examinations. In fact, even though endometrial biopsy is a standard option used to diagnose cellular atypia, non-invasive markers may be useful in minimizing patient discomfort compared to invasive techniques. The present study aims to examine the distribution and regulation of differentially expressed miRNAs (DEMs) and EV-derived substances in EC-affected women and the potential use of these molecules in the clinical management of EC.
2. Materials and Methods
The methods for this work were specified a priori on the basis of the recommendations in the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement [19]. The present review is categorized on the PROSPERO site for the meta-analyses with the following protocol number: ID437914.
2.1. Search Method
A systematic search was performed for articles in the PubMed database, Embase, Cochrane Library, ScienceDirect, and Scopus database in April 2023 using the string “Endometrial Neoplasms AND Exosomes”. There were no restrictions on the year of publication or the country of origin, and only articles published in English were taken into account (Figure 1).
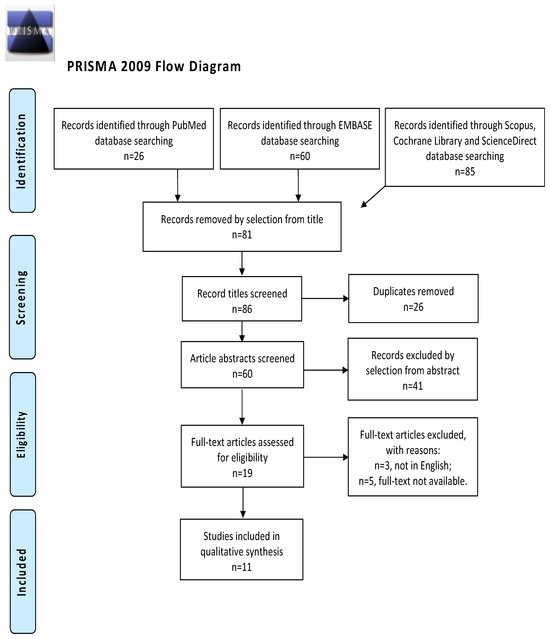
Figure 1.
Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flow diagram.
2.2. Study Selection
The study selection process was carried out by I.I. and R.M. independently. In cases of discrepancies, C.R. established the inclusion or exclusion of the article in question. The inclusion criteria were the following: (1) studies that included patients with EC and described miRNA regulation in the context of that neoplasm; (2) studies reporting the outcome of interest, such as miRNA regulation in EC, and its clinical implication; and (3) all peer-reviewed articles that were originally published. We excluded preclinical trials, unoriginal studies, animal trials, articles in languages other than English, and abstract-only publications. When possible, authors of different studies that were only published as conference abstracts were contacted through e-mail and asked to show their final results. We mentioned all selected studies and the reasons for their exclusion in the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flowchart (Figure 1). We assessed all the studies concerning potential conflicts of interest.
2.3. The Extraction and Quantification of miRNAs
Liquid biopsy is a technique used to isolate microvesicles from serum using a minimally invasive procedure [18]. The isolation, amplification, and quantification of miRNAs were carried out using various methods.
2.3.1. Extracellular Vesicles: Definition and Classification
EVs are small vesicles that can be released by cells in various contexts [20,21,22,23,24,25,26,27,28]. They are subdivided into exosomes, microvesicles, and apoptotic bodies. Exosomes (30 to 100 nm) form endosomes, microvesicles (100 to 1000 nm) come from the plasma membrane, and apoptotic bodies are 0.1 to 5 μm in diameter [29]. Exosomes derive from multivesicular bodies (MVBs), and the “endosomal sorting complex required for transport” protein complex can regulate the process of release [30]. Next, MVBs fuse with the cellular membrane, releasing the particles. Immunoelectron microscopy has been used to assess tetraspanins such as CD9, CD63, and CD81 as crucial elements in exosomes because they can be used as markers [31,32,33,34,35,36,37,38]. In parallel, apoptotic bodies show positivity for caspases 3 and 7 [39]. Recently, EVs have been classified on the basis of diameter as small (30–100 nm) or medium/large (100–200 nm) [40].
2.3.2. Extracellular Vesicles: Methods of Identification and Analysis
EV-specific proteins (e.g., tetraspanins) are detected by immunoblotting to assess EVs in samples [29]. Transmission electron microscopy (TEM), scanning EM, and cryogenic TEM can be used to recognize EV characteristics [30,31,32,33,34,35], whereas atomic force microscopy is used to evaluate the stiffness and elasticity of EVs [36,37,38]. Size distribution and polydispersity in biologic samples are analyzed using dynamic light scattering [39]. Nanoparticle tracking analysis detects dimensions and concentration through Brownian motion and scattered light or emitted fluorescence [40]. Tunable resistive pulse sensing identifies modifications in electricity as each of the EVs passes through an adjustable nanopore [41,42]. Moreover, asymmetric field-flow fractionation separates EVs according to hydrodynamic size down to the nanometer level [43]. The most feasible method for analysis is flow cytometry (FC) [44,45,46,47]. Polychromatic FC employs tracers that stain intact EVs and assesses their immunophenotypic characterization through the use of antibodies [48,49,50,51,52,53].
3. Results
3.1. Study Characteristics
After the database search, 86 studies matched the search criteria. After deleting duplicates and records with no full-text and incorrect study designs (e.g., reviews), 19 were eligible. Of those, 11 matched the inclusion criteria and were included in the final systematic review schema. Those data are summarized in Figure 1. Seven articles were prospective case-control studies evaluating miRNA expression in patients and controls; two records were prospective cohort studies; and two articles were retrospective studies. The countries where the studies were conducted, publication year, study design, number of participants, substance instilled, and procedure characteristics are included in Table 1. The publication years ranged from 2015 to 2021. A total of 371 patients with EC and 273 controls were included. The FIGO stage of disease ranged from I to IV.

Table 1.
Characteristics of included studies.
