Comprehensive Analysis Reveals Prognostic and Therapeutic Immunity-Related Biomarkers for Pediatric Metastatic Osteosarcoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of DEGs
2.2. Protein–Protein Interaction Network Analysis
2.3. Survival-Related DEG Screening
2.4. Expression of Survival-Related DEGs in Sarcomas
2.5. Analysis of Associations between DEGs and Tumor Immunity
3. Results
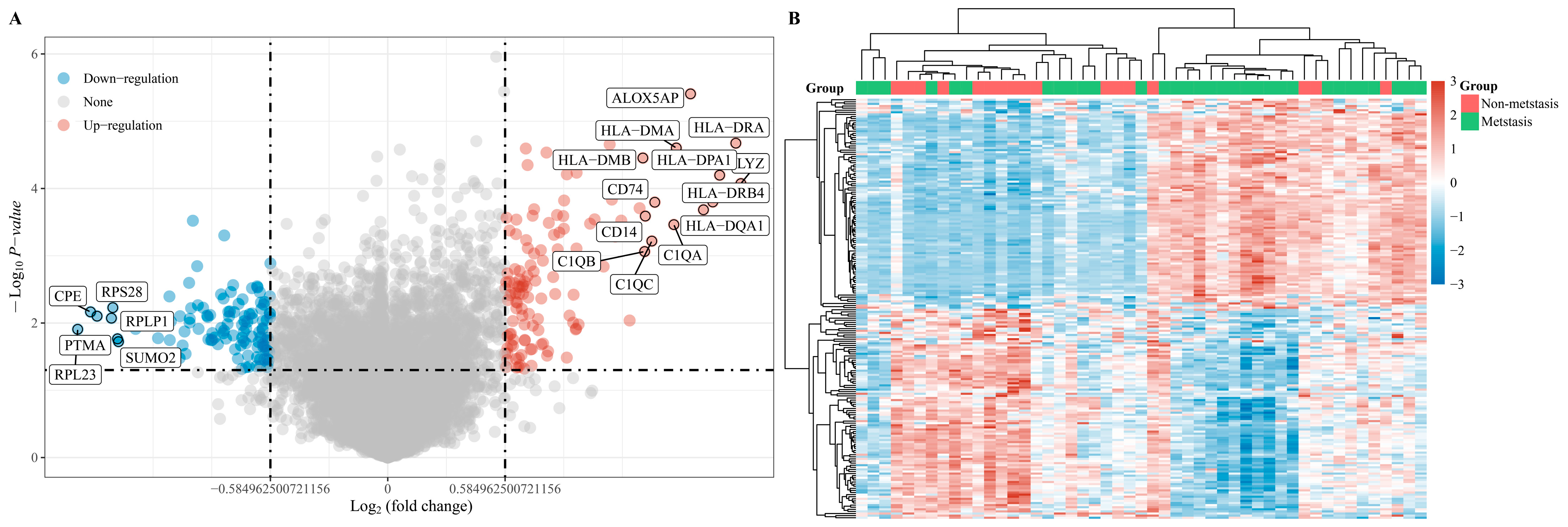
3.1. Sample Characteristics and Identification of DEGs
3.2. Identification of Central DEGs by Constructing a PPI Network
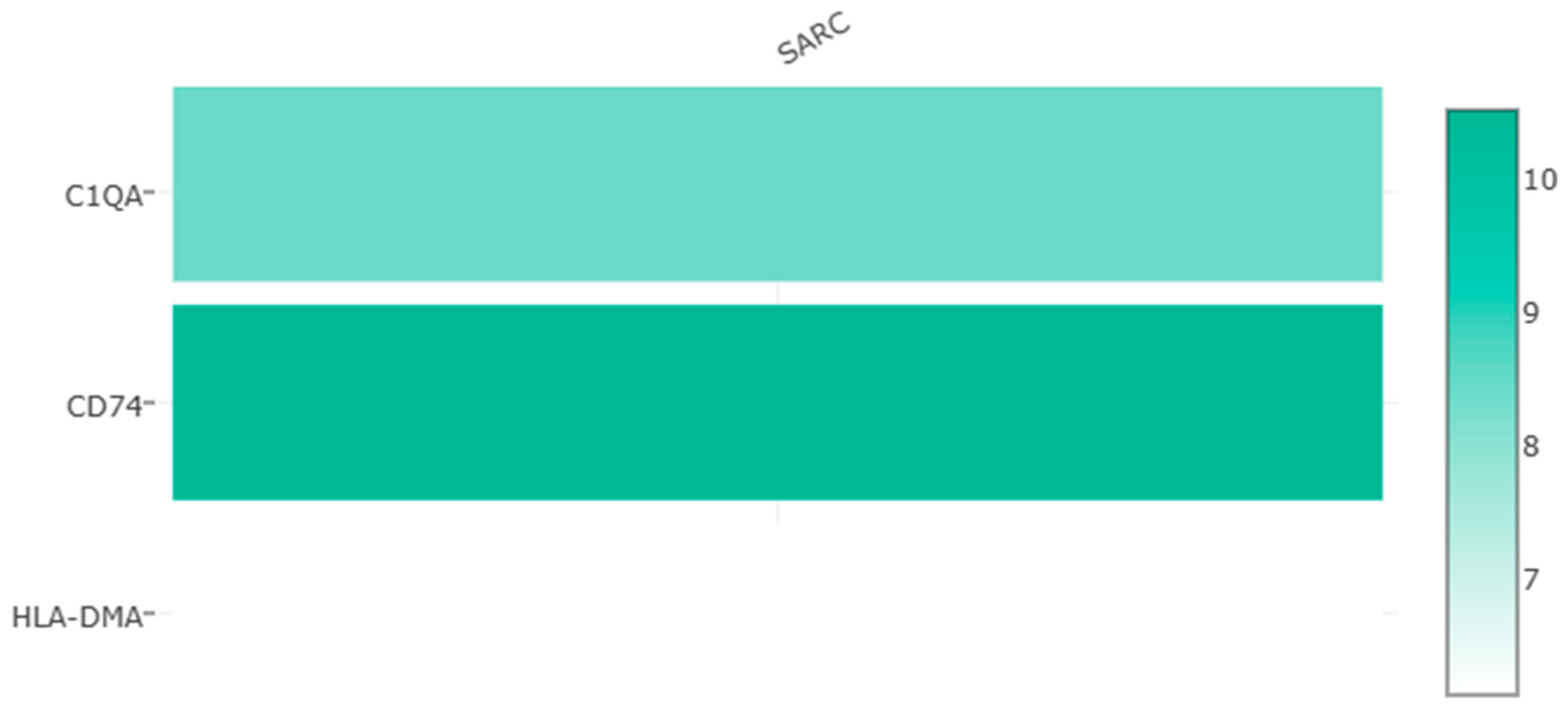
3.3. Identification of Survival-Related Central DEGs Based on the TARGET Database
3.4. Validation and Correlation of Three Survival-Related Central DEGs
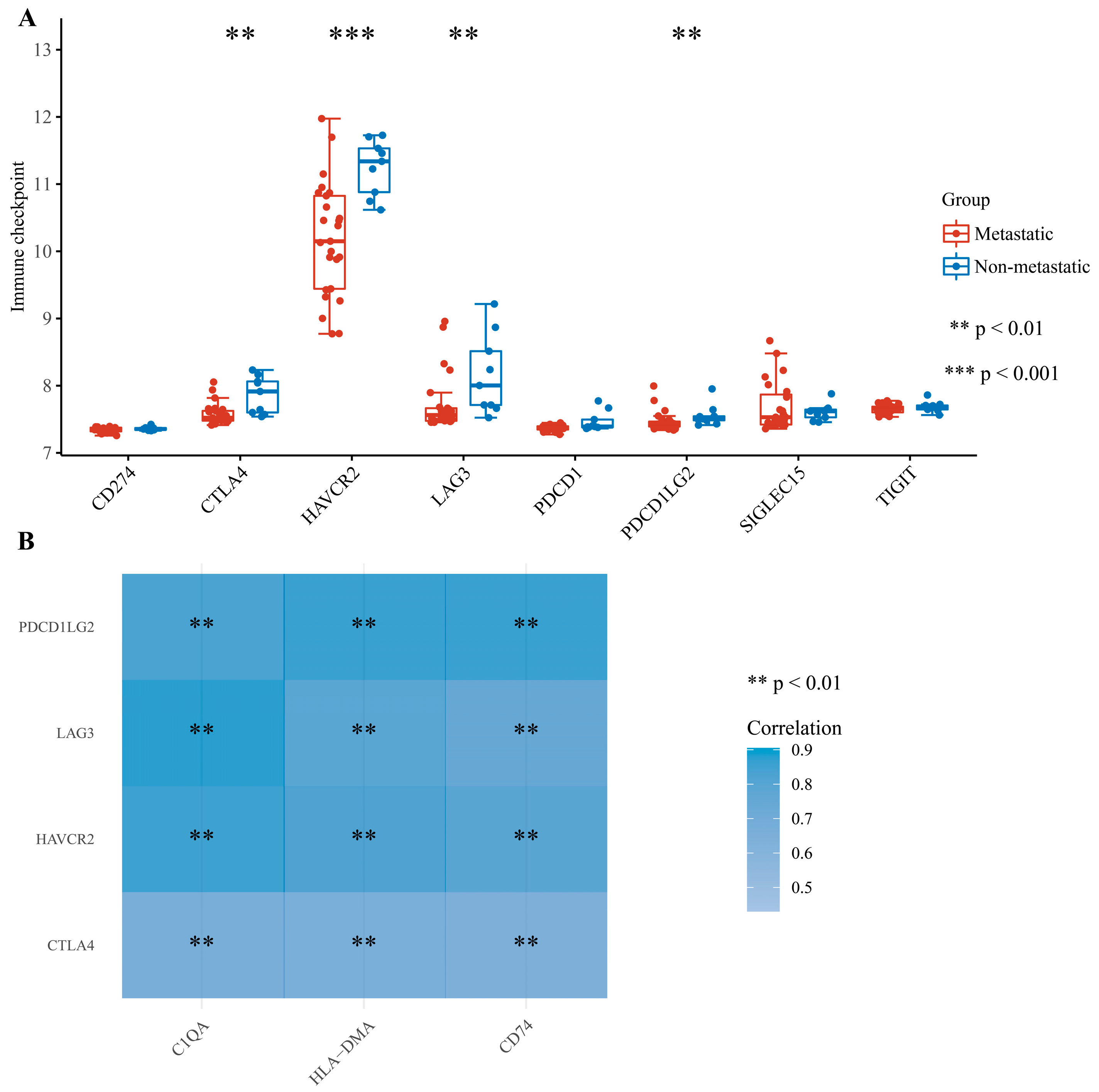
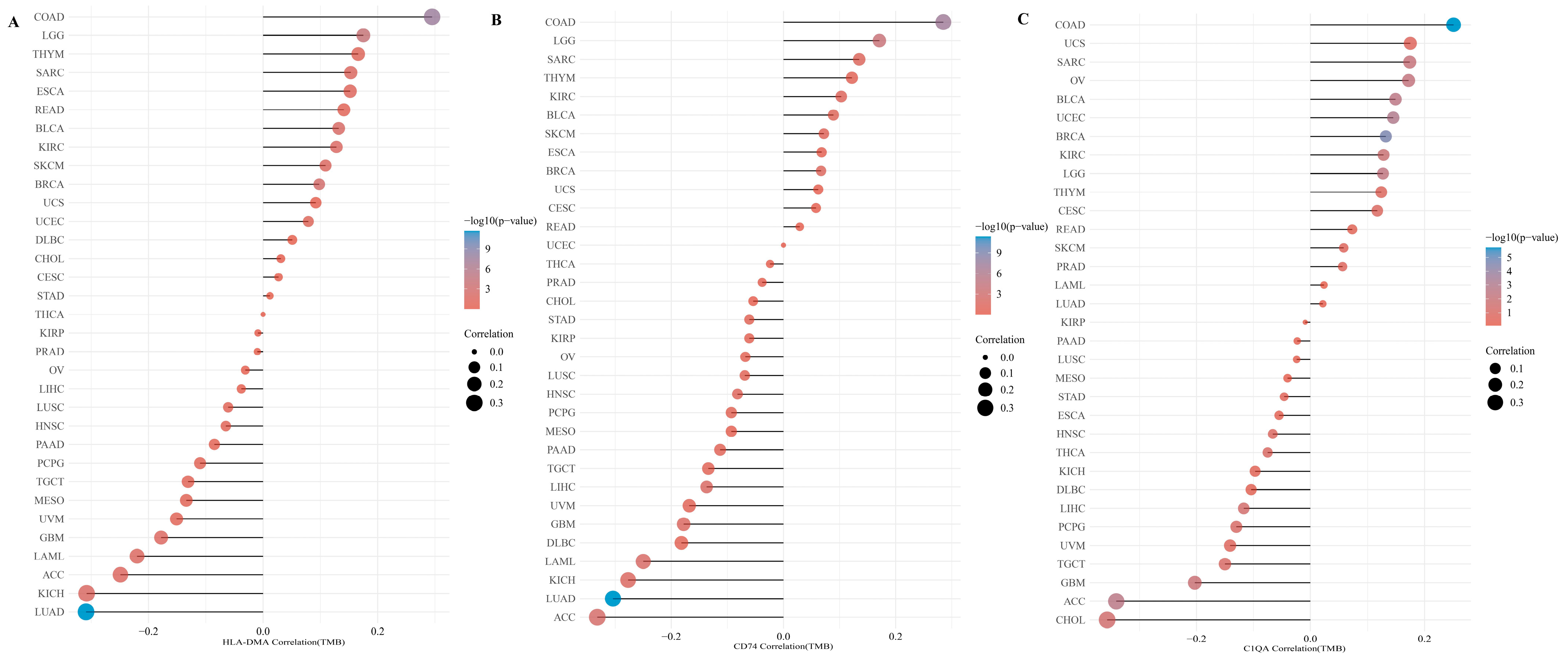
3.5. Correlation of DEGs and Tumor Immunity
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mirabello, L.; Troisi, R.J.; Savage, S.A. Osteosarcoma incidence and survival rates from 1973 to 2004. Cancer 2009, 115, 1531–1543. [Google Scholar] [CrossRef]
- Stiller, C.A.; Bielack, S.S.; Jundt, G.; Steliarova-Foucher, E. Bone tumours in European children and adolescents, 1978–1997. Report from the Automated Childhood Cancer Information System project. Eur. J. Cancer 2006, 42, 2124–2135. [Google Scholar] [CrossRef]
- Stiller, C.A.; Craft, A.W.; Corazziari, I. Survival of children with bone sarcoma in Europe since 1978: Results from the EUROCARE study. Eur. J. Cancer 2001, 37, 760–766. [Google Scholar] [CrossRef]
- Bielack, S.S.; Kempf-Bielack, B.; Delling, G.; Exner, G.U.; Flege, S.; Helmke, K.; Kotz, R.; Salzer-Kuntschik, M.; Werner, M.; Winkelmann, W.; et al. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: An analysis of 1702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J. Clin. Oncol. 2002, 20, 776–790. [Google Scholar] [CrossRef]
