An Ultra-Fast Green UHPLC-MS/MS Method for Assessing the In Vitro Metabolic Stability of Dovitinib: In Silico Study for Absorption, Distribution, Metabolism, Excretion, Metabolic Lability, and DEREK Alerts
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Instruments
2.3. Assessment of DVB Metabolic Lability
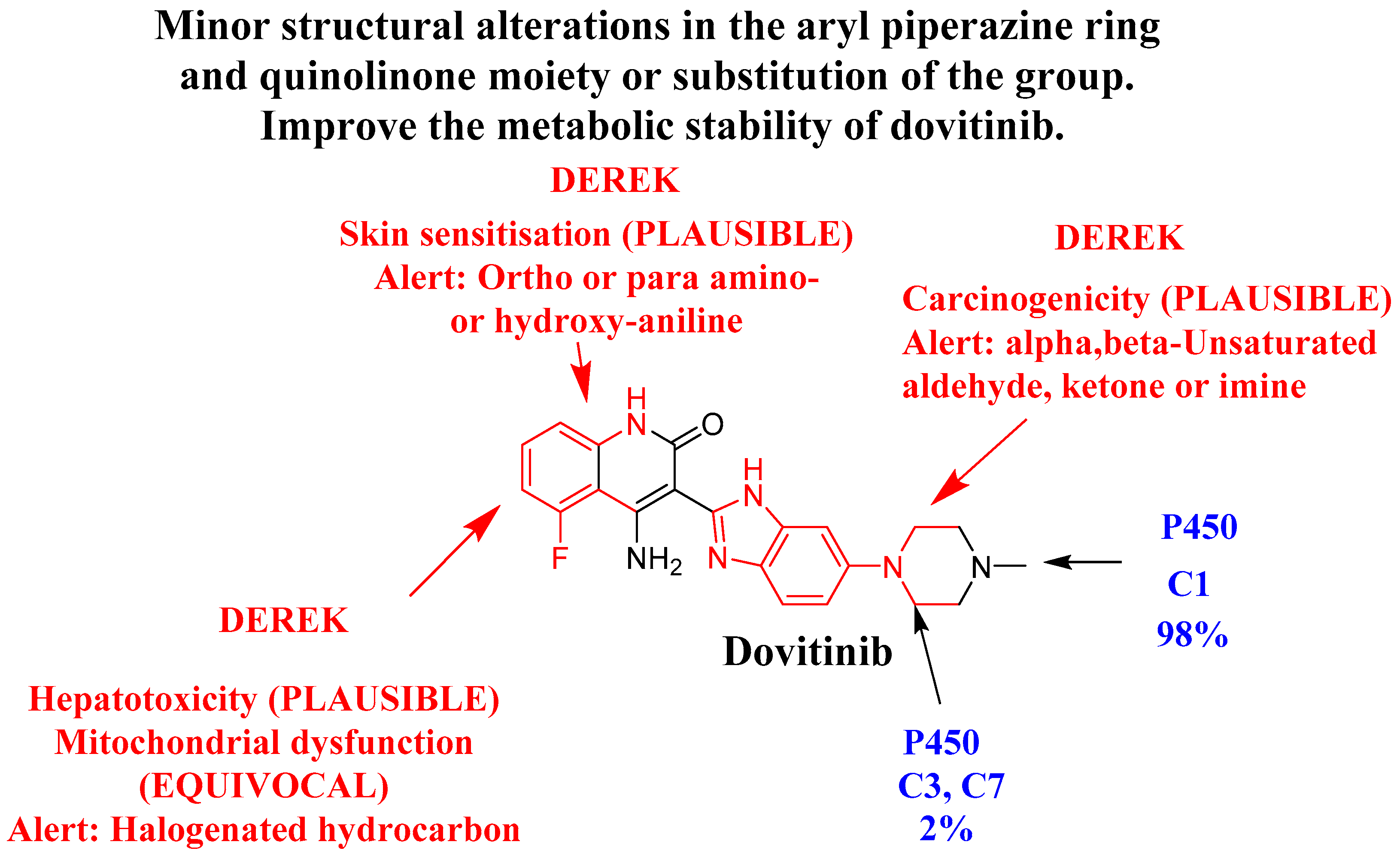
2.4. Screening of the DVB Toxicity Alerts Using DEREK In Silico Software
2.5. DVB In Silico ADME Profile
2.6. UHPLC-MS/MS Analytical Features
2.7. DVB and EFB Working Dilutions
2.8. DVB Calibration Levels
2.9. The Extraction Recovery of DVB and EFB
2.10. Validation Features of the Established UHPLC-MS/MS Approach
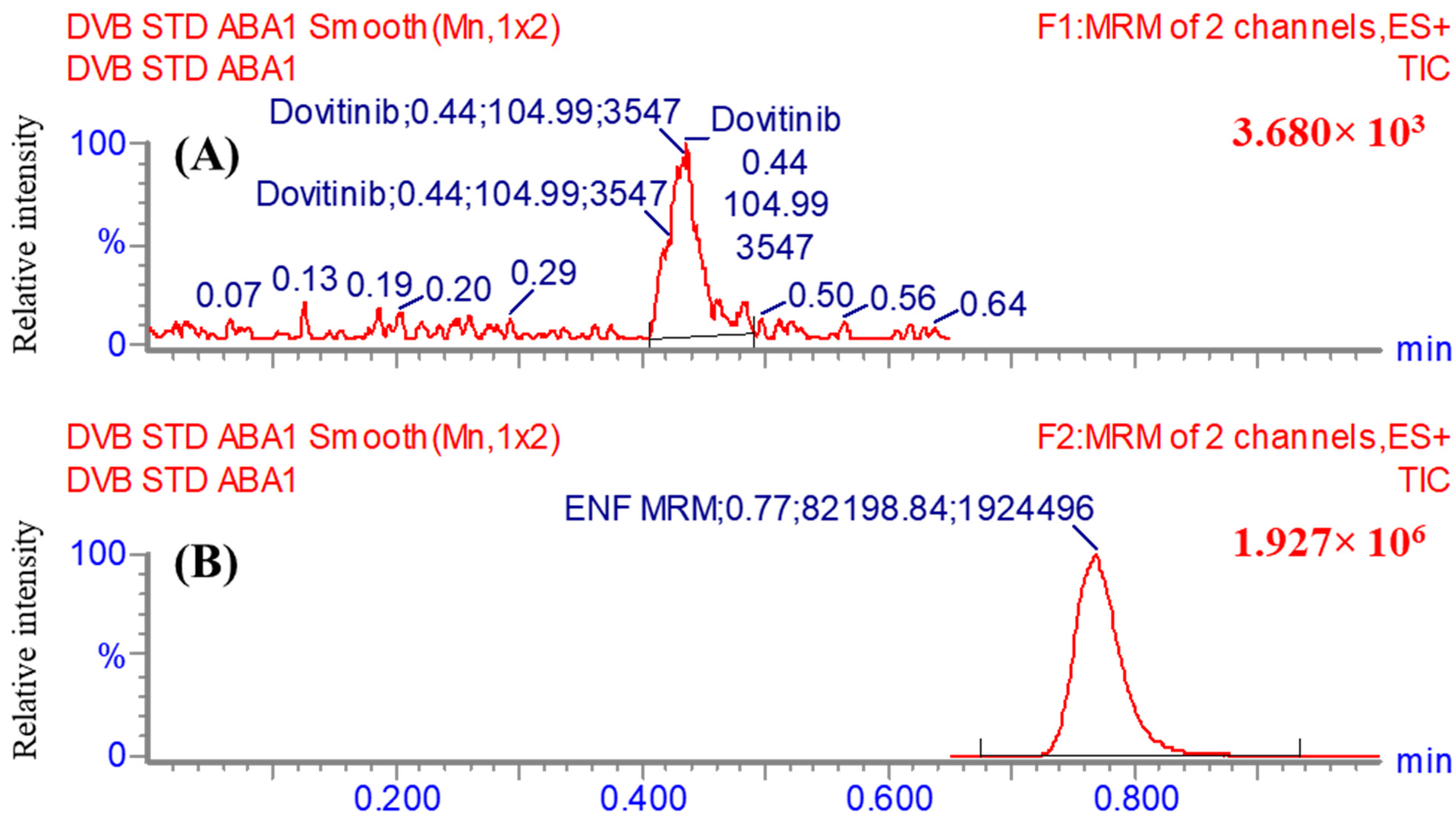
2.10.1. Specificity
2.10.2. Sensitivity and Linearity
2.10.3. Accuracy and Precision
2.10.4. Extraction Recovery and Matrix Effect
2.10.5. Stability
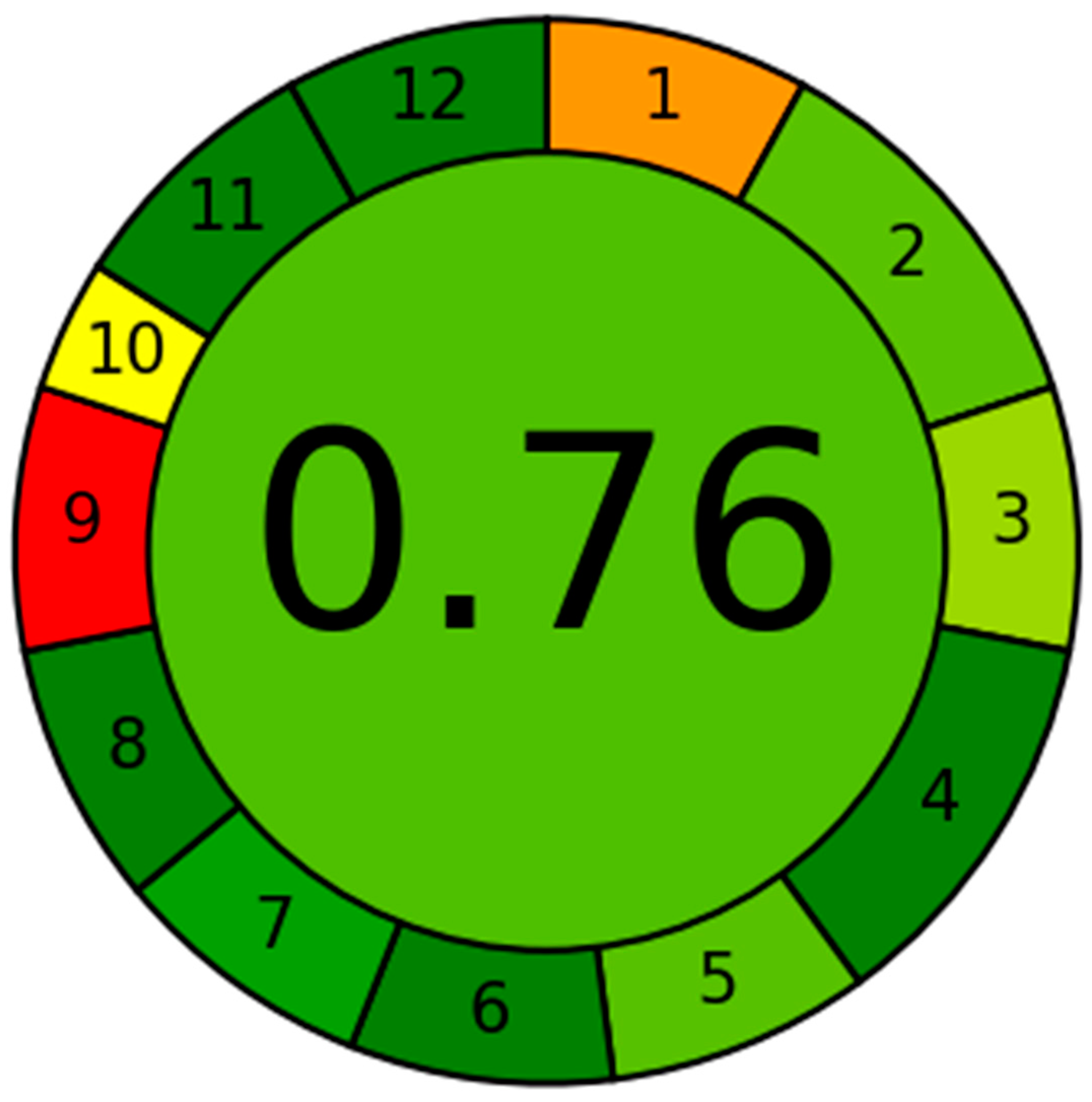
2.11. Evaluation of the System Greenness Employing AGREE Software
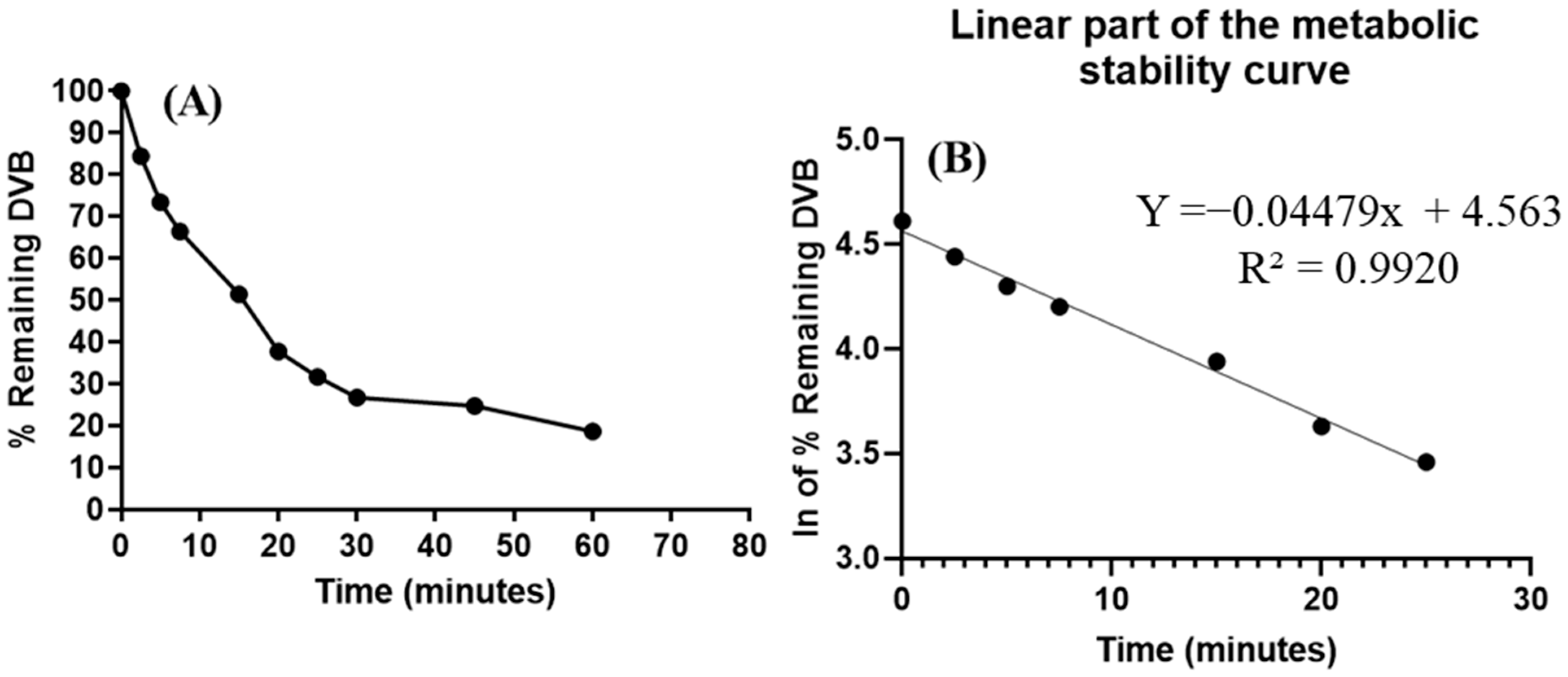
2.12. In Vitro Assessment of the DVB Metabolic Stability
3. Results
3.1. In Silico Metabolic Lability Assessment of DVB
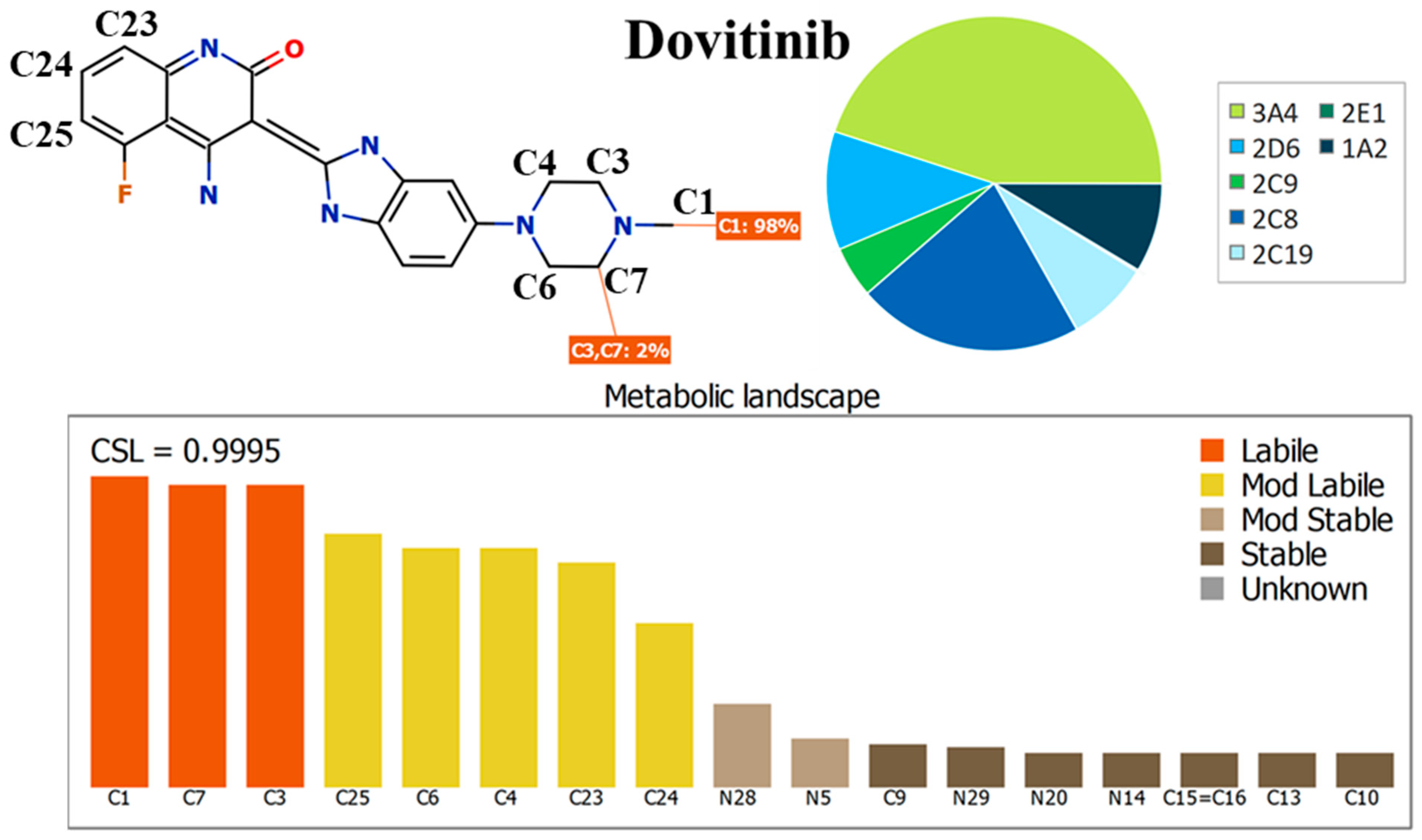
3.2. In Silico Testing of DVB Toxicity Alerts Using DEREK Module
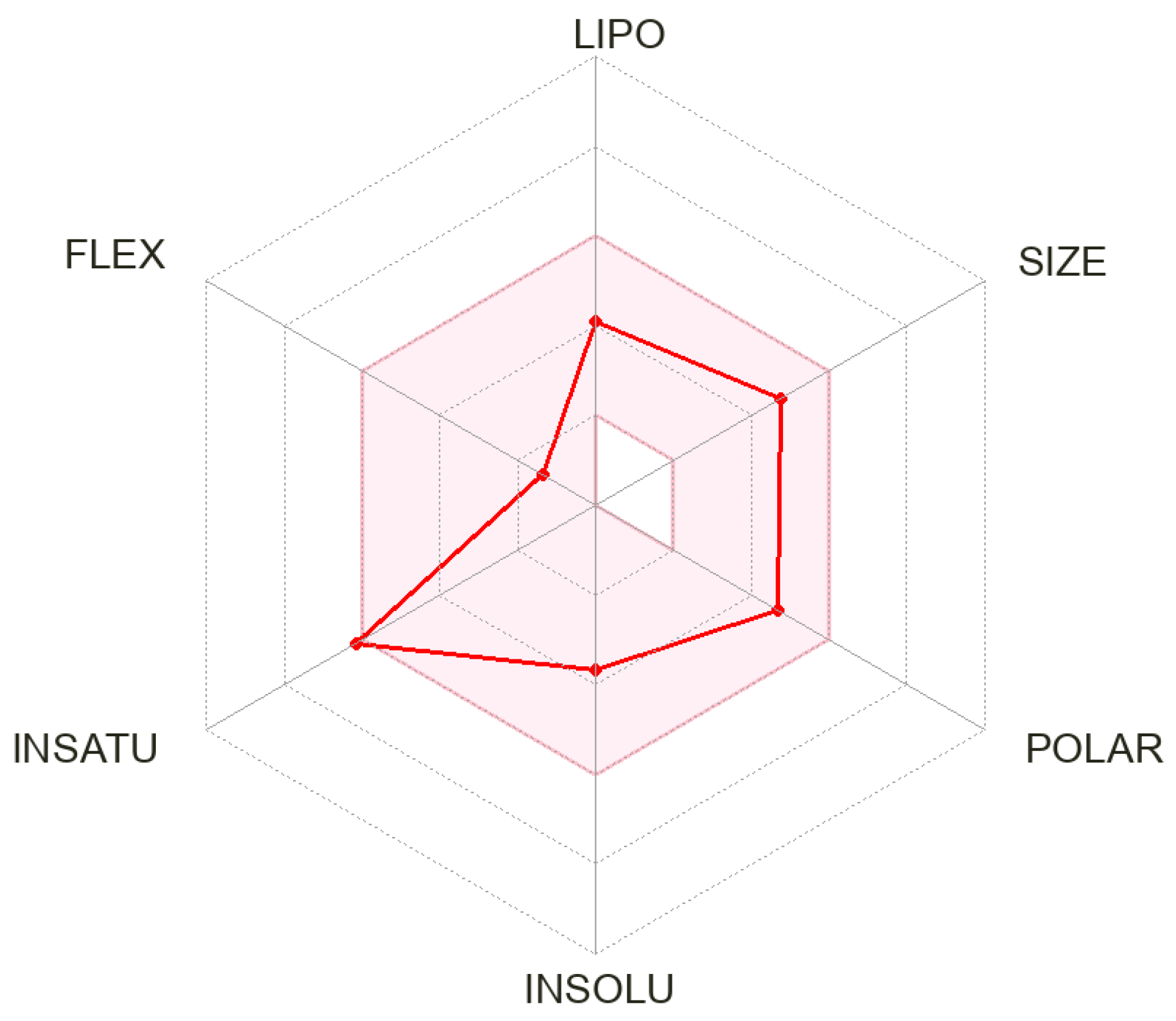
3.3. In Silico ADME Parameters
3.4. UHPLC-MS/MS Approach
3.5. Validation of the LC-MS/MS Approach
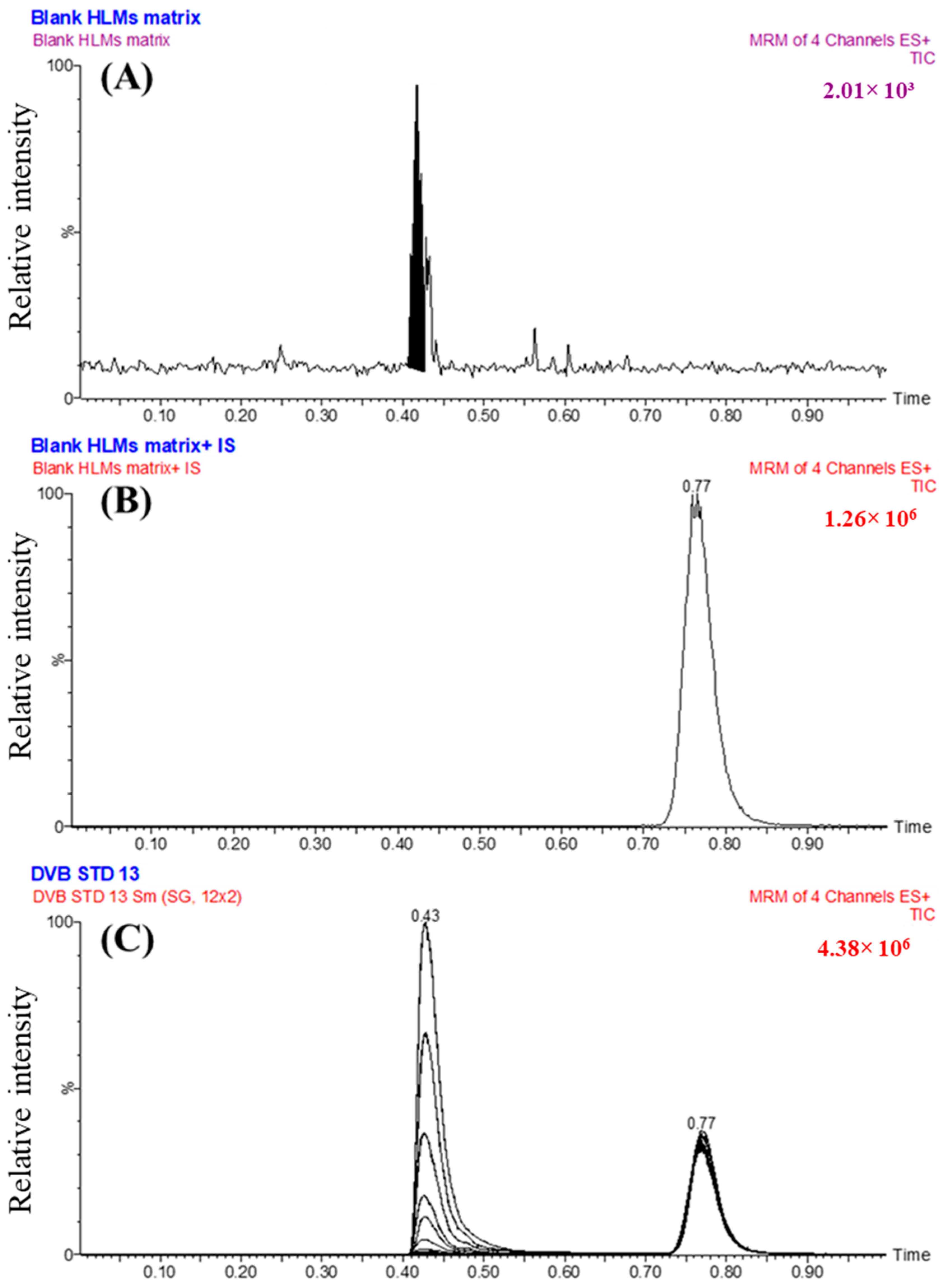
3.5.1. Specificity
3.5.2. Linearity and Sensitivity
3.5.3. Accuracy and Precision Validation Parameters
3.5.4. The HLMs Matrix Does Not Show Any Observed Effect on the Extraction Recovery of DVB in the Proposed UHPLC-MS/MS System
3.5.5. Stability of DVB in the HLMs (The Metabolic Incubation Matrix) and DMSO
3.6. Evaluation of the UHPLC-MS/MS System Greenness Utilizing the AGREE Software
3.7. In Vitro Metabolic Incubations of DVB with HLMs
4. Discussions
5. Limitation of the Current Study
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Trudel, S.; Li, Z.H.; Wei, E.; Wiesmann, M.; Chang, H.; Chen, C.; Reece, D.; Heise, C.; Stewart, A.K. CHIR-258, a novel, multitargeted tyrosine kinase inhibitor for the potential treatment of t (4; 14) multiple myeloma. Blood 2005, 105, 2941–2948. [Google Scholar] [CrossRef] [PubMed]
- Azab, A.K.; Azab, F.; Quang, P.; Maiso, P.; Sacco, A.; Ngo, H.T.; Liu, Y.; Zhang, Y.; Morgan, B.L.; Roccaro, A.M. FGFR3 is overexpressed Waldenström macroglobulinemia and its inhibition by dovitinib induces apoptosis and overcomes stroma-induced proliferation. Clin. Cancer Res. 2011, 17, 4389–4399. [Google Scholar] [CrossRef][Green Version]
- Huynh, H.; Chow, P.K.H.; Tai, W.M.; Choo, S.P.; Chung, A.Y.F.; Ong, H.S.; Soo, K.C.; Ong, R.; Linnartz, R.; Shi, M.M. Dovitinib demonstrates antitumor and antimetastatic activities in xenograft models of hepatocellular carcinoma. J. Hepatol. 2012, 56, 595–601. [Google Scholar] [CrossRef]