3.2. Outcomes
3.2.1. Early Diagnosis
A total of 19 miRNAs were upregulated and 6 were downregulated in women with EC. Those results are included in Table 2 and Table 3. Five of the records did not contain the FIGO stage of disease, but the six remaining studies did: two involved patients with stage I disease, two involved patients with disease in stages II and III, and two included patients with disease in stages I–IV. Particularly, Li et al. enrolled 23 women with stage I disease and showed that miR-148b was downregulated in cancer-associated fibroblasts (CAFs) [55]. In parallel, Zhou L. et al. enrolled 25 patients with stage I disease and showed that miR-765 was downregulated in endometrial specimens [64]. The studies by Záveský et al. and Zhang et al. each contained 10 enrolled patients with stage II–III and showed that miR-106b and miR-320a were downregulated in urine samples and endometrial specimens, respectively [54,60]. In studies involving patients with disease in stages I–IV, ECs were isolated from both blood samples and endometrial specimens and contained the following upregulated miRNAs: miR-381-3p, miR-143-3p, miR-195-5p, miR-20b-5p, miR-204-5p, miR-423-3p, and miR-484 [57,61].

Table 2.
Upregulated miRNA expression profile in patients with endometrial cancer.

Table 3.
Downregulated miRNA expression profile in patients with endometrial cancer.
3.2.2. Endometrial Cancer-Specific miRNAs
Among the DEMs, the miRNAs with the widest delta between patients and controls—relative expression ≥ 1 > 3 log2(ratio)—were miR-20b-5p, miR-204-5p, miR-15a-5p, and miR-320a [60,61,63]. In particular, miR-20b-5p and miR-204-5p were extracted from both serum and endometrial specimens, whereas miR-15a-5p was only isolated from plasma, and miR-320a was only extracted from the endometrial specimens [60,61,63]. The only downregulated miRNA was miR-320a in the Zhang et al. study, and it was extracted from endometrial specimens and had a 1.25 delta between patients and controls [62]. Those results are summarized in Table 2 and Table 3.
4. Discussion
Considering the functional aspects of miRNAs, these molecules participate in intercellular crosstalk in the healthy and neoplastic endometrium [14,15,65]. Evidence has highlighted the potential role of miRNAs as important markers in EC patients. Hypothetically, miRNAs may form in an intracellular environment, leading to the release of ECs from various cells [66,67,68]. Although it is very complicated to establish which specific miRNAs are involved in the genesis of EC, miRNA expression in women undergoing a hysteroscopy or blood test has been underlined because miRNA analysis in these women may identify conditions that are difficult to diagnose [69,70]. These miRNAs may represent the first building blocks in the evolution of cancer pathology. A deep understanding of the tumor microenvironment can help elucidate the actual personal risk of cancer progression to more compromised disease states and can help tailor treatments for these patients. For example, lymph node status is crucial for disease management in these patients. The ascertainment of lymph node status has been made easier through the use of the sentinel lymph node [71,72,73], but the lymph node positivity range in patients with early lesions is highly variable, and little is known about the actual mode of progression of cancer dissemination in lymph nodes [74,75]. MiRNAs might play a role in this. Another application of miRNAs is the elucidation of tumor aggressiveness. The identification of upregulated or downregulated miRNAs may help clinicians monitor the transition from pretumor lesions, such as atypical hyperplasia, to established tumors. Similarly, to date, clinical decisions are made on the basis of postoperative risk factors, making treatment personalization difficult, e.g., for patients who want to undergo fertility sparing treatment (FST). To date, the absence of myometrial infiltration and a low tumor grade are the only permissive conditions for FST, but they do not necessarily represent the actual risk for those women and are difficult to ascertain preoperatively because of the fallibility of current imaging methods and the possibility that the biopsy sample is not representative of the tumor as a whole [6]. Reassuring results were obtained even in patients with grade 2 tumors, which has already been demonstrated by our group. This may be related to an incomplete understanding the microscopic mechanisms of tumor invasiveness in which miRNAs may play a crucial role [75]. Because the data in the scientific literature are heterogeneous, it would be useful to pay attention to the DEMs with the widest delta in terms of relative expression between patients and controls to reduce false positives in the extraction process. Downregulated miR-320a had a 1.25 delta between patients and controls and was isolated in endometrial specimens [60]. Those data show how miRNA modulation is likely influenced by the site of expression. The real focus of miRNA applications should be on increasing the ability of clinicians to diagnose early-stage disease [76]. All the miRNAs and molecules summarized in Table 2 and Table 3 originate from tumor-derived exosomes via RNA sorting and delivery. Those mechanisms allow sampling from other compartments, like serum or plasma. Obviously, as a primary method, liquid biopsy mainly via the serum of patients is more feasible and cost-effective. Liquid biopsy could be part of screening programs in both fertile and postmenopausal women, even in the absence of symptoms [77,78]. However, that needs further investigation into miRNA modulation, particularly in early-stage EC. For all those reasons, the best candidates for early detection of EC should be upregulated miRNAs [61,63]. Unfortunately, those miRNAs can be overexpressed in disease stages I to IV, underlining a potential difficulty in early diagnosis [61,63,79]. In addition, miRNAs could be used during follow-ups for women undergoing different therapeutic regimens. In that context, fluctuations in miRNA expression could clarify patient responses to therapy, allowing clinicians to optimize treatment protocols [79,80]. Furthermore, miRNA expression can help clinicians predict the pattern of recurrence and personalize treatment for patients [81]. The present work examined all the existing literature on this topic. That can be considered both a strength and a weakness because there is great heterogeneity of results in the literature. Another strength is that this topic could spur further studies about the most specific miRNAs that can be used as biomarkers in the treatment of endometrial neoplasms in the clinic. Recently, great emphasis has been placed on the molecular profile of these tumors, highlighting how mismatch repair defects and microsatellite instability may have prognostic weight [82]. MiRNA identification may be a remote signature of the underlying genomic substrate, but no data about it are reported in the literature. Therefore, in our view, future studies should be aimed at a more careful sample stratification to profile the actual risk categories of patients.