- PosthumaDeBoer, J.; Witlox, M.A.; Kaspers, G.J.; van Royen, B.J. Molecular alterations as target for therapy in metastatic osteosarcoma: A review of literature. Clin. Exp. Metastasis 2011, 28, 493–503. [Google Scholar] [CrossRef]
- Hattinger, C.M.; Patrizio, M.P.; Fantoni, L.; Casotti, C.; Riganti, C.; Serra, M. Drug Resistance in Osteosarcoma: Emerging Biomarkers, Therapeutic Targets and Treatment Strategies. Cancers 2021, 13, 2878. [Google Scholar] [CrossRef]
- Wen, Y.; Tang, F.; Tu, C.; Hornicek, F.; Duan, Z.; Min, L. Immune checkpoints in osteosarcoma: Recent advances and therapeutic potential. Cancer Lett. 2022, 547, 215887. [Google Scholar] [CrossRef]
- Letson, G.D.; Muro-Cacho, C.A. Genetic and Molecular Abnormalities in Tumors of the Bone and Soft Tissues. Cancer Control 2001, 8, 239–251. [Google Scholar] [CrossRef]
- Kansara, M.; Teng, M.W.; Smyth, M.J.; Thomas, D.M. Translational biology of osteosarcoma. Nat. Rev. Cancer 2014, 14, 722–735. [Google Scholar] [CrossRef]
- Rothzerg, E.; Xu, J.; Wood, D.; Koks, S. 12 Survival-related differentially expressed genes based on the TARGET-osteosarcoma database. Exp. Biol. Med. 2021, 246, 2072–2081. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ge, F.; Wang, S. Four genes predict the survival of osteosarcoma patients based on TARGET database. J. Bioenerg. Biomembr. 2020, 52, 291–299. [Google Scholar] [CrossRef]
- Chen, L.H.; Liu, J.F.; Lu, Y.; He, X.Y.; Zhang, C.; Zhou, H.H. Complement C1q (C1qA, C1qB, and C1qC) May Be a Potential Prognostic Factor and an Index of Tumor Microenvironment Remodeling in Osteosarcoma. Front. Oncol. 2021, 11, 642144. [Google Scholar] [CrossRef]