- Knudsen, S.; Hansen, A.; Foegh, M.; Petersen, S.; Mekonnen, H.; Jia, L.; Shah, P.; Martin, V.; Frykman, G.; Pili, R. A novel drug specific mRNA biomarker predictor for selection of patients responding to dovitinib treatment of advanced renal cell carcinoma and other solid tumors. PLoS ONE 2023, 18, e0290681. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.T.; Tan, Y.J.; Oon, C.E. Benzimidazole and its derivatives as cancer therapeutics: The potential role from traditional to precision medicine. Acta Pharm. Sin. B 2023, 13, 478–497. [Google Scholar] [CrossRef]
- Pili, R.; Kauffman, E.; Rodriguez, R. 82—Cancer of the Kidney. In Abeloff’s Clinical Oncology, 5th ed.; Niederhuber, J.E., Armitage, J.O., Doroshow, J.H., Kastan, M.B., Tepper, J.E., Eds.; Churchill Livingstone: Philadelphia, PA, USA, 2014; pp. 1416–1444.e1415. [Google Scholar]
- Kang, Y.K.; Yoo, C.; Ryoo, B.Y.; Lee, J.J.; Tan, E.; Park, I.; Park, J.H.; Choi, Y.J.; Jo, J.; Ryu, J.S.; et al. Phase II study of dovitinib in patients with metastatic and/or unresectable gastrointestinal stromal tumours after failure of imatinib and sunitinib. Br. J. Cancer 2013, 109, 2309–2315. [Google Scholar] [CrossRef] [PubMed]
- Milowsky, M.I.; Dittrich, C.; Durán, I.; Jagdev, S.; Millard, F.E.; Sweeney, C.J.; Bajorin, D.; Cerbone, L.; Quinn, D.I.; Stadler, W.M.; et al. Phase 2 trial of dovitinib in patients with progressive FGFR3-mutated or FGFR3 wild-type advanced urothelial carcinoma. Eur. J. Cancer 2014, 50, 3145–3152. [Google Scholar] [CrossRef] [PubMed]
- Cedeno-Serna, J. Chapter 82—Investigational new drug (IND) application. In Translational Sports Medicine; Eltorai, A.E.M., Bakal, J.A., DeFroda, S.F., Owens, B.D., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 419–424. [Google Scholar]
- Tyzack, J.D.; Kirchmair, J. Computational methods and tools to predict cytochrome P450 metabolism for drug discovery. Chem. Biol. Drug Des. 2019, 93, 377–386. [Google Scholar] [CrossRef]
- Liu, M.; Shi, L.; Guo, J.; Gu, Y.; Li, S.; Yi, L.; Ren, D.; Li, B. Determination of organic acids for predicting sourness intensity of tea beverage by liquid chromatography–tandem mass spectrometry and chemometrics methods. J. Sep. Sci. 2024, 47, 2300628. [Google Scholar] [CrossRef]
- Mahajan, B.; Miniyar, P.; Chodankar, R.; Mahajan, A. Liquid chromatography and liquid chromatography coupled with tandem mass spectrometry studies for the identification and characterization of degradation products of lobeglitazone. Sep. Sci. Plus 2024, 7, 2300223. [Google Scholar] [CrossRef]
- Bosco Ackerman, B.; Martín, S.; Espinosa, M.; Chocrón, M.; Babay, P.A. Development and validation of a liquid chromatography-mass spectrometry method for quantification of octadecylamine in the secondary circuit of a nuclear power plant in the presence of other amines. Sep. Sci. Plus 2024, 7, e202400099. [Google Scholar] [CrossRef]
- Darwish, I.A.; Alzoman, N.Z.; Almomen, A.; Almehizia, A.A.; Attwa, M.W.; Darwish, H.W.; Sayed, A.Y. Development and validation of an UPLC-ESI-MS/MS method for quantification of duvelisib in plasma: Application to pharmacokinetic study in rats. RSC Adv. 2023, 13, 7929–7938. [Google Scholar] [CrossRef]