Further strategies to develop methods of early EC diagnosis could involve the study of microbiota in different compartments, which is similar to studies that have been conducted in other contexts, like endometriosis [83]. For example, concerning the microbial composition of patients with gynecological disorders, conclusive results in the scientific literature detail an existing interplay between the immune system, the neuroendocrine system, and the gut, which all participate in this pathogenesis [84,85]. Changes or potential alterations in the gut microbiota that can cause dysbiosis can also modify the pathways regulating immunosurveillance [86]. Microbes in the endometrial mucosa are less frequently studied, and that includes those in the entire female reproductive tract (FRT), even though there is a lack of agreement regarding the microbial composition in the FRT; hence, there are not enough data supporting microbial composition as a feature of cancer genesis, as is the case with other gynecological diseases [86]. On the other hand, the immune response can include imbalances in the pathways of cellular epigenetic regulation and augmented levels of proinflammatory cytokines in the serum of patients. In addition, the replication and diffusion of neoplastic cells can parallel the endurance of the abovementioned contexts, resulting in chronic inflammation in the tumor microenvironment, which will lead to angiogenesis and cell adhesion over time [85]. That is facilitated by the suppression of the cell-mediated immune response and by the participation of the following molecules and interleukins (ILs): IL-6, IL-8, and vascular endothelial growth factor (VEGF). These molecules are able to establish both the primary constitution and the duration of the disease [86]. Moreover, the microbiota regulates proteolysis, for example, serotonin is derived from tryptophan, and dopamine, noradrenaline, and adrenaline are synthesized from tyrosine [87]. The decarboxylation of tryptophan aids in the synthesis of tyrosine. At the same time, the uptake of tryptophan can activate the acute phase reaction (APR), generating inflammation [88,89]. In dysbiosis, the microbial composition changes, especially in terms of microbial enzymatic activity [87,88,89], whereas a eubiotic microenvironment consists of active vitamin (such as cobalamin) biosynthesis and an intense catabolism of xenobiotics, which could be implicated in the inflammatory process [87]. The recognition of key microbiota would pave the way for new diagnostic and therapeutic strategies, including both prebiotics and probiotics, that can be implemented before considering surgical approaches. That would be crucial in helping patients avoid hysterectomy; fertility-sparing techniques can also be considered [90,91,92]. Lastly, more knowledge regarding miRNAs may help clinicians better understand the natural evolution of this disease. The risk of EC spreading via lymphatic, hematologic, or contiguous routes may influence treatment options in terms of molecular profile, adjuvant therapy, and personalizing surgery with procedures such as lymphadenectomy, omentectomy, or adnexectomy [81,92,93,94,95,96]. Moreover, we call attention to other novel molecular signatures, such as circular RNAs and proteins, that are different among EC patients and healthy control subjects [97,98]. In particular, the proteins apolipoprotein A-I (APOA1), hemoglobin subunit beta (HBB), carbonic anhydrase 1 (CA1), hemoglobin subunit delta (HBD), apolipoprotein(a) (LPA), serum amyloid A-4 protein (SAA4), platelet factor 4 variant (PF4V1), and apolipoprotein E (APOE) were upregulated in the serum of EC patients compared to controls [97]. In parallel, the circular RNAs hsa-circ-0109046 and hsa-circ-0002577 were upregulated in the serum of EC-affected women [98], whereas the circular RNA hsa-miR-200c-3p was slightly upregulated in urine [99]. The latter approach could be employed in future studies.
5. Conclusions
Many recent studies have demonstrated that intercellular crosstalk is crucially important in EC, even though data in the literature are heterogeneous. A panel of miRNAs could be feasible for the early detection and progression of disease in EC-affected patients using liquid and endometrial biopsies even though there is a lack of evidence showing that DEMs can be used as biomarkers to guide various treatment protocols. In conclusion, although pathways of epigenetic regulation are unclear, the evaluation of miRNA expression is a valuable and cost-effective option in the diagnosis of EC. Further evidence is needed to clarify miRNA regulation during the remission, relapse, and progression of EC to facilitate the planning of a targeted disease management strategy.
Author Contributions