- Chen, C.; Xie, L.; Ren, T.; Huang, Y.; Xu, J.; Guo, W. Immunotherapy for osteosarcoma: Fundamental mechanism, rationale, and recent breakthroughs. Cancer Lett. 2021, 500, 1–10. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. S4). [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Nagy, A.; Lanczky, A.; Menyhart, O.; Gyorffy, B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 2018, 8, 9227. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830. [Google Scholar] [CrossRef]
- Brown, H.K.; Tellez-Gabriel, M.; Heymann, D. Cancer stem cells in osteosarcoma. Cancer Lett. 2017, 386, 189–195. [Google Scholar] [CrossRef]
- Thielens, N.M.; Tedesco, F.; Bohlson, S.S.; Gaboriaud, C.; Tenner, A.J. C1q: A fresh look upon an old molecule. Mol. Immunol. 2017, 89, 73–83. [Google Scholar] [CrossRef]
- Kemp, S.B.; Steele, N.G.; Carpenter, E.S.; Donahue, K.L.; Bushnell, G.G.; Morris, A.H.; The, S.; Orbach, S.M.; Sirihorachai, V.R.; Nwosu, Z.C.; et al. Pancreatic cancer is marked by complement-high blood monocytes and tumor-associated macrophages. Life Sci. Alliance 2021, 4, e202000935. [Google Scholar] [CrossRef]
- Bonavita, E.; Gentile, S.; Rubino, M.; Maina, V.; Papait, R.; Kunderfranco, P.; Greco, C.; Feruglio, F.; Molgora, M.; Laface, I.; et al. PTX3 is an extrinsic oncosuppressor regulating complement-dependent inflammation in cancer. Cell 2015, 160, 700–714. [Google Scholar] [CrossRef]
- Afshar-Kharghan, V. The role of the complement system in cancer. J. Clin. Investig. 2017, 127, 780–789. [Google Scholar] [CrossRef]