- Attwa, M.W.; Abdelhameed, A.S.; Alsaif, N.A.; Kadi, A.A.; AlRabiah, H. A validated LC-MS/MS analytical method for the quantification of pemigatinib: Metabolic stability evaluation in human liver microsomes. RSC Adv. 2022, 12, 20387–20394. [Google Scholar] [CrossRef]
- Alanazi, M.M.; Alkahtani, H.M.; Almehizia, A.A.; Attwa, M.W.; Bakheit, A.H.; Darwish, H.W. Validated liquid chromatography tandem mass spectrometry for simultaneous quantification of foretinib and lapatinib, and application to metabolic stability investigation. RSC Adv. 2019, 9, 19325–19332. [Google Scholar] [CrossRef]
- Ryu, R.; Hebert, M.F. Chapter 3—Impact of pregnancy on maternal pharmacokinetics of medications. In Clinical Pharmacology during Pregnancy, 2nd ed.; Mattison, D., Halbert, L.-A., Eds.; Academic Press: Boston, MA, USA, 2022; pp. 19–46. [Google Scholar]
- Pena-Pereira, F.; Wojnowski, W.; Tobiszewski, M. AGREE—Analytical GREEnness Metric Approach and Software. Anal. Chem. 2020, 92, 10076–10082. [Google Scholar] [CrossRef]
- Duan, X.; Liu, X.; Dong, Y.; Yang, J.; Zhang, J.; He, S.; Yang, F.; Wang, Z.; Dong, Y. A Green HPLC Method for Determination of Nine Sulfonamides in Milk and Beef, and Its Greenness Assessment with Analytical Eco-Scale and Greenness Profile. J. AOAC Int. 2020, 103, 1181–1189. [Google Scholar] [CrossRef]
- Attwa, M.W.; Abdelhameed, A.S.; Kadi, A.A. An ultra-fast ultra-high-performance liquid chromatography-tandem mass spectrometry method for estimating the in vitro metabolic stability of palbociclib in human liver microsomes: In silico study for metabolic lability, absorption, distribution, metabolism, and excretion features, and DEREK alerts screening. J. Sep. Sci. 2024, 47, 2400346. [Google Scholar] [CrossRef]
- Goldner, D.M.; do Nascimento, F.H.; Masini, J.C. A Green Liquid Chromatography Method for Simultaneous Quantification of Caffeine and its Three Major Metabolites in Urine, Drinks, and Herbal Products. Sep. Sci. Plus 2024, 7, e202400098. [Google Scholar] [CrossRef]
- Alrabiah, H.; Kadi, A.A.; Attwa, M.W.; Abdelhameed, A.S. A simple liquid chromatography-tandem mass spectrometry method to accurately determine the novel third-generation EGFR-TKI naquotinib with its applicability to metabolic stability assessment. RSC Adv. 2019, 9, 4862–4869. [Google Scholar] [CrossRef]
- Alsubi, T.A.; Attwa, M.W.; Bakheit, A.H.; Darwish, H.W.; Abuelizz, H.A.; Kadi, A.A. In silico and in vitro metabolism of ribociclib: A mass spectrometric approach to bioactivation pathway elucidation and metabolite profiling. RSC Adv. 2020, 10, 22668–22683. [Google Scholar] [CrossRef]
- Attwa, M.W.; Kadi, A.A.; Abdelhameed, A.S. Characterization of reactive intermediates formation in dacomitinib metabolism and bioactivation pathways elucidation by LC-MS/MS: In vitro phase I metabolic investigation. RSC Adv. 2018, 8, 38733–38744. [Google Scholar] [CrossRef] [PubMed]