I.I.: data curation, writing—original draft, and writing—review and editing; R.M.: writing—original draft; S.N.: validation; P.F.: data curation; M.G.V.: data curation; M.T.V.: methodology and validation; P.D.F.: validation; C.R.: methodology and conceptualization. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data supporting the findings of the study are available in the References section.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Henley, S.J.; Ward, E.M.; Scott, S.; Ma, J.; Anderson, R.N.; Firth, A.U.; Thomas, C.C.; Islami, F.; Weir, H.K.; Lewis, D.R.; et al. Annual report to the nation on the status of cancer, part I: National cancer statistics. Cancer 2020, 126, 2225–2249. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Morice, P.; Leary, A.; Creutzberg, C.; Abu-Rustum, N.; Darai, E. Endometrial cancer. Lancet 2016, 387, 1094–1108. [Google Scholar] [CrossRef] [PubMed]
- Clarke, M.A.; Long, B.J.; Del Mar Morillo, A.; Arbyn, M.; Bakkum-Gamez, J.N.; Wentzensen, N. Association of Endometrial Cancer Risk With Postmenopausal Bleeding in Women: A Systematic Review and Meta-analysis. JAMA Intern. Med. 2018, 178, 1210–1222. [Google Scholar] [CrossRef] [PubMed]
- Brooks, R.A.; Fleming, G.F.; Lastra, R.R.; Lee, N.K.; Moroney, J.W.; Son, C.H.; Tatebe, K.; Veneris, J.L. Current recommendations and recent progress in endometrial cancer. CA Cancer J. Clin. 2019, 69, 258–279. [Google Scholar] [CrossRef] [PubMed]
- Concin, N.; Matias-Guiu, X.; Vergote, I.; Cibula, D.; Mirza, M.R.; Marnitz, S.; Ledermann, J.; Bosse, T.; Chargari, C.; Fagotti, A.; et al. ESGO/ESTRO/ESP guidelines for the management of patients with endometrial carcinoma. Int. J. Gynecol. Cancer 2021, 31, 12–39. [Google Scholar] [CrossRef]
- Gueli Alletti, S.; Restaino, S.; Finelli, A.; Ronsini, C.; Lucidi, A.; Scambia, G.; Fanfani, F. Step by Step Total Laparoscopic Hysterectomy with Uterine Arteries Ligation at the Origin. J. Minim. Invasive Gynecol. 2020, 27, 22–23. [Google Scholar] [CrossRef]
- Gaia, G.; Holloway, R.W.; Santoro, L.; Ahmad, S.; Di Silverio, E.; Spinillo, A. Robotic-assisted hysterectomy for endometrial cancer compared with traditional laparoscopic and laparotomy approaches: A systematic review. Obstet. Gynecol. 2010, 116, 1422–1431. [Google Scholar] [CrossRef]
- Walker, J.L.; Piedmonte, M.R.; Spirtos, N.M.; Eisenkop, S.M.; Schlaerth, J.B.; Mannel, R.S.; Spiegel, G.; Barakat, R.; Pearl, M.L.; Sharma, S.K. Laparoscopy compared with laparotomy for comprehensive surgical staging of uterine cancer: Gynecologic Oncology Group Study LAP2. J. Clin. Oncol. 2009, 27, 5331–5336. [Google Scholar] [CrossRef]
- Restaino, S.; Ronsini, C.; Finelli, A.; Perrone, E.; Scambia, G.; Fanfani, F. Role of blue dye for sentinel lymph node detection in early endometrial cancer. Gynecol. Surg. 2017, 14, 23. [Google Scholar] [CrossRef]
- Makker, V.; MacKay, H.; Ray-Coquard, I.; Levine, D.A.; Westin, S.N.; Aoki, D.; Oaknin, A. Endometrial cancer. Nat. Rev. Dis. Primers 2021, 7, 88. [Google Scholar] [CrossRef]
- Kommoss, S.; McConechy, M.K.; Kommoss, F.; Leung, S.; Bunz, A.; Magrill, J.; Britton, H.; Grevenkamp, F.; Karnezis, A.; Yang, W.; et al. Final validation of the ProMisE molecular classifier for endometrial carcinoma in a large population-based case series. Ann. Oncol. 2018, 29, 1180–1188. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Liang, J.; Wang, S.; Wang, Z. Role of microRNAs in embryo implantation. Reprod. Biol. Endocrinol. 2017, 15, 90. [Google Scholar] [CrossRef]
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Pathol. 2014, 9, 287–314. [Google Scholar] [CrossRef]
- Ronsini, C.; Fumiento, P.; Iavarone, I.; Greco, P.F.; Cobellis, L.; De Franciscis, P. Liquid Biopsy in Endometriosis: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 6116. [Google Scholar] [CrossRef] [PubMed]
- Legge, F.; Restaino, S.; Leone, L.; Carone, V.; Ronsini, C.; Di Fiore, G.L.M.; Pasciuto, T.; Pelligra, S.; Ciccarone, F.; Scambia, G.; et al. Clinical outcome of recurrent endometrial cancer: Analysis of post-relapse survival by pattern of recurrence and secondary treatment. Int. J. Gynecol. Cancer 2020, 30, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Ritter, A.; Hirschfeld, M.; Berner, K.; Jaeger, M.; Grundner-Culemann, F.; Schlosser, P.; Asberger, J.; Weiss, D.; Noethling, C.; Mayer, S.; et al. Discovery of potential serum and urine-based microRNA as minimally-invasive biomarkers for breast and gynecological cancer. Cancer Biomark. 2020, 27, 225–242. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed]
- Franzago, M.; Lanuti, P.; Fraticelli, F.; Marchioni, M.; Buca, D.; Di Nicola, M.; Liberati, M.; Miscia, S.; Stuppia, L.; Vitacolonna, E. Biological insight into the extracellular vesicles in women with and without gestational diabetes. J. Endocrinol. Investig. 2021, 44, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Buca, D.; Bologna, G.; D’Amico, A.; Cugini, S.; Musca, F.; Febbo, M.; D’arcangelo, D.; Buca, D.; Simeone, P.; Liberati, M.; et al. Extracellular Vesicles in Feto-Maternal Crosstalk and Pregnancy Disorders. Int. J. Mol. Sci. 2020, 21, 2120. [Google Scholar] [CrossRef] [PubMed]
- Goossens, N.; Nakagawa, S.; Sun, X.; Hoshida, Y. Cancer biomarker discovery and validation. Transl. Cancer Res. 2015, 4, 256–269. [Google Scholar] [CrossRef] [PubMed]