- Mangogna, A.; Belmonte, B.; Agostinis, C.; Zacchi, P.; Iacopino, D.G.; Martorana, A.; Rodolico, V.; Bonazza, D.; Zanconati, F.; Kishore, U.; et al. Prognostic Implications of the Complement Protein C1q in Gliomas. Front. Immunol. 2019, 10, 2366. [Google Scholar] [CrossRef]
- Bulla, R.; Tripodo, C.; Rami, D.; Ling, G.S.; Agostinis, C.; Guarnotta, C.; Zorzet, S.; Durigutto, P.; Botto, M.; Tedesco, F. C1q acts in the tumour microenvironment as a cancer-promoting factor independently of complement activation. Nat. Commun. 2016, 7, 10346. [Google Scholar] [CrossRef]
- Bohlson, S.S.; O’Conner, S.D.; Hulsebus, H.J.; Ho, M.M.; Fraser, D.A. Complement, c1q, and c1q-related molecules regulate macrophage polarization. Front. Immunol. 2014, 5, 402. [Google Scholar] [CrossRef]
- Buddingh, E.P.; Kuijjer, M.L.; Duim, R.A.; Burger, H.; Agelopoulos, K.; Myklebost, O.; Serra, M.; Mertens, F.; Hogendoorn, P.C.; Lankester, A.C.; et al. Tumor-infiltrating macrophages are associated with metastasis suppression in high-grade osteosarcoma: A rationale for treatment with macrophage activating agents. Clin. Cancer Res. 2011, 17, 2110–2119. [Google Scholar] [CrossRef]
- Mezheyeuski, A.; Backman, M.; Mattsson, J.; Martín-Bernabé, A.; Larsson, C.; Hrynchyk, I.; Hammarström, K.; Ström, S.; Ekström, J.; Mauchanski, S.; et al. An immune score reflecting pro- and anti-tumoural balance of tumour microenvironment has major prognostic impact and predicts immunotherapy response in solid cancers. EBioMedicine 2023, 88, 104452. [Google Scholar] [CrossRef]
- Zeiner, P.S.; Zinke, J.; Kowalewski, D.J.; Bernatz, S.; Tichy, J.; Ronellenfitsch, M.W.; Thorsen, F.; Berger, A.; Forster, M.T.; Muller, A.; et al. CD74 regulates complexity of tumor cell HLA class II peptidome in brain metastasis and is a positive prognostic marker for patient survival. Acta Neuropathol. Commun. 2018, 6, 18. [Google Scholar] [CrossRef]