- Marothu Vamsi, K.; Kantamaneni, P.; Gorrepati, M. In vitro Metabolic Stability of Drugs and Applications of LC-MS in Metabolite Profiling. In Drug Metabolism; Katherine, D., Ed.; IntechOpen: Rijeka, Croatia, 2021; p. Ch. 5. [Google Scholar]
- Attwa, M.W.; AlRabiah, H.; Alsibaee, A.M.; Abdelhameed, A.S.; Kadi, A.A. An UPLC–ESI–MS/MS Bioanalytical Methodology for the Quantification of Gilteritinib in Human Liver Microsomes: Application to In Vitro and In Silico Metabolic Stability Estimation. Separations 2023, 10, 278. [Google Scholar] [CrossRef]
- Attwa, M.W.; Alsibaee, A.M.; Aljohar, H.I.; Abdelhameed, A.S.; Kadi, A.A. Development of a Fast and Sensitive UPLC–MS/MS Analytical Methodology for Fenebrutinib Estimation in Human Liver Microsomes: In Vitro and In Silico Metabolic Stability Evaluation. Separations 2023, 10, 302. [Google Scholar] [CrossRef]
- Attwa, M.W.; Mostafa, G.A.E.; AlRabiah, H.; Kadi, A.A. An LC–MS/MS Analytical Method for Quantifying Tepotinib in Human Liver Microsomes: Application to In Vitro and In Silico Metabolic Stability Estimation. Separations 2023, 10, 330. [Google Scholar] [CrossRef]
- Houston, J.B. Utility of in vitro drug metabolism data in predicting in vivo metabolic clearance. Biochem. Pharmacol. 1994, 47, 1469–1479. [Google Scholar] [CrossRef] [PubMed]
- Obach, R.S.; Baxter, J.G.; Liston, T.E.; Silber, B.M.; Jones, B.C.; MacIntyre, F.; Rance, D.J.; Wastall, P. The prediction of human pharmacokinetic parameters from preclinical and in vitro metabolism data. J. Pharmacol. Exp. Ther. 1997, 283, 46–58. [Google Scholar]
- Marchant, C.A.; Briggs, K.A.; Long, A. In Silico Tools for Sharing Data and Knowledge on Toxicity and Metabolism: Derek for Windows, Meteor, and Vitic. Toxicol. Mech. Methods 2008, 18, 177–187. [Google Scholar] [CrossRef]
- AlRabiah, H.; Kadi, A.A.; Attwa, M.W.; Mostafa, G.A.E. Development and validation of an HPLC-MS/MS method for the determination of filgotinib, a selective Janus kinase 1 inhibitor: Application to a metabolic stability study. J. Chromatogr. B 2020, 1154, 122195. [Google Scholar] [CrossRef]
- Attwa, M.W.; Abdelhameed, A.S.; Kadi, A.A. An ultra-fast green ultra-high-performance liquid chromatography-tandem mass spectrometry method for estimating the in vitro metabolic stability of zotizalkib in human liver microsomes. J. Sep. Sci. 2024, 47, 2400393. [Google Scholar] [CrossRef]
- Attwa, M.W.; Abdelhameed, A.S.; Kadi, A.A. Characterization of the in vitro metabolic profile of nazartinib in HLMs using UPLC-MS/MS method: In silico metabolic lability and DEREK structural alerts screening using StarDrop software. Heliyon 2024, 10, e34109. [Google Scholar] [CrossRef]
- Busby, W.F., Jr.; Ackermann, J.M.; Crespi, C.L. Effect of methanol, ethanol, dimethyl sulfoxide, and acetonitrile on in vitro activities of cDNA-expressed human cytochromes P-450. Drug Metab. Dispos. 1999, 27, 246–249. [Google Scholar] [PubMed]
- Störmer, E.; Roots, I.; Brockmöller, J. Benzydamine N-oxidation as an index reaction reflecting FMO activity in human liver microsomes and impact of FMO3 polymorphisms on enzyme activity. Br. J. Clin. Pharmacol. 2000, 50, 553–561. [Google Scholar] [CrossRef]