- Di Tomo, P.; Lanuti, P.; Di Pietro, N.; Baldassarre, M.P.A.; Marchisio, M.; Pandolfi, A.; Consoli, A.; Formoso, G. Liraglutide mitigates TNF-α induced pro-atherogenic changes and microvesicle release in HUVEC from diabetic women. Diabetes Metab. Res. Rev. 2017, 33, e2925. [Google Scholar] [CrossRef] [PubMed]
- Pieragostino, D.; Lanuti, P.; Cicalini, I.; Cufaro, M.C.; Ciccocioppo, F.; Ronci, M.; Simeone, P.; Onofrj, M.; van der Pol, E.; Fontana, A.; et al. Proteomics characterization of extracellular vesicles sorted by flow cytometry reveals a disease-specific molecular cross-talk from cerebrospinal fluid and tears in multiple sclerosis. J. Proteomics. 2019, 204, 103403. [Google Scholar] [CrossRef]
- Grande, R.; Dovizio, M.; Marcone, S.; Szklanna, P.B.; Bruno, A.; Ebhardt, H.A.; Cassidy, H.; Ní Áinle, F.; Caprodossi, A.; Lanuti, P.; et al. Platelet-Derived Microparticles from Obese Individuals: Characterization of Number, Size, Proteomics, and Crosstalk with Cancer and Endothelial Cells. Front. Pharmacol. 2019, 10, 7. [Google Scholar] [CrossRef] [PubMed]
- Pieragostino, D.; Cicalini, I.; Lanuti, P.; Ercolino, E.; Di Ioia, M.; Zucchelli, M.; Zappacosta, R.; Miscia, S.; Marchisio, M.; Sacchetta, P.; et al. Enhanced release of acid sphingomyelinase-enriched exosomes generates a lipidomics signature in CSF of Multiple Sclerosis patients. Sci. Rep. 2018, 8, 3071. [Google Scholar] [CrossRef]
- Codagnone, M.; Recchiuti, A.; Lanuti, P.; Pierdomenico, A.M.; Cianci, E.; Patruno, S.; Mari, V.C.; Simiele, F.; Di Tomo, P.; Pandolfi, A.; et al. Lipoxin A4 stimulates endothelial miR-126-5p expression and its transfer via microvesicles. FASEB J. 2017, 31, 1856–1866. [Google Scholar] [CrossRef]
- Zhang, W.; Lu, S.; Pu, D.; Zhang, H.; Yang, L.; Zeng, P.; Su, F.; Chen, Z.; Guo, M.; Gu, Y.; et al. Detection of fetal trisomy and single gene disease by massively parallel sequencing of extracellular vesicle DNA in maternal plasma: A proof-of-concept validation. BMC Med. Genom. 2019, 12, 151. [Google Scholar] [CrossRef]
- Cufaro, M.C.; Pieragostino, D.; Lanuti, P.; Rossi, C.; Cicalini, I.; Federici, L.; De Laurenzi, V.; Del Boccio, P. Extracellular Vesicles and Their Potential Use in Monitoring Cancer Progression and Therapy: The Contribution of Proteomics. J. Oncol. 2019, 2019, 1639854. [Google Scholar] [CrossRef]
- Andreu, Z.; Yáñez-Mó, M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014, 5, 442. [Google Scholar] [CrossRef]
- Krause, M.; Rak-Raszewska, A.; Naillat, F.; Saarela, U.; Schmidt, C.; Ronkainen, V.; Bart, G.; Ylä-Herttuala, S.; Vainio, S.J. Exosomes as secondary inductive signals involved in kidney organogenesis. J. Extracell. Vesicles 2018, 7, 1422675. [Google Scholar] [CrossRef] [PubMed]
- Helwa, I.; Cai, J.; Drewry, M.D.; Zimmerman, A.; Dinkins, M.B.; Khaled, M.L.; Seremwe, M.; Dismuke, W.M.; Bieberich, E.; Stamer, W.D.; et al. A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents. PLoS ONE 2017, 12, e0170628. [Google Scholar] [CrossRef]
- Chairoungdua, A.; Smith, D.L.; Pochard, P.; Hull, M.; Caplan, M.J. Exosome release of β-catenin: A novel mechanism that antagonizes Wnt signaling. J. Cell. Biol. 2010, 190, 1079–1091. [Google Scholar] [CrossRef] [PubMed]
- Buschow, S.I.; Nolte-’t Hoen, E.N.; van Niel, G.; Pols, M.S.; ten Broeke, T.T.; Lauwen, M.; Ossendorp, F.; Melief, C.J.M.; Raposo, G.; Wubbolts, R.; et al. MHC II in dendritic cells is targeted to lysosomes or T cell-induced exosomes via distinct multivesicular body pathways. Traffic 2009, 10, 1528–1542. [Google Scholar] [CrossRef] [PubMed]
- Escola, J.M.; Kleijmeer, M.J.; Stoorvogel, W.; Griffith, J.M.; Yoshie, O.; Geuze, H.J. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 1998, 273, 20121–20127. [Google Scholar] [CrossRef] [PubMed]
- Nazarenko, I.; Rana, S.; Baumann, A.; McAlear, J.; Hellwig, A.; Trendelenburg, M.; Lochnit, G.; Preissner, K.T.; Zöller, M. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 2010, 70, 1668–1678. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Baixauli, F.; Gutiérrez-Vázquez, C.; Sánchez-Madrid, F.; Mittelbrunn, M. Sorting it out: Regulation of exosome loading. Semin. Cancer Biol. 2014, 28, 3–13. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Poon, I.K.H.; Parkes, M.A.F.; Jiang, L.; Atkin-Smith, G.K.; Tixeira, R.; Gregory, C.D.; Ozkocak, D.C.; Rutter, S.F.; Caruso, S.; Santavanond, J.P.; et al. Moving beyond size and phosphatidylserine exposure: Evidence for a diversity of apoptotic cell-derived extracellular vesicles in vitro. J. Extracell. Vesicles 2019, 8, 1608786. [Google Scholar] [CrossRef]
- Ciardiello, C.; Leone, A.; Lanuti, P.; Roca, M.S.; Moccia, T.; Minciacchi, V.R.; Minopoli, M.; Gigantino, V.; De Cecio, R.; Rippa, M.; et al. Large oncosomes overexpressing integrin alpha-V promote prostate cancer adhesion and invasion via AKT activation. J. Exp. Clin. Cancer Res. 2019, 38, 317. [Google Scholar] [CrossRef]
- Hartjes, T.A.; Mytnyk, S.; Jenster, G.W.; van Steijn, V.; van Royen, M.E. Extracellular Vesicle Quantification and Characterization: Common Methods and Emerging Approaches. Bioengineering 2019, 6, 7. [Google Scholar] [CrossRef]