- Zhang, J.F.; Tao, L.Y.; Yang, M.W.; Xu, D.P.; Jiang, S.H.; Fu, X.L.; Liu, D.J.; Huo, Y.M.; Liu, W.; Yang, J.Y.; et al. CD74 promotes perineural invasion of cancer cells and mediates neuroplasticity via the AKT/EGR-1/GDNF axis in pancreatic ductal adenocarcinoma. Cancer Lett. 2021, 508, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Farr, L.; Ghosh, S.; Moonah, S. Role of MIF Cytokine/CD74 Receptor Pathway in Protecting Against Injury and Promoting Repair. Front. Immunol. 2020, 11, 1273. [Google Scholar] [CrossRef] [PubMed]
- Tanese, K.; Hashimoto, Y.; Berkova, Z.; Wang, Y.; Samaniego, F.; Lee, J.E.; Ekmekcioglu, S.; Grimm, E.A. Cell Surface CD74-MIF Interactions Drive Melanoma Survival in Response to Interferon-gamma. J. Investig. Dermatol. 2015, 135, 2775–2784. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, C.R.; Azevedo, R.A.; Mousdell, S.; Resende-Lara, P.T.; Ireland, L.; Santos, A.; Girola, N.; Cunha, R.; Schmid, M.C.; Polonelli, L.; et al. Blockade of MIF-CD74 Signalling on Macrophages and Dendritic Cells Restores the Antitumour Immune Response Against Metastatic Melanoma. Front. Immunol. 2018, 9, 1132. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wei, B.; Hu, G.; Wang, L.; Bi, M.; Sun, Z.; Jin, Y. Screening of differentially expressed genes associated with human glioblastoma and functional analysis using a DNA microarray. Mol. Med. Rep. 2015, 12, 1991–1996. [Google Scholar] [CrossRef] [PubMed]
- Hang, X.; Li, D.; Wang, J.; Wang, G. Prognostic significance of microsatellite instability-associated pathways and genes in gastric cancer. Int. J. Mol. Med. 2018, 42, 149–160. [Google Scholar] [CrossRef]