- Fouin-Fortunet, H.; Tinel, M.; Descatoire, V.; Letteron, P.; Larrey, D.; Geneve, J.; Pessayre, D. Inactivation of cytochrome P-450 by the drug methoxsalen. J. Pharmacol. Exp. Ther. 1986, 236, 237–247. [Google Scholar] [PubMed]
- Meesters, R.; Voswinkel, S. Bioanalytical method development and validation: From the USFDA 2001 to the USFDA 2018 guidance for industry. J. Appl. Bioanal. 2018, 4, 67–73. [Google Scholar] [CrossRef]
- Sankar, P.R.; Geethika, A.S.; Rachana, G.; Babu, P.S.; Bhargavi, J. Bioanalytical method validation: A comprehensive review. Int. J. Pharm. Sci. Rev. Res 2019, 9, 50–58. [Google Scholar]
- Walfish, S. Analytical methods: A statistical perspective on the ICH Q2A and Q2B guidelines for validation of analytical methods. BioPharm Int. 2006, 19, 1–6. [Google Scholar]
- González, O.; Alonso, R.M. Chapter 6—Validation of bioanalytical chromatographic methods for the quantification of drugs in biological fluids. In Handbook of Analytical Separations; Hempel, G., Ed.; Elsevier Science B.V.: Amsterdam, The Netherlands, 2020; Volume 7, pp. 115–134. [Google Scholar]
- De Nicolò, A.; Cantù, M.; D’Avolio, A. Matrix Effect Management in Liquid Chromatography Mass Spectrometry: The Internal Standard Normalized Matrix Effect. Bioanalysis 2017, 9, 1093–1105. [Google Scholar] [CrossRef]
- Steiner, D.; Krska, R.; Malachová, A.; Taschl, I.; Sulyok, M. Evaluation of Matrix Effects and Extraction Efficiencies of LC–MS/MS Methods as the Essential Part for Proper Validation of Multiclass Contaminants in Complex Feed. J. Agric. Food Chem. 2020, 68, 3868–3880. [Google Scholar] [CrossRef]
- Matuszewski, B.K.; Constanzer, M.L.; Chavez-Eng, C.M. Strategies for the Assessment of Matrix Effect in Quantitative Bioanalytical Methods Based on HPLC−MS/MS. Anal. Chem. 2003, 75, 3019–3030. [Google Scholar] [CrossRef]
- McNaney, C.A.; Drexler, D.M.; Hnatyshyn, S.Y.; Zvyaga, T.A.; Knipe, J.O.; Belcastro, J.V.; Sanders, M. An automated liquid chromatography-mass spectrometry process to determine metabolic stability half-life and intrinsic clearance of drug candidates by substrate depletion. Assay Drug Dev. Technol. 2008, 6, 121–129. [Google Scholar] [CrossRef]
- Słoczyńska, K.; Gunia-Krzyżak, A.; Koczurkiewicz, P.; Wójcik-Pszczoła, K.; Żelaszczyk, D.; Popiół, J.; Pękala, E. Metabolic stability and its role in the discovery of new chemical entities. Acta Pharm. 2019, 69, 345–361. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Liu, G.; Wang, J.; Aubry, A.F.; Arnold, M.E. Selecting the correct weighting factors for linear and quadratic calibration curves with least-squares regression algorithm in bioanalytical LC-MS/MS assays and impacts of using incorrect weighting factors on curve stability, data quality, and assay performance. Anal. Chem. 2014, 86, 8959–8966. [Google Scholar] [CrossRef] [PubMed]