- Park, Y.H.; Shin, H.W.; Jung, A.R.; Kwon, O.S.; Choi, Y.-J.; Park, J.; Lee, J.Y. Author Correction: Prostate-specific extracellular vesicles as a novel biomarker in human prostate cancer. Sci. Rep. 2019, 9, 6051. [Google Scholar] [CrossRef]
- Linares, R.; Tan, S.; Gounou, C.; Brisson, A.R. Imaging and Quantification of Extracellular Vesicles by Transmission Electron Microscopy. Methods Mol. Biol. 2017, 1545, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, V.; Ludwig, A.K.; Hornung, S.; Rotan, O.; Horn, P.A.; Epple, M.; Giebel, B. Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids Surf. B Biointerfaces 2011, 87, 146–150. [Google Scholar] [CrossRef]
- Casado, S.; Lobo, M.d.V.T.; Paíno, C. Dynamics of plasma membrane surface related to the release of extracellular vesicles by mesenchymal stem cells in culture. Sci. Rep. 2017, 7, 6767. [Google Scholar] [CrossRef]
- Nanou, A.; Crespo, M.; Flohr, P.; De Bono, J.S.; Terstappen, L.W.M.M. Scanning Electron Microscopy of Circulating Tumor Cells and Tumor-Derived Extracellular Vesicles. Cancers 2018, 10, 416. [Google Scholar] [CrossRef]
- Biggs, C.N.; Siddiqui, K.M.; Al-Zahrani, A.A.; Pardhan, S.; Brett, S.I.; Guo, Q.Q.; Yang, J.; Wolf, P.; Power, N.E.; Durfee, P.N.; et al. Prostate extracellular vesicles in patient plasma as a liquid biopsy platform for prostate cancer using nanoscale flow cytometry. Oncotarget 2016, 7, 8839–8849. [Google Scholar] [CrossRef] [PubMed]
- Hardij, J.; Cecchet, F.; Berquand, A.; Gheldof, D.; Chatelain, C.; Mullier, F.; Chatelain, B.; Dogné, J. Characterisation of tissue factor-bearing extracellular vesicles with AFM: Comparison of air-tapping-mode AFM and liquid Peak Force AFM. J. Extracell. Vesicles 2013, 2, 21045. [Google Scholar] [CrossRef] [PubMed]
- Vorselen, D.; Marchetti, M.; López-Iglesias, C.; Peters, P.J.; Roos, W.H.; Wuite, G.J.L. Multilamellar nanovesicles show distinct mechanical properties depending on their degree of lamellarity. Nanoscale 2018, 10, 5318–5324. [Google Scholar] [CrossRef]
- Gardiner, C.; Di Vizio, D.; Sahoo, S.; Théry, C.; Witwer, K.W.; Wauben, M.; Hill, A.F. Techniques used for the isolation and characterization of extracellular vesicles: Results of a worldwide survey. J. Extracell. Vesicles 2016, 5, 32945. [Google Scholar] [CrossRef]
- Poncelet, P.; Robert, S.; Bailly, N.; Garnache-Ottou, F.; Bouriche, T.; Devalet, B.; Segatchian, J.H.; Saas, P.; Mullier, F. Tips and tricks for flow cytometry-based analysis and counting of microparticles. Transfus. Apher. Sci. 2015, 53, 110–126. [Google Scholar] [CrossRef]
- Chandler, W.L. Measurement of microvesicle levels in human blood using flow cytometry. Cytom. B Clin. Cytom. 2016, 90, 326–336. [Google Scholar] [CrossRef]
- Deshmukh, A.; Kim, G.; Burke, M.; Anyanwu, E.; Jeevanandam, V.; Uriel, N.; Tung, R.; Ozcan, C. Atrial Arrhythmias and Electroanatomical Remodeling in Patients with Left Ventricular Assist Devices. J. Am. Heart Assoc. 2017, 6, e005340. [Google Scholar] [CrossRef]
- Záveský, L.; Jandáková, E.; Turyna, R.; Langmeierová, L.; Weinberger, V.; Drábková, L.Z.; Hůlková, M.; Hořínek, A.; Dušková, D.; Feyereisl, J.; et al. Evaluation of Cell-Free Urine microRNAs Expression for the Use in Diagnosis of Ovarian and Endometrial Cancers. A Pilot Study. Pathol. Oncol. Res. 2015, 21, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Li, B.L.; Lu, W.; Qu, J.J.; Ye, L.; Du, G.Q.; Wan, X.P. Loss of exosomal miR-148b from cancer-associated fibroblasts promotes endometrial cancer cell invasion and cancer metastasis. J. Cell. Physiol. 2019, 234, 2943–2953. [Google Scholar] [CrossRef]
- Roman-Canal, B.; Moiola, C.P.; Gatius, S.; Bonnin, S.; Ruiz-Miró, M.; González, E.; González-Tallada, X.; Llordella, I.; Hernández, I.; Porcel, J.M.; et al. EV-Associated miRNAs from Peritoneal Lavage are a Source of Biomarkers in Endometrial Cancer. Cancers 2019, 11, 839. [Google Scholar] [CrossRef]
- Jia, J.; Guo, S.; Zhang, D.; Tian, X.; Xie, X. Exosomal-lncRNA DLEU1 Accelerates the Proliferation, Migration, and Invasion of Endometrial Carcinoma Cells by Regulating microRNA-E2F3. OncoTargets Ther. 2020, 13, 8651–8663. [Google Scholar] [CrossRef] [PubMed]
- Jing, L.; Hua, X.; Yuanna, D.; Rukun, Z.; Junjun, M. Exosomal miR-499a-5p Inhibits Endometrial Cancer Growth and Metastasis via Targeting VAV3. Cancer Manag. Res. 2020, 12, 13541–13552. [Google Scholar] [CrossRef]
- Shi, S.; Tan, Q.; Feng, F.; Huang, H.; Liang, J.; Cao, D.; Wang, Z. Identification of core genes in the progression of endometrial cancer and cancer cell-derived exosomes by an integrative analysis. Sci. Rep. 2020, 10, 9862. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Wang, Y.; Liu, H.; Shen, W. Extracellular vesicle encapsulated microRNA-320a inhibits endometrial cancer by suppression of the HIF1α/VEGFA axis. Exp. Cell. Res. 2020, 394, 112113. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.T.; Zhou, Z.Y.; Luo, Y.L.; Luo, Q.; Chen, S.-B.; Zhao, J.-C.; Chen, Q.-R. Exosomal lncRNA NEAT1 from cancer-associated fibroblasts facilitates endometrial cancer progression via miR-26a/b-5p-mediated STAT3/YKL-40 signaling pathway. Neoplasia 2021, 23, 692–703. [Google Scholar] [CrossRef]