- Huang, Y.-j.; He, J.-k.; Duan, X.; Hou, R.; Shi, J. Prognostic gene HLA-DMA associated with cell cycle and immune infiltrates in LUAD. Clin. Respir. J. 2023, 17, 1286–1300. [Google Scholar] [CrossRef]
- Liu, D.; Hofman, P. Expression of NOTCH1, NOTCH4, HLA-DMA and HLA-DRA is synergistically associated with T cell exclusion, immune checkpoint blockade efficacy and recurrence risk in ER-negative breast cancer. Cell. Oncol. 2022, 45, 463–477. [Google Scholar] [CrossRef]



| Accession | Gender | Age (Months) | Metastasis within 5 Years | Location | Histological Subtype |
|---|---|---|---|---|---|
| GSM825626 | F | 192 | no | tibia/fibula | osteoblastic |
| GSM825627 | F | 111 | no | femur | pleomorphic |
| GSM825631 | M | 200 | yes | tibia/fibula | osteoblastic |
| GSM825633 | M | 177 | yes | femur | osteoblastic |
| GSM825634 | F | 212 | yes | femur | osteoblastic |
| GSM825635 | M | 216 | no | femur | osteoblastic |
| GSM825636 | M | 200 | yes | femur | telangiectatic |
| GSM825637 | F | 164 | yes | tibia/fibula | telangiectatic |
| GSM825639 | M | 175 | no | tibia/fibula | osteoblastic |
| GSM825640 | M | 200 | yes | humerus | osteoblastic |
| GSM825641 | M | 101 | no | femur | osteoblastic |
| GSM825644 | M | 136 | no | tibia/fibula | osteoblastic |
| GSM825646 | M | 198 | yes | tibia/fibula | osteoblastic |
| GSM825647 | F | 137 | no | tibia/fibula | osteoblastic |
| GSM825654 | M | 165 | yes | femur | osteoblastic |
| GSM825658 | M | 96 | yes | femur | osteoblastic |
| GSM825659 | F | 128 | yes | femur | osteoblastic |
| GSM825661 | F | 81 | yes | humerus | osteoblastic |
| GSM825662 | M | 144 | yes | tibia/fibula | chondroblastic |
| GSM825663 | F | 204 | yes | femur | osteoblastic |