- Abdelhameed, A.S.; Kadi, A.A.; Attwa, M.W.; AlRabiah, H. Validated LC-MS/MS assay for quantification of the newly approved tyrosine kinase inhibitor, dacomitinib, and application to investigating its metabolic stability. PLoS ONE 2019, 14, e0214598. [Google Scholar] [CrossRef] [PubMed]
- Leahy, D.E. Integrating invitro ADMET data through generic physiologically based pharmacokinetic models. Expert Opin. Drug Metab. Toxicol. 2006, 2, 619–628. [Google Scholar] [CrossRef] [PubMed]
- De Simone, M.; Conti, V.; Palermo, G.; De Maria, L.; Iaconetta, G. Advancements in Glioma Care: Focus on Emerging Neurosurgical Techniques. Biomedicines 2024, 12, 8. [Google Scholar] [CrossRef]
- De Simone, M.; Choucha, A.; Dannhoff, G.; Kong, D.-S.; Zoia, C.; Iaconetta, G. Treating Trigeminal Schwannoma through a Transorbital Approach: A Systematic Review. J. Clin. Med. 2024, 13, 3701. [Google Scholar] [CrossRef]

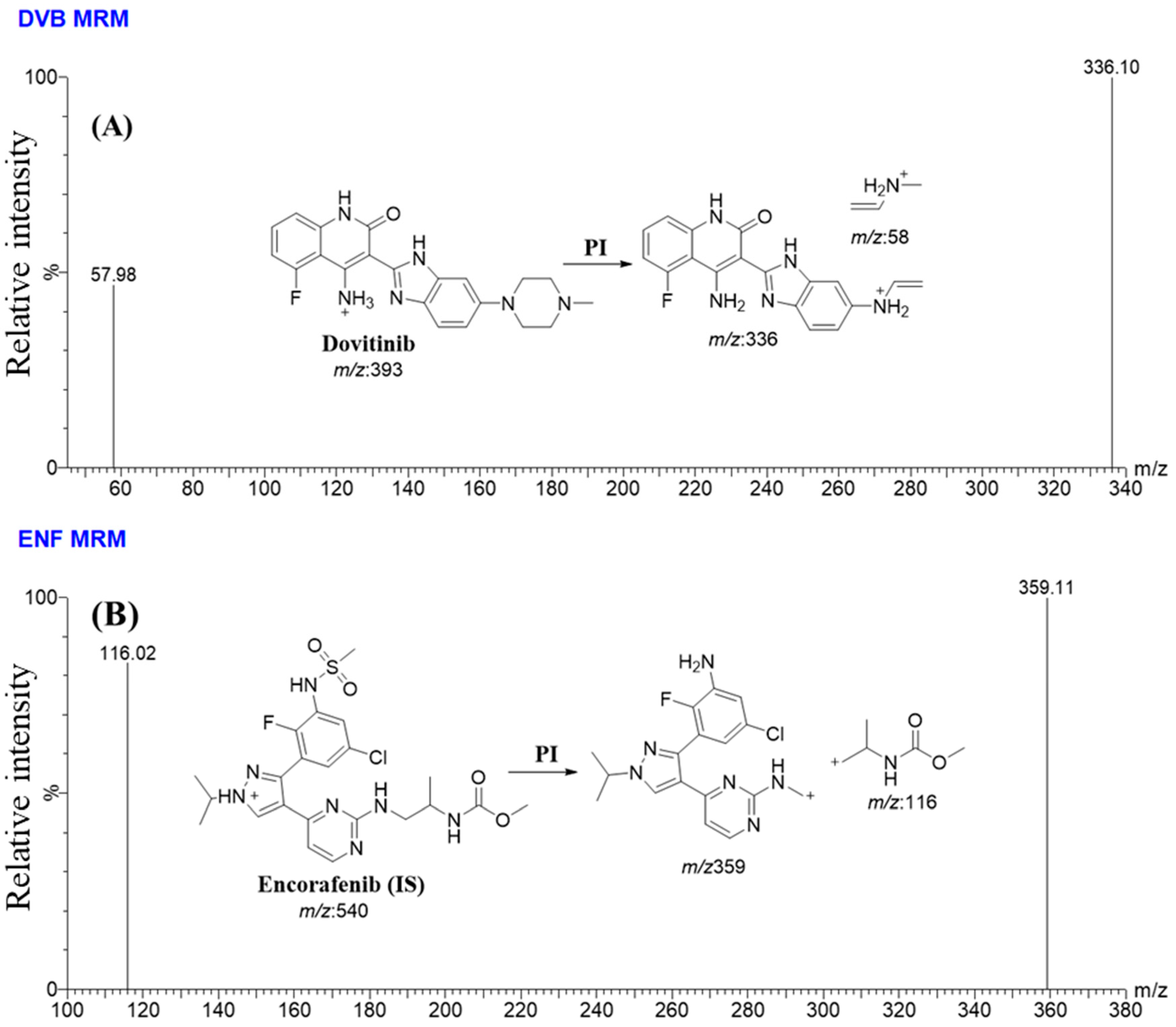
| UPLC | TQD MS | ||||
|---|---|---|---|---|---|
| Eclipse plus-C8 reversed column | Particle size | 3.5 μm | ESI | Positive ESI source | |
| Internal diameter | 2.1 mm | The extractor voltage | 3.0 (V) | ||
| Length | 50.0 mm | Cone gas rate | 100 L/hr | ||
| Column T | 22.0 ± 2.0 °C | The RF lens voltage | 0.1 (V) | ||
| Isocratic mobile phase system | Aqueous part | 0.1% Formic acid in H2O | Nitrogen gas | Drying gas | |
| 55% | 100 L/hr | ||||
| pH: 3.2 | 350 °C | ||||
| Organic part | 45% ACN | Capillary voltage: 4 KV | |||
| Injection volume: | 5.0 μL | Argon gas | 0.14 mL/min | ||
| Flow rate | 0.5 mL/min. | Mode | MRM | ||
| Time Segment | Rt | Mass Transitions (m/z) | |
|---|---|---|---|
| MRM detection segments | 0.0 to 0.65 min | DVB (0.44 min) | 393→336 (CE a:32 and CV b: 28) |
| 393→58 (CE:36 and CV: 46) | |||
| 0.65 to 1.0 min | EFB (IS; 0.77 min) | 540→359 (CE: 56 and CV: 36) | |
| 540→116 (CE: 56 and CV: 32) |
| Physicochemical Properties | Water Solubility | ||
|---|---|---|---|
| Formula | C21H21FN6O | Solubility | 8.60 × 10−2 mg/mL; 2.19 × 10−4 mol/L |
| Molecular weight | 392.43 g/mol | Log S (ESOL) | −3.66 |
| Heavy atoms num. | 29 | Class | Soluble |
| Arom. heavy atoms num. | 19 | Solubility | 2.32 × 10−1 mg/mL; 5.92 × 10−4 mol/L |
| Rotatable bonds num. | 2 | Log S (Ali) | −3.64 |
| Fraction Csp3 | 0.24 | Class | Soluble |
| Solubility | 8.66 × 10−5 mg/mL; 2.21 × 10−7 mol/L | ||
| Num. H-bond donors | 3 | Log S (SILICOS-IT) | −6.66 |
| TPSA | 94.04 Å2 | Class | Poorly soluble |
| Num. H-bond acceptors | 4 | Medicinal Chemistry | |
| Molar refractivity | 120.28 | Brenk | 0 alert |
| Lipophilicity features | PAINS | 0 alert | |
| Log Po/w (XLOGP3) | 1.64 | Leadlikeness | No; 1 violations: MW > 350 |
| Log Po/w (iLOGP) | 2.26 | Synthetic accessibility | 3.20 |
| Log Po/w (MLOGP) | 2.31 | Pharmacokinetics | |
| Log Po/w (SILICOS-IT) | 3.27 | GI absorption | High |
| Log Po/w (WLOGP) | 2.21 | P-gp substrate | Yes |
| Consensus Log Po/w | 2.34 | Permeant to BBB | No |
| Druglikeness features | Inhibiton of CYP1A2 | Yes | |
| Ghose | Yes | Inhibiton of CYP2D6 | Yes |