- Gu, X.; Shi, Y.; Dong, M.; Jiang, L.; Yang, J.; Liu, Z. Exosomal transfer of tumor-associated macrophage-derived hsa_circ_0001610 reduces radiosensitivity in endometrial cancer. Cell. Death Dis. 2021, 12, 818. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.J.; Zhang, J.; Xie, F.; Wu, J.-N.; Ye, J.-F.; Wang, J.; Wu, K.; Li, M.-Q. CD45RO-CD8+ T cell-derived exosomes restrict estrogen-driven endometrial cancer development via the ERβ/miR-765/PLP2/Notch axis. Theranostics 2021, 11, 5330–5345. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, W.; Wang, F.; Yang, S.; Hu, J.; Lu, B.; Pan, Z.; Ma, Y.; Zheng, M.; Zhou, L.; et al. Plasma-derived exosomal miR-15a-5p as a promising diagnostic biomarker for early detection of endometrial carcinoma. Mol. Cancer. 2021, 20, 57. [Google Scholar] [CrossRef] [PubMed]
- Hiroki, E.; Akahira, J.; Suzuki, F.; Nagase, S.; Ito, K.; Suzuki, T.; Sasano, H.; Yaegashi, N. Changes in microRNA expression levels correlate with clinicopathological features and prognoses in endometrial serous adenocarcinomas. Cancer Sci. 2010, 101, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Misso, G.; Di Martino, M.T.; De Rosa, G.; Farooqi, A.A.; Lombardi, A.; Campani, V.; Zarone, M.R.; Gullà, A.; Tagliaferri, P.; Tassone, P.; et al. Mir-34: A new weapon against cancer? Mol. Ther. Nucleic Acids 2014, 3, e194. [Google Scholar] [CrossRef] [PubMed]
- Korpal, M.; Kang, Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 2008, 5, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Garcia, E.; Lesur, A.; Devis, L.; Cabrera, S.; Matias-Guiu, X.; Hirschfeld, M.; Asberger, J.; van Oostrum, J.; Cal, M.d.L.C.d.; Gómez-Tato, A.; et al. Targeted Proteomics Identifies Proteomic Signatures in Liquid Biopsies of the Endometrium to Diagnose Endometrial Cancer and Assist in the Prediction of the Optimal Surgical Treatment. Clin. Cancer Res. 2017, 23, 6458–6467. [Google Scholar] [CrossRef] [PubMed]
- Banno, K.; Yanokura, M.; Kisu, I.; Yamagami, W.; Susumu, N.; Aoki, D. MicroRNAs in endometrial cancer. Int. J. Clin. Oncol. 2013, 18, 186–192. [Google Scholar] [CrossRef]
- Montagnana, M.; Benati, M.; Danese, E.; Giudici, S.; Perfranceschi, M.; Ruzzenenete, O.; Salvagno, G.L.; Bassi, A.; Gelati, M.; Paviati, E.; et al. Aberrant MicroRNA Expression in Patients with Endometrial Cancer. Int. J. Gynecol. Cancer 2017, 27, 459–466. [Google Scholar] [CrossRef]
- Rossi, E.C.; Kowalski, L.D.; Scalici, J.; Cantrell, L.; Schuler, K.; Hanna, R.K.; Method, M.; Ade, M.; Ivanova, A.; Boggess, J.F. A comparison of sentinel lymph node biopsy to lymphadenectomy for endometrial cancer staging (FIRES trial): A multicentre, prospective, cohort study. Lancet Oncol. 2017, 18, 384–392. [Google Scholar] [CrossRef]
- Fanfani, F.; Monterossi, G.; Di Meo, M.L.; La Fera, E.; Dell’Orto, F.; Gioè, A.; Lamanna, M.; Ferrari, D.; De Ponti, E.; Perego, P.; et al. Standard ultra-staging compared to one-step nucleic acid amplification for the detection of sentinel lymph node metastasis in endometrial cancer patients: A retrospective cohort comparison. Int. J. Gynecol. Cancer 2020, 30, 372–377. [Google Scholar] [CrossRef]
- Rozenholc, A.; Samouelian, V.; Warkus, T.; Gauthier, P.; Provencher, D.; Sauthier, P.; Gauthier, F.; Drakopoulos, P.; Cormier, B. Green versus blue: Randomized controlled trial comparing indocyanine green with methylene blue for sentinel lymph node detection in endometrial cancer. Gynecol. Oncol. 2019, 153, 500–504. [Google Scholar] [CrossRef] [PubMed]
- Restaino, S.; Buda, A.; Puppo, A.; Capozzi, V.A.; Sozzi, G.; Casarin, J.; Gallitelli, V.; Murgia, F.; Vizzielli, G.; Baroni, A.; et al. Anatomical distribution of sentinel lymph nodes in patients with endometrial cancer: A multicenter study. Int. J. Gynecol. Cancer 2022, 32, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Ronsini, C.; Mosca, L.; Iavarone, I.; Nicoletti, R.; Vinci, D.; Carotenuto, R.M.; Pasanisi, F.; Solazzo, M.C.; De Franciscis, P.; Torella, M.; et al. Oncological outcomes in fertility-sparing treatment in stage IA-G2 endometrial cancer. Front. Oncol. 2022, 12, 965029. [Google Scholar] [CrossRef] [PubMed]
- Lucidi, A.; Buca, D.; Ronsini, C.; Tinari, S.; Bologna, G.; Buca, D.; Leombroni, M.; Liberati, M.; D’antonio, F.; Scambia, G.; et al. Role of Extracellular Vesicles in Epithelial Ovarian Cancer: A Systematic Review. Int. J. Mol. Sci. 2020, 21, 8762. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.; Torres, K.; Pesci, A.; Ceccaroni, M.; Paszkowski, T.; Cassandrini, P.; Zamboni, G.; Maciejewski, R. Deregulation of miR-100, miR-99a and miR-199b in tissues and plasma coexists with increased expression of mTOR kinase in endometrioid endometrial carcinoma. BMC Cancer 2012, 12, 369. [Google Scholar] [CrossRef] [PubMed]
- Muinelo-Romay, L.; Casas-Arozamena, C.; Abal, M. Liquid Biopsy in Endometrial Cancer: New Opportunities for Personalized Oncology. Int. J. Mol. Sci. 2018, 19, 2311. [Google Scholar] [CrossRef]
- Vasilatou, D.; Sioulas, V.D.; Pappa, V.; Papageorgiou, S.G.; Vlahos, N.F. The role of miRNAs in endometrial cancer. Epigenomics 2015, 7, 951–959. [Google Scholar] [CrossRef]