| GSM825664 | M | 181 | yes | tibia/fibula | sclerosing |
| GSM825665 | M | 200 | yes | femur | osteoblastic |
| GSM825666 | M | 205 | yes | femur | chondroblastic |
| GSM825667 | M | 183 | yes | femur | osteoblastic |
| GSM825672 | F | 144 | yes | femur | osteoblastic |
| GSM825673 | F | 204 | no | humerus | fibroblastic |
| GSM825676 | M | 174 | yes | femur | giant cell-rich |
| GSM825677 | M | 162 | yes | humerus | osteoblastic |
| GSM825678 | M | 170 | yes | femur | osteoblastic |
| GSM825679 | F | 133 | yes | femur | fibroblastic |
| GSM825687 | M | 180 | yes | tibia/fibula | chondroblastic |
| GSM825692 | F | 129 | yes | femur | chondroblastic |
| GSM825704 | M | 192 | no | femur | osteoblastic |
| GSM825706 | F | 180 | yes | femur | sclerosing |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, J.; Yu, S. Comprehensive Analysis Reveals Prognostic and Therapeutic Immunity-Related Biomarkers for Pediatric Metastatic Osteosarcoma. Medicina 2024, 60, 95. https://doi.org/10.3390/medicina60010095
Yuan J, Yu S. Comprehensive Analysis Reveals Prognostic and Therapeutic Immunity-Related Biomarkers for Pediatric Metastatic Osteosarcoma. Medicina. 2024; 60(1):95. https://doi.org/10.3390/medicina60010095
Chicago/Turabian StyleYuan, Jin, and Shengji Yu. 2024. "Comprehensive Analysis Reveals Prognostic and Therapeutic Immunity-Related Biomarkers for Pediatric Metastatic Osteosarcoma" Medicina 60, no. 1: 95. https://doi.org/10.3390/medicina60010095
APA StyleYuan, J., & Yu, S. (2024). Comprehensive Analysis Reveals Prognostic and Therapeutic Immunity-Related Biomarkers for Pediatric Metastatic Osteosarcoma. Medicina, 60(1), 95. https://doi.org/10.3390/medicina60010095