| Lipinski | Yes; 0 violation | Inhibiton of CYP3A4 | No |
| Egan | Yes | Inhibiton of CYP2C9 | No |
| Veber | Yes | Inhibiton of CYP2C19 | No |
| Muegge | Yes | Skin permeation (Log Kp) | −7.53 cm/s |
| The score of bioavailability | 0.55 | ||
| Analytes | Recovery | Stationary System | Stationary System | |||
|---|---|---|---|---|---|---|
| Solid Phase Extraction | Protein Precipitation Using ACN | Methanol | ACN | C18 Column | C8 Column | |
| DVB | Low (88.27%) | High (102.62 ± 3.73%) | 0.52 min | 0.43 min | 0.74 min | 0.43 min |
| Not precise | Precise (RSD < 3.63%) | Tailed | Good peak | Tailed peaks | Perfect shape | |
| EFB | Good (89.89%) | High (101.61 ± 3.23% | 0.87 min | 0.77 min | 1.25 min | 0.77 min |
| Not precise | Precise (RSD < 3.18%) | Overlapped | Optimum peak shape | Perfect shape | Optimum shape | |
| DVB (ng/mL) | Mean | SD | Accuracy (%) | RSD (%) | Recovery |
|---|---|---|---|---|---|
| 1.00 | 1.07 | 0.08 | 7.18 | 7.28 | 107.18 |
| 15.00 | 14.73 | 0.09 | −1.83 | 0.64 | 98.17 |
| 40.00 | 40.93 | 0.28 | 2.32 | 0.69 | 102.32 |
| 100.00 | 100.96 | 1.57 | 0.96 | 1.56 | 100.96 |
| 250.00 | 254.41 | 1.90 | 1.77 | 0.75 | 101.77 |
| 500.00 | 508.02 | 5.97 | 1.60 | 1.18 | 101.60 |
| 2000.00 | 1966.90 | 21.08 | −1.65 | 1.07 | 98.35 |
| 3000.00 | 2997.38 | 13.60 | −0.09 | 0.45 | 99.91 |
| % Recovery | 101.28 ± 2.84 |
| DVB (ng/mL) | Intra-Day (12 Sets in 1 Day) | Inter-Day (6 Sets in 3 Days) | ||||||
|---|---|---|---|---|---|---|---|---|
| QCs | 1.00 | 3.00 | 900.00 | 2400.00 | 1.00 | 3.00 | 900.00 | 2400.00 |
| Mean | 1.07 | 3.10 | 902.61 | 2406.71 | 1.09 | 3.05 | 894.99 | 2389.57 |
| SD | 0.08 | 0.03 | 7.54 | 10.10 | 0.03 | 0.06 | 5.18 | 5.74 |
| Precision (%RSD) | 7.28 | 0.81 | 0.83 | 0.42 | 2.30 | 2.00 | 0.58 | 0.24 |
| % Accuracy | 7.18 | 3.19 | 0.29 | 0.28 | 9.33 | 1.68 | −0.56 | −0.43 |
| Recovery (%) | 107.18 | 103.19 | 100.29 | 100.28 | 109.33 | 101.68 | 99.44 | 99.57 |
| Stability as Validation Features | Mean | SD | Precision (%RSD) | Accuracy (%E) | ||||
|---|---|---|---|---|---|---|---|---|
| 3.0 | 2400.0 | 3.0 | 2400.0 | 3.0 | 2400.0 | 3.0 | 2400.0 | |
| Long-Term (−80 °C for 28 d) | 2.91 | 2388.44 | 0.05 | 5.52 | 1.82 | 0.23 | −3.00 | −0.48 |
| Auto-Sampler (15 °C for 24 h) | 3.01 | 2406.68 | 0.12 | 7.77 | 4.13 | 0.32 | 0.22 | 0.28 |
| Freeze–Thaw (3 cycles at −80 °C) | 3.06 | 2414.45 | 0.08 | 14.51 | 2.72 | 0.60 | 2.11 | 0.60 |
| Short-Term (4 h at room T) | 2.95 | 2390.13 | 0.11 | 6.38 | 3.59 | 0.27 | −1.56 | −0.41 |
| Principles | Score | Weight |
|---|---|---|
| 1. To avoid the necessity for sample treatment, it is recommended to use direct analytical methods. | 0.3 | 2 |
| 2. The targets of this research are to achieve a minimal amount and a small sample size of specimens. | 0.75 | 3 |
| 3. Ideally, it is advisable to perform assessments on-site whenever it is feasible. | 0.66 | 2 |
| 4. Studies have shown that the integration of analytical steps and activities leads to positive results concerning energy preservation and reducing the use of reagents. | 1.0 | 3 |