- Wilczynski, M.; Danielska, J.; Dzieniecka, M.; Szymanska, B.; Wojciechowski, M.; Malinowski, A. Prognostic and Clinical Significance of miRNA-205 in Endometrioid Endometrial Cancer. PLoS ONE 2016, 11, e0164687. [Google Scholar] [CrossRef]
- Ronsini, C.; Iavarone, I.; Reino, A.; Vastarella, M.G.; De Franciscis, P.; Sangiovanni, A.; Della Corte, L. Radiotherapy Chemotherapy Features in the Treatment for Locoregional Recurrence of Endometrial Cancer: A Systematic Review. J. Pers. Med. 2023, 13, 886. [Google Scholar] [CrossRef]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Yang, W.; Lum, A.; Senz, J.; Boyd, N.; Pike, J.; Anglesio, M.; Kwon, J.S.; et al. Confirmation of ProMisE: A simple, genomics-based clinical classifier for endometrial cancer. Cancer 2017, 123, 802–813. [Google Scholar] [CrossRef]
- Iavarone, I.; Greco, P.F.; La Verde, M.; Morlando, M.; Torella, M.; de Franciscis, P.; Ronsini, C. Correlations between Gut Microbial Composition, Pathophysiological and Surgical Aspects in Endometriosis: A Review of the Literature. Medicina 2023, 59, 347. [Google Scholar] [CrossRef]
- Ser, H.; You Jing Wong, J.; Letchumanan, V.; Law, J.W.; Tan, L.T.; Lee, L.H. Moving beyond the gastrointestinal tract: The involvement of gut microbiome in endometriosis. Gut 2021, 70, A46–A47. [Google Scholar] [CrossRef]
- Jiang, I.; Yong, P.J.; Allaire, C.; Bedaiwy, M.A. Intricate Connections between the Microbiota and Endometriosis. Int. J. Mol. Sci. 2021, 22, 5644. [Google Scholar] [CrossRef] [PubMed]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Aarts, J.W.; Nieboer, T.E.; Johnson, N.; Tavender, E.; Garry, R.; Mol, B.W.J.; Kluivers, K.B. Surgical approach to hysterectomy for benign gynaecological disease. Cochrane Database Syst Rev. 2015, 2015, CD003677. [Google Scholar] [CrossRef] [PubMed]
- Montiel-Castro, A.J.; González-Cervantes, R.M.; Bravo-Ruiseco, G.; Pacheco-López, G. The microbiota-gut-brain axis: Neurobehavioral correlates, health and sociality. Front. Integr. Neurosci. 2013, 7, 70. [Google Scholar] [CrossRef] [PubMed]
- Lago, V.; Marina, T.; Laseca Modrego, M.; Gil-Ibañez, B.; Rodriguez, J.R.; Domingo, J.; Minig, L.; Padilla-Iserte, P.; Sánchez, O.A.; Ferichola, M.S.; et al. Fertility sparing treatment in patients with endometrial cancer (FERT-ENC): A multicentric retrospective study from the Spanish Investigational Network Gynecologic Oncology Group (SPAIN-GOG). Arch Gynecol. Obstet. 2022, 306, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Ronsini, C.; Pasanisi, F.; Molitierno, R.; Iavarone, I.; Vastarella, M.G.; De Franciscis, P.; Conte, C. Minimally Invasive Staging of Early-Stage Epithelial Ovarian Cancer versus Open Surgery in Terms of Feasibility and Safety: A Systematic Review and Meta-Analysis. J. Clin. Med. 2023, 12, 3831. [Google Scholar] [CrossRef] [PubMed]
- Restaino, S.; Ronsini, C.; Finelli, A.; Santarelli, A.; Scambia, G.; Fanfani, F. Laparoscopic Approach for Shull Repair of Pelvic Floor Defects. J. Minim. Invasive Gynecol. 2018, 25, 954. [Google Scholar] [CrossRef]
- Riemma, G.; Pasanisi, F.; Reino, A.; Solazzo, M.C.; Ronsini, C. Robotic Single-Site Hysterectomy in Gynecologic Benign Pathology: A Systematic Review of the Literature. Medicina 2023, 59, 411. [Google Scholar] [CrossRef]
- Ronsini, C.; Reino, A.; Molitierno, R.; Vastarella, M.G.; La Mantia, E.; De Franciscis, P. Critical Overview of Serous Endometrial Intraepithelial Cancer Treatment: Systematic Review of Adjuvant Options. Life 2023, 13, 1429. [Google Scholar] [CrossRef] [PubMed]
- Ronsini, C.; Foresta, A.; Giudice, M.; Reino, A.; La Verde, M.; della Corte, L.; Bifulco, G.; de Franciscis, P.; Cianci, S.; Capozzi, V.A. Is Adnexectomy Mandatory at the Time of Hysterectomy for Uterine Sarcomas? A Systematic Review and Meta-Analysis. Medicina 2022, 58, 1140. [Google Scholar] [CrossRef] [PubMed]
- Vietri, M.T.; D’Elia, G.; Benincasa, G.; Ferraro, G.; Caliendo, G.; Nicoletti, G.F.; Napoli, C. DNA methylation and breast cancer: A way forward (Review). Int. J. Oncol. 2021, 59, 98. [Google Scholar] [CrossRef] [PubMed]
- Vietri, M.T.; D’Elia, G.; Caliendo, G.; Casamassimi, A.; Federico, A.; Passariello, L.; Cioffi, M.; Molinari, A.M. Prevalence of mutations in BRCA and MMR genes in patients affected with hereditary endometrial cancer. Med. Oncol. 2021, 38, 13. [Google Scholar] [CrossRef]
- Sommella, E.; Capaci, V.; Aloisio, M.; Salviati, E.; Campiglia, P.; Molinario, G.; Licastro, D.; Di Lorenzo, G.; Romano, F.; Ricci, G.; et al. A Label-Free Proteomic Approach for the Identification of Biomarkers in the Exosome of Endometrial Cancer Serum. Cancers 2022, 14, 6262. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Gong, Z.; Shen, Y.; Fang, Y.; Zhong, S. Circular RNA expression in extracellular vesicles isolated from serum of patients with endometrial cancer. Epigenomics 2018, 10, 187–197. [Google Scholar] [CrossRef]
- Srivastava, A.; Moxley, K.; Ruskin, R.; Dhanasekaran, D.N.; Zhao, Y.D.; Ramesh, R. A Non-invasive Liquid Biopsy Screening of Urine-Derived Exosomes for miRNAs as Biomarkers in Endometrial Cancer Patients. AAPS J. 2018, 20, 82. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).