| 5. It is advisable to choose automated and streamlined operations. | 0.75 | 2 |
| 6. Avoiding the implementation of derivatization procedures is prudent. | 1.0 | 2 |
| 7. To reduce the causing of a noteworthy amount of analytical surplus and implement effective disposal methods is paramount. | 0.88 | 2 |
| 8. Within the field of analytical chemistry, there is a predilection for employing multi-analyte or multi-parameter methodologies as opposed to those that just concentrate on a single analyte. | 1.0 | 2 |
| 9. Attempts should be considered to reduce energy use. | 0.0 | 2 |
| 10. It is sensible to give importance to the usage of reagents produced from renewable sources. | 0.5 | 1 |
| 11. The necessity of eliminating or replacing detrimental substances is of paramount significance. | 1.0 | 2 |
| 12. There is a necessity to improve the safety regulations for workers. | 1.0 | 2 |
| Time (min.) | Mean a (ng/mL) | X b | LN X | The Linear Segment Features |
|---|---|---|---|---|
| 0.00 | 380.45 | 100 | 4.61 | Regression line equation: y = −0.04479x + 4.563 |
| 2.50 | 321.44 | 84.49 | 4.44 | |
| 5.00 | 279.33 | 73.42 | 4.30 | R2 = 0.9920 |
| 7.50 | 252.96 | 66.49 | 4.20 | |
| 15.00 | 195.86 | 51.48 | 3.94 | Slope: −0.04479 |
| 20.00 | 143.62 | 37.75 | 3.63 | |
| 25.00 | 120.75 | 31.74 | 3.46 | t1/2: 15.48 min and |
| 30.00 | 101.88 | 26.78 | 3.29 | Clint: 52.39 mL/min/kg |
| 45.00 | 94.28 | 24.78 | 3.21 | |
| 60.00 | 71.30 | 18.74 | 2.93 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Published by MDPI on behalf of the Lithuanian University of Health Sciences. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Attwa, M.W.; Abdelhameed, A.S.; Kadi, A.A. An Ultra-Fast Green UHPLC-MS/MS Method for Assessing the In Vitro Metabolic Stability of Dovitinib: In Silico Study for Absorption, Distribution, Metabolism, Excretion, Metabolic Lability, and DEREK Alerts. Medicina 2024, 60, 1626. https://doi.org/10.3390/medicina60101626
Attwa MW, Abdelhameed AS, Kadi AA. An Ultra-Fast Green UHPLC-MS/MS Method for Assessing the In Vitro Metabolic Stability of Dovitinib: In Silico Study for Absorption, Distribution, Metabolism, Excretion, Metabolic Lability, and DEREK Alerts. Medicina. 2024; 60(10):1626. https://doi.org/10.3390/medicina60101626
Chicago/Turabian StyleAttwa, Mohamed W., Ali S. Abdelhameed, and Adnan A. Kadi. 2024. "An Ultra-Fast Green UHPLC-MS/MS Method for Assessing the In Vitro Metabolic Stability of Dovitinib: In Silico Study for Absorption, Distribution, Metabolism, Excretion, Metabolic Lability, and DEREK Alerts" Medicina 60, no. 10: 1626. https://doi.org/10.3390/medicina60101626
APA StyleAttwa, M. W., Abdelhameed, A. S., & Kadi, A. A. (2024). An Ultra-Fast Green UHPLC-MS/MS Method for Assessing the In Vitro Metabolic Stability of Dovitinib: In Silico Study for Absorption, Distribution, Metabolism, Excretion, Metabolic Lability, and DEREK Alerts. Medicina, 60(10), 1626. https://doi.org/10.3390/medicina60101626






